Antiproliferative Ruthenium Complexes Containing Curcuminoid Ligands Tested In Vitro on Human Ovarian Tumor Cell Line A2780, towards Their Capability to Modulate the NF-κBTranscription Factor, FGF-2 Growth Factor, and MMP-9 Pathway
Abstract
:1. Introduction
2. Results and Discussion
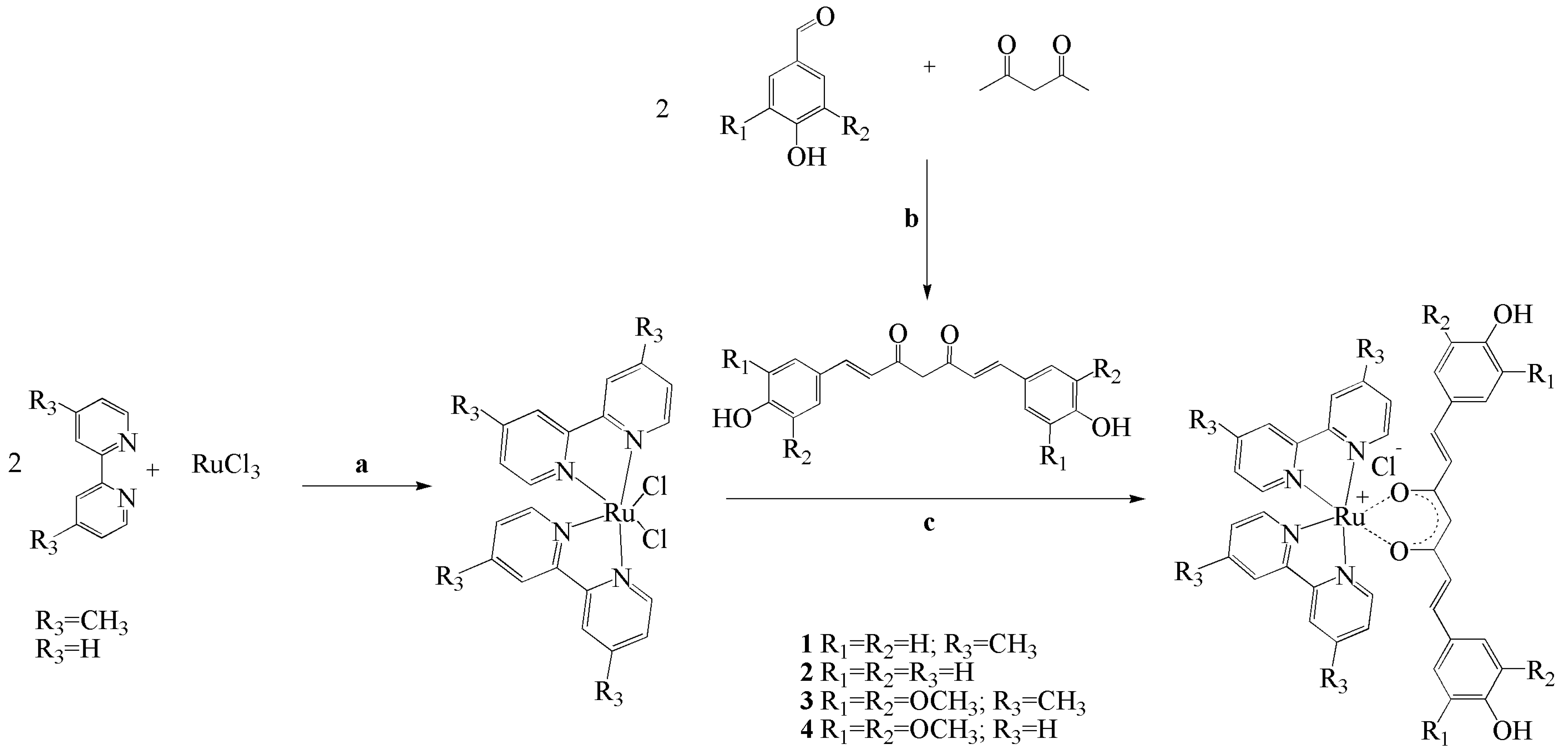
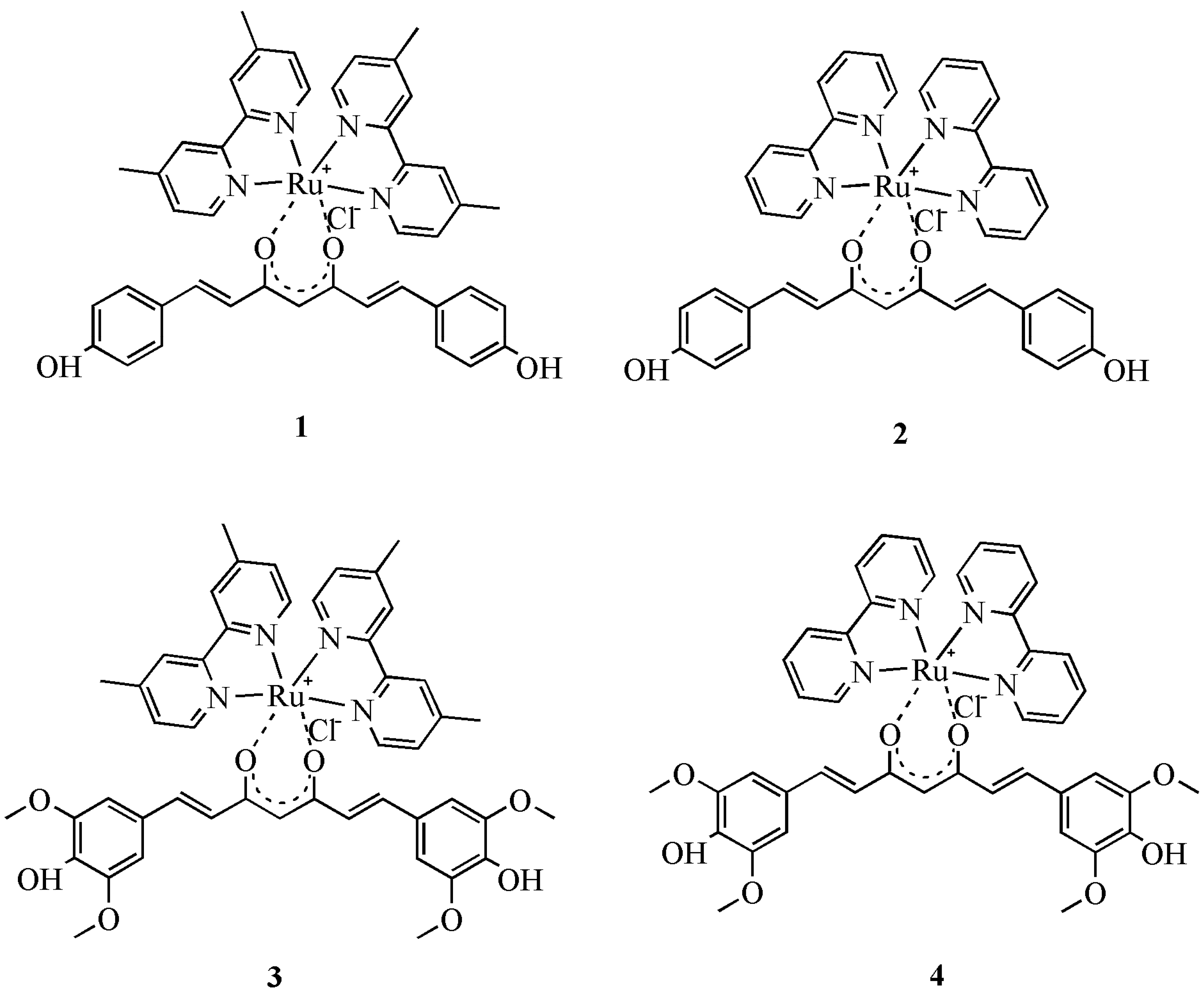
2.1. Synthesis and Structural Characterization of Ruthenium Complexes
2.2. Biological Attempts-Cytotoxicity
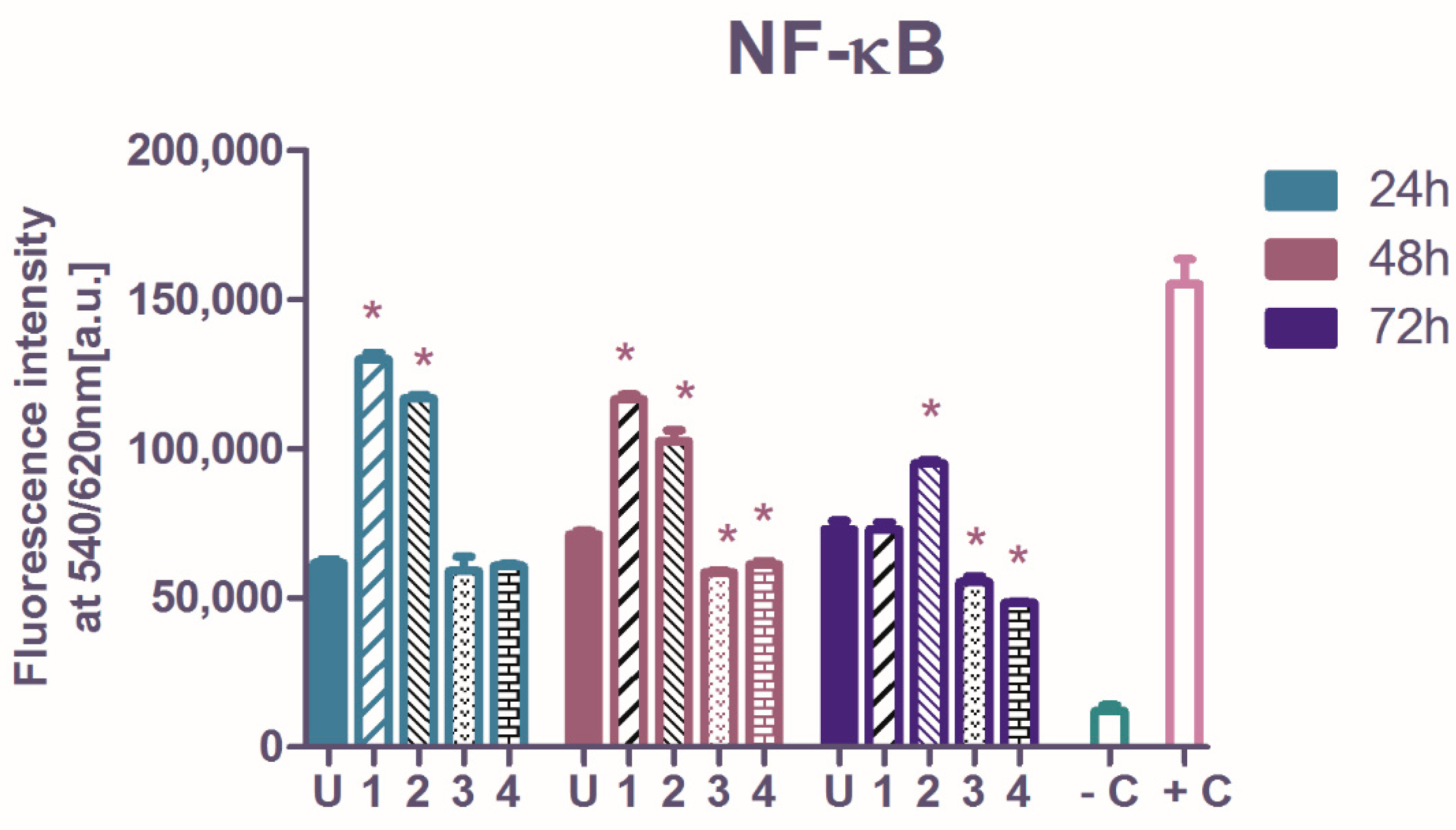
2.3. The Influence of Ruthenium Complexes on Nuclear Factor Kappa-Light-Chain-Enhancer of Activated B Cells (NF-κB Transcription Factor) p65 Subunit
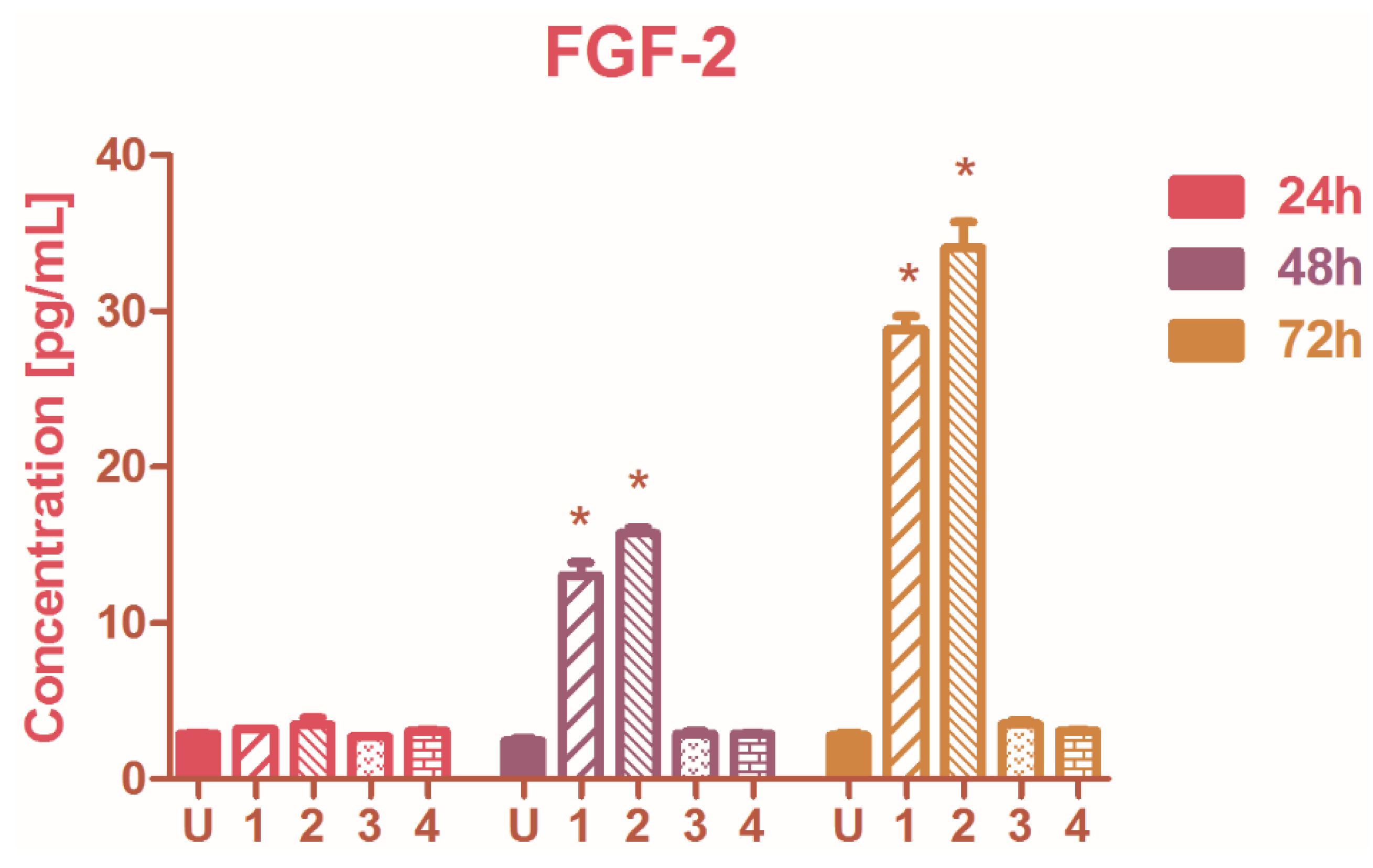
2.4. The Modulation of Basic Fibroblast Growth Factor (FGF-2)
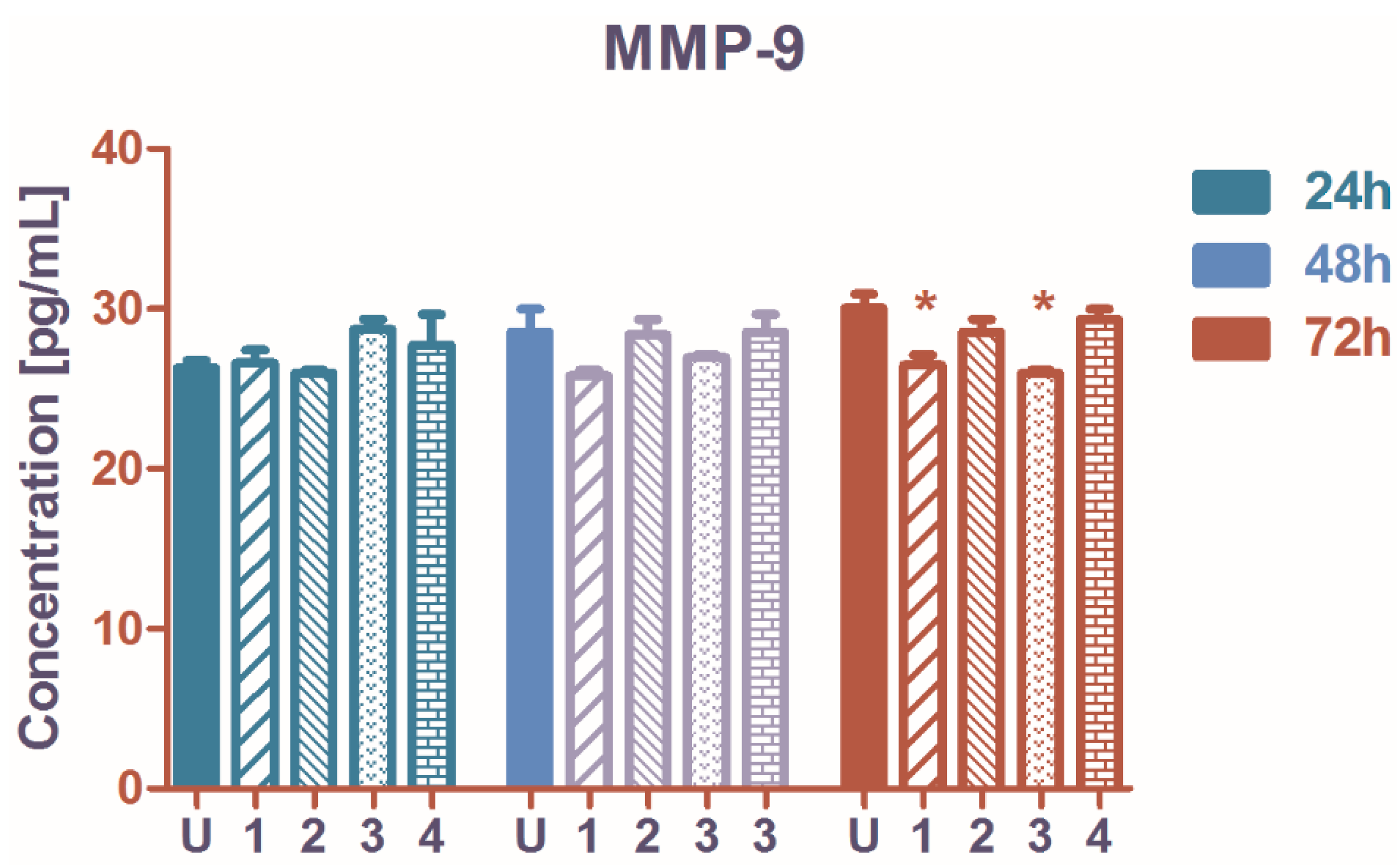
2.5. The Influence on Matrix Metalloproteinase-9 (MMP-9) Secretion
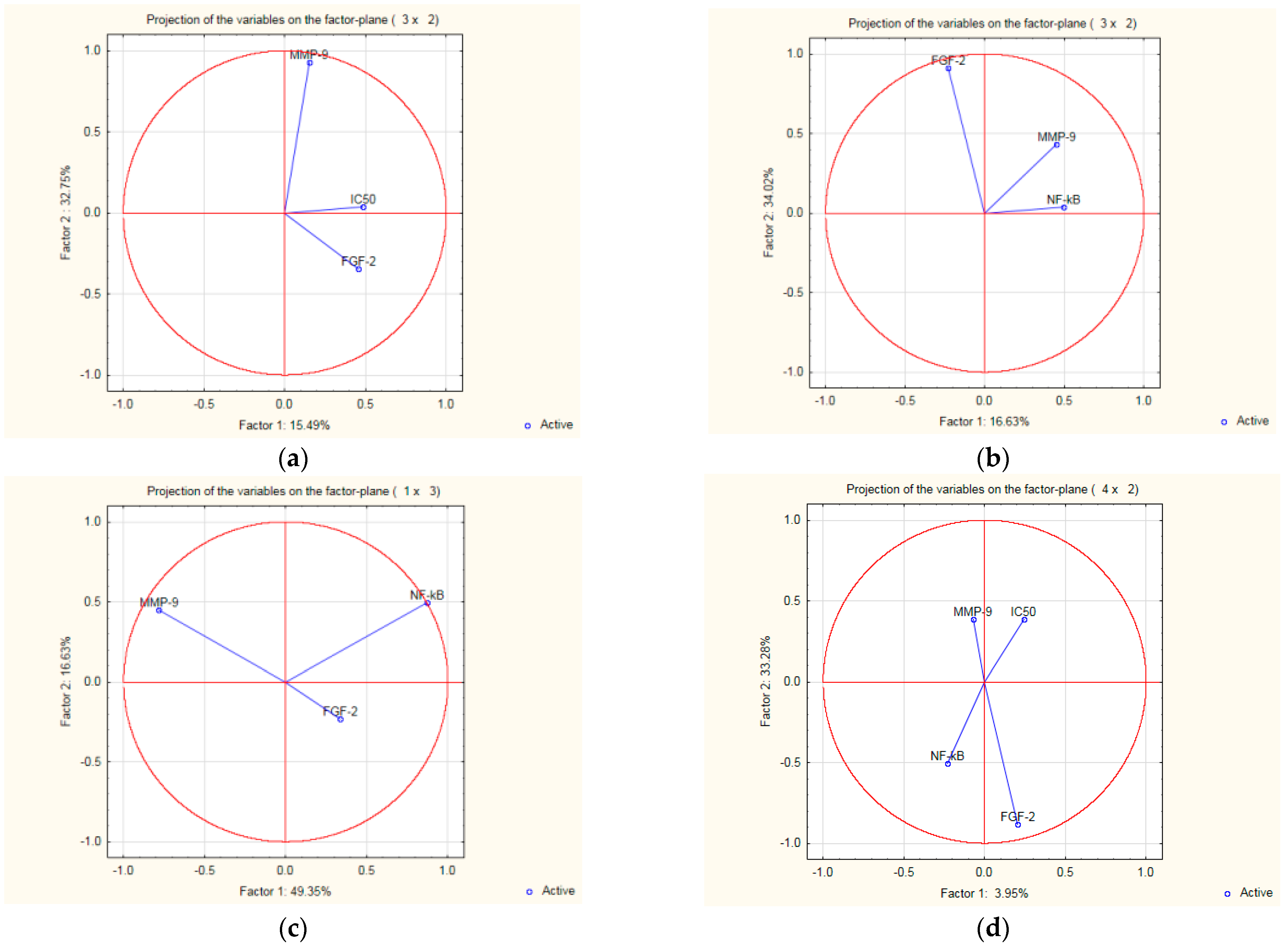
2.6. Interdependence and Connections between the Biologic Parameters
3. Materials and Methods
3.1. Synthesis of Ruthenium Complexes
3.2. Biological Testing—Cell Growth Inhibition
3.3. Soluble Basic Fibroblast Growth Factor (FGF-2) Production
3.4. Matrix Metalloproteinase-9 (MMP-9)
3.5. The Intracellular Nuclear Factor Kappa-Light-Chain-Enhancer of Activated B Cells (NF-κB)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Oyagbemi, A.A.; Saba, A.B.; Ibraheem, A.O. Mini-Review Curcumin: From Food Spice to Cancer Prevention. Cancer 2009, 10, 963–968. [Google Scholar]
- Ammon, H.P.T.; Wahl, M.A. Pharmacology of Curcuma Longa. Planta Med. 1991, 57, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayakumar, T.; Hsu, C.Y.; Khamrang, T.; Hsia, C.H.; Hsia, C.W.; Manubolu, M.; Sheu, J.R. Possible Molecular Targets of Novel Ruthenium Complexes in Antiplatelet Therapy. Int. J. Mol. Sci. 2018, 19, 1818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Khan, A.R.; Fu, M.; Zhai, Y.; Yu, A.; Zhai, G. The Progresses in Curcuminoids-Based Metal Complexes: Especially in Cancer Therapy. Future Med. Chem. 2019, 11, 1035–1056. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, H.; Naseri, G.; Rezaee, R.; Mohammadi, M.; Banikazemi, Z.; Mirzaei, H.R.; Salehi, H.; Peyvandi, M.; Pawelek, J.M.; Sahebkar, A. Curcumin: A New Candidate for Melanoma Therapy? Int. J. Cancer 2016, 139, 1683–1695. [Google Scholar] [CrossRef]
- Panahi, Y.; Khalili, N.; Sahebi, E.; Namazi, S.; Karimian, M.S.; Majeed, M.; Sahebkar, A. Antioxidant Effects of Curcuminoids in Patients with Type 2 Diabetes Mellitus: A Randomized Controlled Trial. Inflammopharmacology 2017, 25, 25–31. [Google Scholar] [CrossRef]
- Sharma, K.K.; Chandra, S.; Basu, D.K. Synthesis and Antiarthritic Study of a New Orally Active Diferuloylmethane (Curcumin) Gold Complex. Inorg. Chim. Acta 1987, 135, 47–48. [Google Scholar] [CrossRef]
- Panahi, Y.; Khalili, N.; Hosseini, M.S.; Abbasinazari, M.; Sahebkar, A. Lipid-Modifying Effects of Adjunctive Therapy with Curcuminoids-Piperine Combination in Patients with Metabolic Syndrome: Results of a Randomized Controlled Trial. Complement. Ther. Med. 2014, 22, 851–857. [Google Scholar] [CrossRef]
- Kalaycıoğlu, Z.; Gazioğlu, I.; Erim, F.B. Comparison of Antioxidant, Anticholinesterase, and Antidiabetic Activities of Three Curcuminoids Isolated from Curcuma Longa L. Nat. Prod. Res. 2017, 31, 2914–2917. [Google Scholar] [CrossRef]
- Deogade, S.C.; Ghate, S. Curcumin: Therapeutic applications in systemic and oral health. Int. J. Biol. Pharm. Res. Int. J. Biol. Pharm. Res. J. 2015, 6, 281–290. [Google Scholar]
- Zambre, A.P.; Jamadar, A.; Padhye, S.; Kulkarni, V.M. Copper Conjugates of Knoevenagel Condensates of Curcumin and Their Schiff Base Derivatives: Synthesis, Spectroscopy, Magnetism, ESR, and Electrochemistry. Synth. React. Inorg. Met. Nano-Metal Chem. 2007, 37, 19–27. [Google Scholar] [CrossRef]
- Wanninger, S.; Lorenz, V.; Subhan, A.; Edelmann, F.T. Metal Complexes of Curcumin—Synthetic Strategies, Structures and Medicinal Applications. Chem. Soc. Rev. 2015, 44, 4986–5002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pongrakhananon, V.; Rojanasakul, Y. Anticancer Properties of Curcumin. In Advances in Cancer Therapy; InTech: Rijeka, Croatia, 2011; ISBN 978-953-307-703-1. [Google Scholar]
- Anand, P.; Kunnumakkara, A.B.; Newman, R.A.; Aggarwal, B.B. Bioavailability of Curcumin: Problems and Promises. Mol. Pharm. 2007, 4, 807–818. [Google Scholar] [CrossRef]
- Ramezani, M.; Hatamipour, M.; Sahebkar, A. Promising Anti-Tumor Properties of Bisdemethoxycurcumin: A Naturally Occurring Curcumin Analogue. J. Cell. Physiol. 2018, 233, 880–887. [Google Scholar] [CrossRef] [PubMed]
- Gao, S. Bisdemethoxycurcumin Inhibits Hepatocellular Carcinoma Proliferation Through Akt Inactivation via CYLD-Mediated Deubiquitination. Drug Des. Dev. Ther. 2020, 14, 993. [Google Scholar]
- Liao, C.L.; Chu, Y.L.; Lin, H.Y.; Chen, C.Y.; Hsu, M.J.; Liu, K.C.; Lai, K.C.; Huang, A.C.; Chung, J.G. Bisdemethoxycurcumin Suppresses Migration and Invasion of Human Cervical Cancer Hela Cells via Inhibition of NF-ĸB, MMP-2 and -9 Pathways. Anticancer Res. 2018, 38, 3989–3997. [Google Scholar] [CrossRef] [PubMed]
- Miklášová, N.; Herich, P.; Dávila-Becerril, J.C.; Barroso-Flores, J.; Fischer-Fodor, E.; Valentová, J.; Leskovská, J.; Kožíšek, J.; Takáč, P.; Mojžiš, J. Evaluation of Antiproliferative Palladium(II) Complexes of Synthetic Bisdemethoxycurcumin towards in Vitro Cytotoxicity and Molecular Docking on DNA Sequence. Molecules 2021, 26, 4369. [Google Scholar] [CrossRef]
- Jin, F.; Chen, X.; Yan, H.; Xu, Z.; Yang, B.; Luo, P.; He, Q. Bisdemethoxycurcumin Attenuates Cisplatin-Induced Renal Injury through Anti-Apoptosis, Anti-Oxidant and Anti-Inflammatory. Eur. J. Pharmacol. 2020, 874, 173026. [Google Scholar] [CrossRef]
- Borsari, M.; Ferrari, E.; Grandi, R.; Saladini, M. Curcuminoids as Potential New Iron-Chelating Agents: Spectroscopic, Polarographic and Potentiometric Study on Their Fe(III) Complexing Ability. Inorganica Chim. Acta 2002, 328, 61–68. [Google Scholar] [CrossRef]
- Pröhl, M.; Schubert, U.S.; Weigand, W.; Gottschaldt, M. Metal Complexes of Curcumin and Curcumin Derivatives for Molecular Imaging and Anticancer Therapy. Coord. Chem. Rev. 2016, 307, 32–41. [Google Scholar] [CrossRef]
- Pallikkavil, R.; Ummathur, M.B.; Sreedharan, S.; Krishnankutty, K. Synthesis, Characterization and Antimicrobial Studies of Cd(II), Hg(II), Pb(II), Sn(II) and Ca(II) Complexes of Curcumin. Main Gr. Met. Chem. 2013, 36, 123–127. [Google Scholar] [CrossRef]
- Subhan, M.A.; Alam, K.; Rahaman, M.S.; Rahman, M.A.; Awal, R. Synthesis and Characterization of Metal Complexes Containing Curcumin (C21H20O6) and Study of Their Anti-Microbial Activities and DNA-Binding Properties. J. Sci. Res. 2013, 6, 97–109. [Google Scholar] [CrossRef] [Green Version]
- Caruso, F.; Pettinari, R.; Rossi, M.; Monti, E.; Gariboldi, M.B.; Marchetti, F.; Pettinari, C.; Caruso, A.; Ramani, M.V.; Subbaraju, G.V. The in Vitro Antitumor Activity of Arene-Ruthenium(II) Curcuminoid Complexes Improves When Decreasing Curcumin Polarity. J. Inorg. Biochem. 2016, 162, 44–51. [Google Scholar] [CrossRef]
- Pröhl, M.; Bus, T.; Czaplewska, J.A.; Traeger, A.; Deicke, M.; Weiss, H.; Weigand, W.; Schubert, U.S.; Gottschaldt, M. Synthesis and in Vitro Toxicity of d-Glucose and d-Fructose Conjugated Curcumin–Ruthenium Complexes. Eur. J. Inorg. Chem. 2016, 2016, 5197–5204. [Google Scholar] [CrossRef]
- Li, S.; Xu, G.; Zhu, Y.; Zhao, J.; Gou, S. Bifunctional Ruthenium(Ii) Polypyridyl Complexes of Curcumin as Potential Anticancer Agents. Dalt. Trans. 2020, 49, 9454–9463. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lai, H.; Xiong, Z.; Chen, B.; Chen, T. Functionalization and Cancer-Targeting Design of Ruthenium Complexes for Precise Cancer Therapy. Chem. Commun. 2019, 55, 9904–9914. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, K.M.; Niloy, M.S.; Shakil, M.S.; Islam, M.A. Ruthenium Complexes: An Alternative to Platinum Drugs in Colorectal Cancer Treatment. Pharmaceutics 2021, 13, 1295. [Google Scholar] [CrossRef]
- Allison, M.; Caramés-Méndez, P.; Pask, C.M.; Phillips, R.M.; Lord, R.M.; McGowan, P.C. Bis(Bipyridine)Ruthenium(II) Ferrocenyl β-Diketonate Complexes: Exhibiting Nanomolar Potency against Human Cancer Cell Lines. Chem. A Eur. J. 2021, 27, 3737–3744. [Google Scholar] [CrossRef]
- Welsh, A.; Rylands, L.I.; Arion, V.B.; Prince, S.; Smith, G.S. Synthesis and Antiproliferative Activity of Benzimidazole-Based, Trinuclear Neutral Cyclometallated and Cationic, N^N-Chelated Ruthenium(II) Complexes. Dalt. Trans. 2020, 49, 1143–1156. [Google Scholar] [CrossRef]
- Grozav, A.; Balacescu, O.; Balacescu, L.; Cheminel, T.; Berindan-Neagoe, I.; Therrien, B. Synthesis, Anticancer Activity, and Genome Profiling of Thiazolo Arene Ruthenium Complexes. J. Med. Chem. 2015, 58, 8475–8490. [Google Scholar] [CrossRef] [Green Version]
- Riccardi, L.; Genna, V.; De Vivo, M. Metal–Ligand Interactions in Drug Design. Nat. Rev. Chem. 2018, 2, 100–112. [Google Scholar] [CrossRef]
- Zhao, S.; Pi, C.; Ye, Y.; Zhao, L.; Wei, Y. Recent Advances of Analogues of Curcumin for Treatment of Cancer. Eur. J. Med. Chem. 2019, 180, 524–535. [Google Scholar] [CrossRef] [PubMed]
- Kazantzis, K.T.; Koutsonikoli, K.; Mavroidi, B.; Zachariadis, M.; Alexiou, P.; Pelecanou, M.; Politopoulos, K.; Alexandratou, E.; Sagnou, M. Curcumin Derivatives as Photosensitizers in Photodynamic Therapy: Photophysical Properties and: In Vitro Studies with Prostate Cancer Cells. Photochem. Photobiol. Sci. 2020, 19, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Tomeh, M.A.; Hadianamrei, R.; Zhao, X. A Review of Curcumin and Its Derivatives as Anticancer Agents. Int. J. Mol. Sci. 2019, 20, 1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeo, C.I.; Ooi, K.K.; Tiekink, E.R.T. Gold-Based Medicine: A Paradigm Shift in Anti-Cancer Therapy? Molecules 2018, 23, 1410. [Google Scholar] [CrossRef] [Green Version]
- Nováková, O.; Kašpárková, J.; Vrána, O.; van Vliet, P.M.; Reedijk, J.; Brabec, V. Correlation between Cytotoxicity and DNA Binding of Polypyridyl Ruthenium Complexes. Biochemistry 1995, 34, 12369–12378. [Google Scholar] [CrossRef]
- Habtemariam, A.; Melchart, M.; Fernández, R.; Parsons, S.; Oswald, I.D.H.; Parkin, A.; Fabbiani, F.P.A.; Davidson, J.E.; Dawson, A.; Aird, R.E.; et al. Structure-Activity Relationships for Cytotoxic Ruthenium(II) Arene Complexes Containing N,N-, N,O-, and O,O-Chelating Ligands. J. Med. Chem. 2006, 49, 6858–6868. [Google Scholar] [CrossRef]
- Sun, Y.; Heidary, D.K.; Zhang, Z.; Richards, C.I.; Glazer, E.C. Bacterial Cytological Profiling Reveals the Mechanism of Action of Anticancer Metal Complexes. Mol. Pharm. 2018, 15, 3404–3416. [Google Scholar] [CrossRef]
- Shi, H.; Fang, T.; Tian, Y.; Huang, H.; Liu, Y. A Dual-Fluorescent Nano-Carrier for Delivering Photoactive Ruthenium Polypyridyl Complexes. J. Mater. Chem. B 2016, 4, 4746–4753. [Google Scholar] [CrossRef]
- Groessl, M.; Zava, O.; Dyson, P.J. Cellular Uptake and Subcellular Distribution of Ruthenium-Based Metallodrugs under Clinical Investigation versus Cisplatin. Metallomics 2011, 3, 591–599. [Google Scholar] [CrossRef]
- Casini, A.; Edafe, F.; Erlandsson, M.; Gonsalvi, L.; Ciancetta, A.; Re, N.; Ienco, A.; Messori, L.; Peruzzini, M.; Dyson, P.J. Rationalization of the Inhibition Activity of Structurally Related Organometallic Compounds against the Drug Target Cathepsin B by DFT. Dalt. Trans. 2010, 39, 5556–5563. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Musumeci, D.; Trifuoggi, M.; Irace, C.; Paduano, L.; Montesarchio, D. Anticancer Ruthenium(III) Complexes and Ru(III)-Containing Nanoformulations: An Update on the Mechanism of Action and Biological Activity. Pharmaceuticals 2019, 12, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrington, B.S.; Annunziata, C.M. Nf-Κb Signaling in Ovarian Cancer. Cancers 2019, 11, 1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aggarwal, B.B.; Sung, B. NF-ΚB in Cancer: A Matter of Life and Death. Cancer Discov. 2011, 1, 469–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Page, C.; Ouellet, V.; Madore, J.; Hudson, T.J.; Tonin, P.N.; Provencher, D.M.; Mes-Masson, A.M. From Gene Profiling to Diagnostic Markers: IL-18 and FGF-2 Complement CA125 as Serum-Based Markers in Epithelial Ovarian Cancer. Int. J. Cancer 2006, 118, 1750–1758. [Google Scholar] [CrossRef]
- Dias, M.H.; Fonseca, C.S.; Zeidler, J.D.; Albuquerque, L.L.; da Silva, M.S.; Cararo-Lopes, E.; Reis, M.S.; Noël, V.; dos Santos, E.O.; Prior, I.A.; et al. Fibroblast Growth Factor 2 Lethally Sensitizes Cancer Cells to Stress-Targeted Therapeutic Inhibitors. Mol. Oncol. 2019, 13, 290–306. [Google Scholar] [CrossRef] [Green Version]
- Laios, A.; Mohamed, B.M.; Kelly, L.; Flavin, R.; Finn, S.; McEvoy, L.; Gallagher, M.; Martin, C.; Sheils, O.; Ring, M.; et al. Pre-Treatment of Platinum Resistant Ovarian Cancer Cells with an MMP-9/MMP-2 Inhibitor Prior to Cisplatin Enhances Cytotoxicity as Determined by High Content Screening. Int. J. Mol. Sci. 2013, 14, 2085–2103. [Google Scholar] [CrossRef] [Green Version]
- Khaleel, E.F.; Badi, R.M.; Satti, H.H.; Mostafa, D.G. Exendin-4 Exhibits a Tumour Suppressor Effect in SKOVR-3 and OVACR-3 Ovarian Cancer Cells Lines by the Activation of SIRT1 and Inhibition of NF-ΚB. Clin. Exp. Pharmacol. Physiol. 2020, 47, 1092–1102. [Google Scholar] [CrossRef]
- Changtam, C.; de Koning, H.P.; Ibrahim, H.; Sajid, M.S.; Gould, M.K.; Suksamrarn, A. Curcuminoid Analogs with Potent Activity against Trypanosoma and Leishmania Species. Eur. J. Med. Chem. 2010, 45, 941–956. [Google Scholar] [CrossRef]
- Al-Rawashdeh, N.A.F.; Chatterjee, S.; Krause, J.A.; Connick, W.B. Ruthenium Bis-Diimine Complexes with a Chelating Thioether Ligand: Delineating 1,10-Phenanthrolinyl and 2,2′-Bipyridyl Ligand Substituent Effects. Inorg. Chem. 2014, 53, 294–307. [Google Scholar] [CrossRef]
- Andreicu, A.D.; Fischer-Fodor, E.; Pârvu, A.E.; Ţigu, A.B.; Cenariu, M.; Pârvu, M.; Cǎtoi, F.A.; Irimie, A. Antitumoral and Immunomodulatory Effect of Mahonia Aquifolium Extracts. Oxid. Med. Cell. Longev. 2019, 2019, 6439021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golbaghi, G.; Castonguay, A. Rationally Designed Ruthenium Complexes for Breast Cancer Therapy. Molecules 2020, 25, 265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotcherlakota, R.; Barui, A.K.; Prashar, S.; Fajardo, M.; Briones, D.; Rodríguez-Diéguez, A.; Patra, C.R.; Gómez-Ruiz, S. Curcumin Loaded Mesoporous Silica: An Effective Drug Delivery System for Cancer Treatment. Biomater. Sci. 2016, 4, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Karges, J.; Díaz-García, D.; Prashar, S.; Gómez-Ruiz, S.; Gasser, G. Ru(II) Polypyridine Complex-Functionalized Mesoporous Silica Nanoparticles as Photosensitizers for Cancer Targeted Photodynamic Therapy. ACS Appl. Bio Mater. 2021, 4, 4394–4405. [Google Scholar] [CrossRef]


| IC50Concentration (µM) | L1 | L2 | Complex 1 | Complex 2 | Complex 3 | Complex 4 | Carboplatin | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Duration of Treatment | IC50 | SEM | IC50 | SEM | IC50 | SEM | IC50 | SEM | IC50 | SEM | IC50 | SEM | IC50 | SEM |
| 24 h | 210.29 | 8.12 | 226.91 | 7.61 | 196.72 | 8.49 | 143.55 | 6.948 | 82.81 | 13.10 | 113.36 | 4.69 | 103.78 | 6.51 |
| 48 h | 130.63 | 3.45 | 184.12 | 6.50 | 48.33 | 4.06 | 3.12 | 0.050 | 5.29 | 0.41 | 78.87 | 2.99 | 37.04 | 2.24 |
| 72 h | 107.85 | 3.34 | 126.96 | 14.76 | 1.32 | 0.11 | 0.50 | 0.064 | 3.35 | 0.05 | 22.92 | 1.26 | 9.52 | 0.13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leskovská, J.; Miklášová, N.; Kubelac, P.M.; Achimaş-Cadariu, P.; Valentová, J.; Markuliak, M.; Fischer-Fodor, E. Antiproliferative Ruthenium Complexes Containing Curcuminoid Ligands Tested In Vitro on Human Ovarian Tumor Cell Line A2780, towards Their Capability to Modulate the NF-κBTranscription Factor, FGF-2 Growth Factor, and MMP-9 Pathway. Molecules 2022, 27, 4565. https://doi.org/10.3390/molecules27144565
Leskovská J, Miklášová N, Kubelac PM, Achimaş-Cadariu P, Valentová J, Markuliak M, Fischer-Fodor E. Antiproliferative Ruthenium Complexes Containing Curcuminoid Ligands Tested In Vitro on Human Ovarian Tumor Cell Line A2780, towards Their Capability to Modulate the NF-κBTranscription Factor, FGF-2 Growth Factor, and MMP-9 Pathway. Molecules. 2022; 27(14):4565. https://doi.org/10.3390/molecules27144565
Chicago/Turabian StyleLeskovská, Janka, Natalia Miklášová, Paul Milan Kubelac, Patriciu Achimaş-Cadariu, Jindra Valentová, Mário Markuliak, and Eva Fischer-Fodor. 2022. "Antiproliferative Ruthenium Complexes Containing Curcuminoid Ligands Tested In Vitro on Human Ovarian Tumor Cell Line A2780, towards Their Capability to Modulate the NF-κBTranscription Factor, FGF-2 Growth Factor, and MMP-9 Pathway" Molecules 27, no. 14: 4565. https://doi.org/10.3390/molecules27144565
APA StyleLeskovská, J., Miklášová, N., Kubelac, P. M., Achimaş-Cadariu, P., Valentová, J., Markuliak, M., & Fischer-Fodor, E. (2022). Antiproliferative Ruthenium Complexes Containing Curcuminoid Ligands Tested In Vitro on Human Ovarian Tumor Cell Line A2780, towards Their Capability to Modulate the NF-κBTranscription Factor, FGF-2 Growth Factor, and MMP-9 Pathway. Molecules, 27(14), 4565. https://doi.org/10.3390/molecules27144565






