Improvement of an Automated Sample Injection System for Pillar Array Columns to Increase Analytical Reproducibility
Abstract
:1. Introduction
2. Results and Discussion
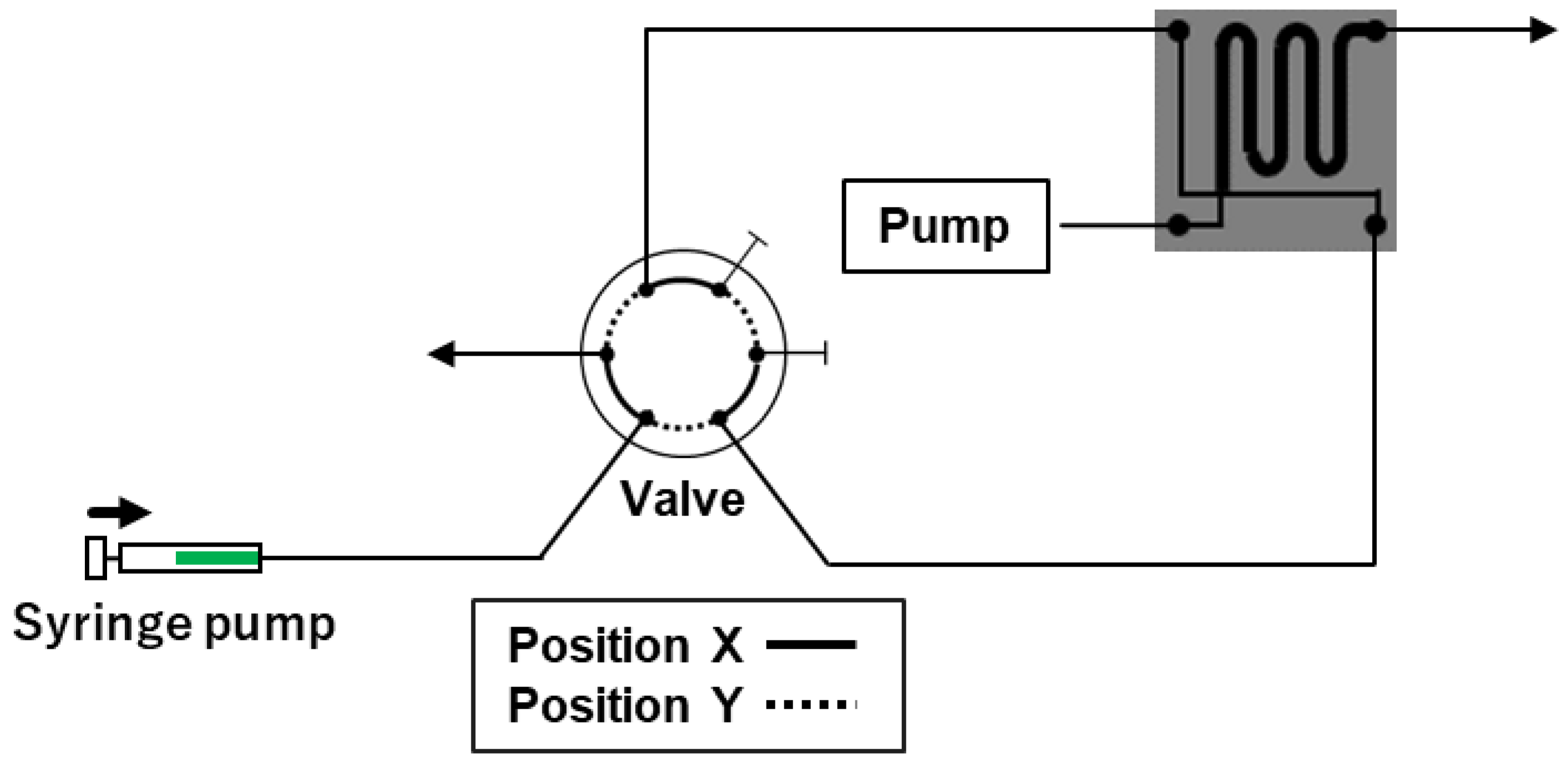
2.1. Previously Developed Automated Sample Injection System
2.2. Improved Automated Sample Injection System
2.3. Optimization of the Improved Automated Sample Injection System
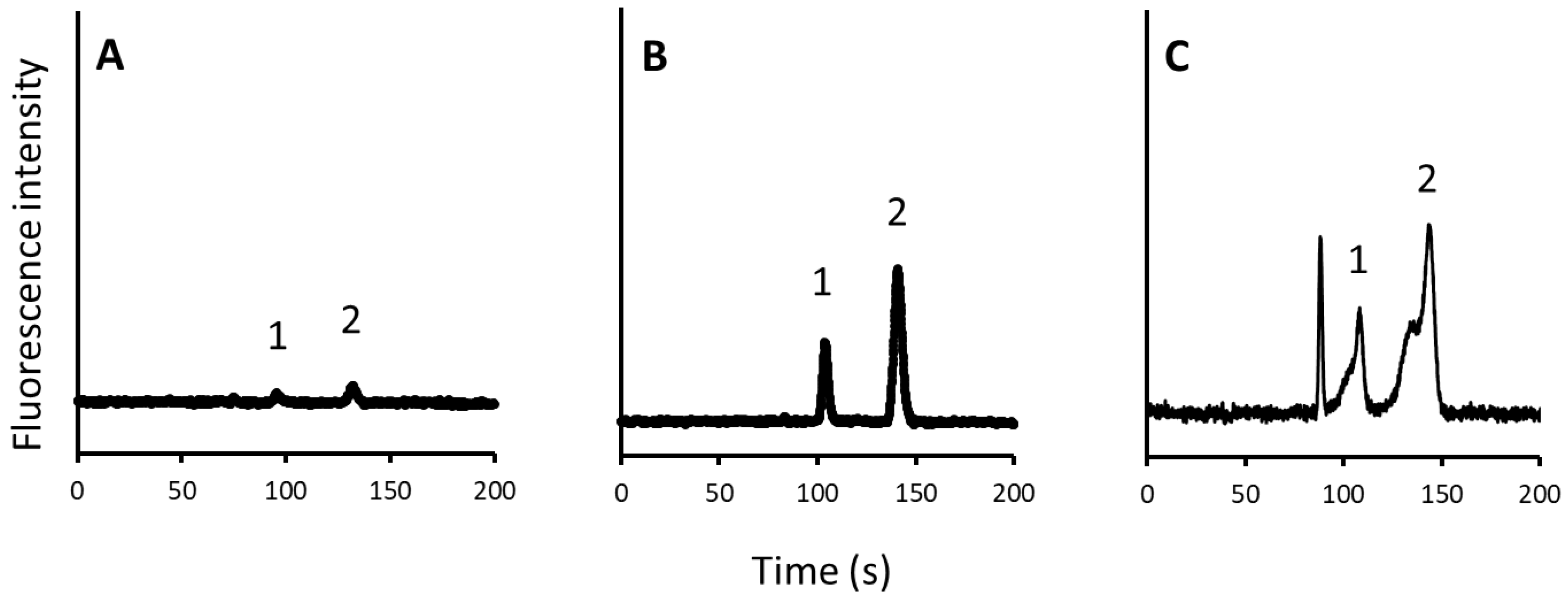
2.4. Evaluation of the Improved Automated Sample Injection System
2.5. Application of the System to Analyze Amino Acids in Human Plasma
3. Conclusions
4. Materials and Methods
4.1. Chemicals
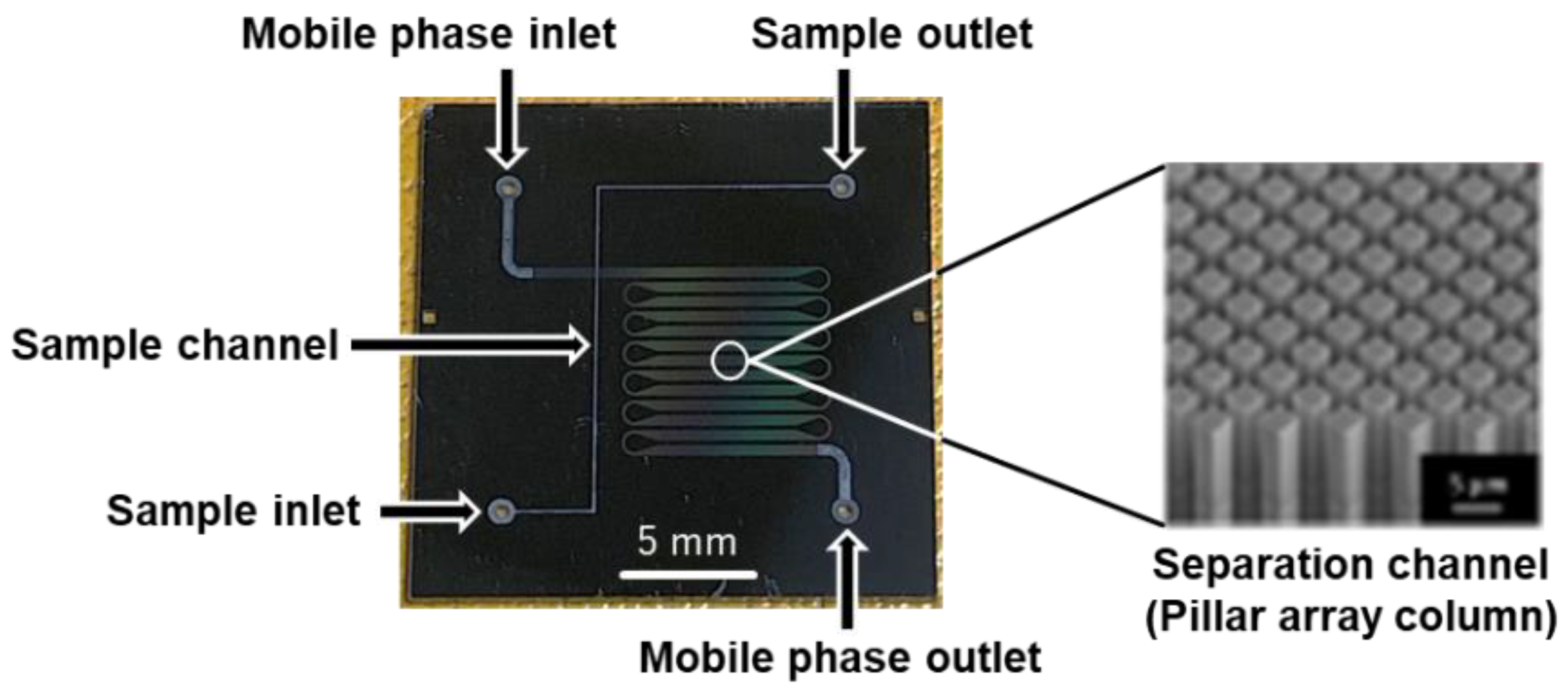
4.2. Microchip Fabrication and Modification
4.3. Derivatization Conditions for Amino Acids with NBD-F
4.4. Pretreatment of Human Plasma
4.5. Chromatographic Conditions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Cheng, G.; Zhao, J.; Wang, X.; Yang, C.; Li, S.; Lu, T.; Li, X.; Wang, X.; Zhu, G. A Highly Sensitive and Selective Method for the Determination of Ceftiofur Sodium in Milk and Animal-Origin Food Based on Molecularly Imprinted Solid-Phase Extraction Coupled with HPLC-UV. Food Chem. 2021, 347, 129013. [Google Scholar] [CrossRef] [PubMed]
- Guggilla, S.; Karthik, M.; Shylendra, B. Sensitive and Stereospecific High-Performance Liquid Chromatographic Method for Flurbiprofen in Human Plasma. Adv. Exp. Med. Biol. 2021, 1339, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Hattori, A.; Ito, T.; Funatsu, T.; Tsunoda, M. Analysis of Intracellular α-Keto Acids by HPLC with Fluorescence Detection. Anal. Methods 2020, 12, 2555–2559. [Google Scholar] [CrossRef] [PubMed]
- Appleby, S.; Chew-Harris, J.; Troughton, R.W.; Richards, A.M.; Pemberton, C.J. Analytical and Biological Assessment of Circulating Human Erythroferrone. Clin. Biochem. 2020, 79, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Karanikolopoulos, G.; Gerakis, A.; Papadopoulou, K.; Mastrantoni, I. Determination of Synthetic Food Colorants in Fish Products by an HPLC-DAD Method. Food Chem. 2015, 177, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Shishovska, M.A.; Stefova, M.T. Fast and Universal HPLC Method for Determination of Permethrin in Formulations Using 1.8-μm Particle-Packed Column and Performance Comparison with Other Column Types. J. Chromatogr. Sci. 2012, 50, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Knox, J.H. Band Dispersion in Chromatography—A Universal Expression for the Contribution from the Mobile Zone. J. Chromatogr. A 2002, 960, 7–18. [Google Scholar] [CrossRef]
- Knox, J.H. Band Dispersion in Chromatography—A New View of A-Term Dispersion. J. Chromatogr. A 1999, 831, 3–15. [Google Scholar] [CrossRef]
- Tsunoda, M. On-Chip Liquid Chromatography. Encyclopedia 2022, 2, 41. [Google Scholar] [CrossRef]
- Fujiwara, T.; Funatsu, T.; Tsunoda, M. Fast Analysis Using Pillar Array Columns: Quantification of Branched-Chain α-Keto Acids in Human Plasma Samples. J. Pharm. Biomed. Anal. 2021, 198, 114019. [Google Scholar] [CrossRef] [PubMed]
- Kuroki, H.; Koyama, H.; Nakatani, Y.; Funatsu, T.; Horiike, S.; Tsunoda, M. Development of an Automated Sample Injection System for Pillar Array Columns. Chromatography 2020, 41, 59–62. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Takatsuki, K.; Sekiguchi, T.; Funatsu, T.; Shoji, S.; Tsunoda, M. Rapid Quantitative Method for the Detection of Phenylalanine and Tyrosine in Human Plasma Using Pillar Array Columns and Gradient Elution. Amino. Acids 2016, 48, 1731–1735. [Google Scholar] [CrossRef] [PubMed]
- Isokawa, M.; Takatsuki, K.; Song, Y.; Shih, K.; Nakanishi, K.; Xie, Z.; Yoon, D.H.; Sekiguchi, T.; Funatsu, T.; Shoji, S.; et al. Liquid Chromatography Chip with Low-Dispersion and Low-Pressure-Drop Turn Structure Utilizing a Distribution-Controlled Pillar Array. Anal. Chem. 2016, 88, 6485–6491. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Takatsuki, K.; Isokawa, M.; Sekiguchi, T.; Mizuno, J.; Funatsu, T.; Shoji, S.; Tsunoda, M. Fast and Quantitative Analysis of Branched-Chain Amino Acids in Biological Samples Using a Pillar Array Column. Anal. Bioanal. Chem. 2013, 405, 7993–7999. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Noguchi, M.; Takatsuki, K.; Sekiguchi, T.; Mizuno, J.; Funatsu, T.; Shoji, S.; Tsunoda, M. Integration of Pillar Array Columns into a Gradient Elution System for Pressure-Driven Liquid Chromatography. Anal. Chem. 2012, 84, 4739–4745. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, C.; Saeki, A.; Noguchi, M.; Shirasaki, Y.; Shoji, S.; Funatsu, T.; Mizuno, J.; Tsunoda, M. Use of Folded Micromachined Pillar Array Column with Low-Dispersion Turns for Pressure-Driven Liquid Chromatography. Anal. Chem. 2010, 82, 1420–1426. [Google Scholar] [CrossRef] [PubMed]
- De Malsche, W.; Eghbali, H.; Clicq, D.; Vangelooven, J.; Gardeniers, H.; Desmet, G. Pressure-Driven Reverse-Phase Liquid Chromatography Separations in Ordered Nonporous Pillar Array Columns. Anal. Chem. 2007, 79, 5915–5926. [Google Scholar] [CrossRef] [PubMed]
- Gzil, P.; Vervoort, N.; Baron, G.V.; Desmet, G. Advantages of Perfectly Ordered 2-D Porous Pillar Arrays over Packed Bed Columns for LC Separations: A Theoretical Analysis. Anal. Chem. 2003, 75, 6244–6250. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Tait, N.; Regnier, F. Fabrication of Nanocolumns for Liquid Chromatography. Anal. Chem. 1998, 70, 3790–3797. [Google Scholar] [CrossRef] [PubMed]
- Hattori, A.; Tsunoda, M.; Konuma, T.; Kobayashi, M.; Nagy, T.; Glushka, J.; Tayyari, F.; McSkimming, D.; Kannan, N.; Tojo, A.; et al. Cancer Progression by Reprogrammed BCAA Metabolism in Myeloid Leukaemia. Nature 2017, 545, 500–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuroki, H.; Koyama, H.; Tsunoda, M. Improvement of an Automated Sample Injection System for Pillar Array Columns to Increase Analytical Reproducibility. Molecules 2022, 27, 4715. https://doi.org/10.3390/molecules27154715
Kuroki H, Koyama H, Tsunoda M. Improvement of an Automated Sample Injection System for Pillar Array Columns to Increase Analytical Reproducibility. Molecules. 2022; 27(15):4715. https://doi.org/10.3390/molecules27154715
Chicago/Turabian StyleKuroki, Hiroshi, Hirotaka Koyama, and Makoto Tsunoda. 2022. "Improvement of an Automated Sample Injection System for Pillar Array Columns to Increase Analytical Reproducibility" Molecules 27, no. 15: 4715. https://doi.org/10.3390/molecules27154715
APA StyleKuroki, H., Koyama, H., & Tsunoda, M. (2022). Improvement of an Automated Sample Injection System for Pillar Array Columns to Increase Analytical Reproducibility. Molecules, 27(15), 4715. https://doi.org/10.3390/molecules27154715





