Analysis and Evaluation of the Flagellin Activity of Bacillus amyloliquefaciens Ba168 Antimicrobial Proteins against Penicillium expansum
Abstract
1. Introduction
2. Results
2.1. Identification of Antimicrobial Proteins in Ba168
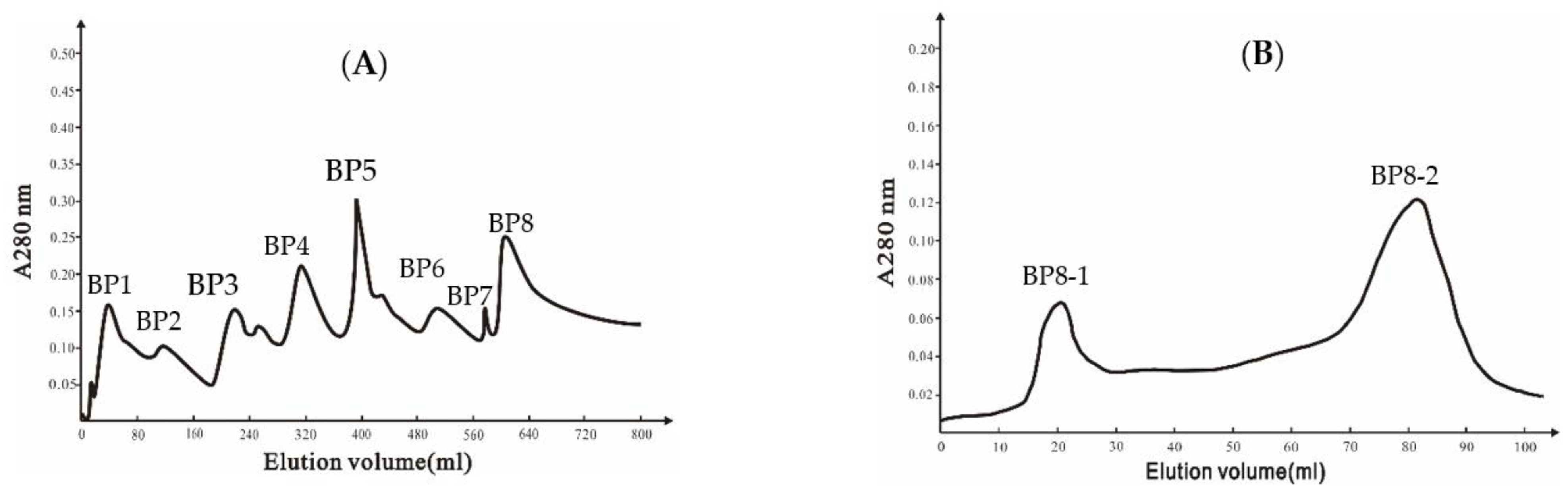
2.2. Purification of the Antifungal Protein in Ba168
2.3. Inhibitory Effects of the Antifungal Protein BP8-2 on P. expansum In Vitro
2.4. Control Effect of the Antifungal Protein BP8-2 on P. expansum In Vivo
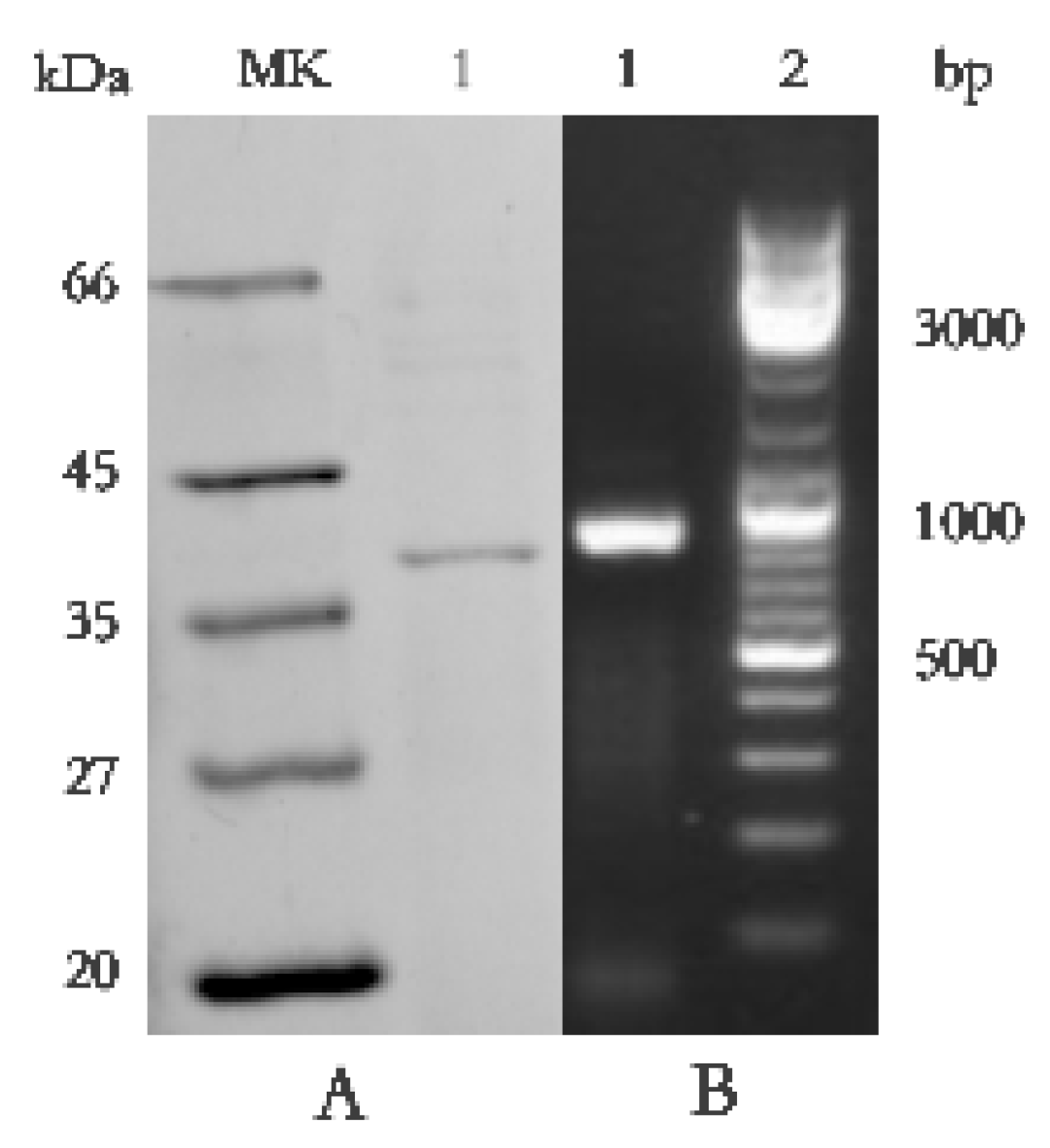
2.5. Identification of the Antifungal Protein BP8-2 and Amplification of Its Genomes
3. Discussion
4. Materials and Methods
4.1. Strain and Cultivation Conditions
4.2. Protein Extraction
4.3. Preparation of Proteins for LC-MS/MS
4.4. LC-MS/MS Analysis
4.5. Data Analysis
4.6. Purification of the Antifungal Protein
4.7. Inhibitory Effect of the Antifungal Protein BP8-2 on P. expansum In Vitro
4.8. Determination of Minimum Inhibitory Concentration (MIC) of the Antifungal Protein BP8-2 on P. expansum
4.9. Control Effect of the Antifungal Protein BP8-2 on P. expansum In Vivo
4.10. Identification of the Antifungal Protein BP8-2 and Cloning of Hag GENE in Ba168
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Droby, S. Improving quality and safety of fresh fruits and vegetables after harvest by the use of biocontrol agents and natural materials. Acta Hortic. 2006, 709, 45–51. [Google Scholar] [CrossRef]
- El-Ghaouth, A.; Smilanick, J.L.; Brown, G.E.; Ippolito, A.; Wilson, C.L. Control of decay of apple and citrus fruits in semicommercial tests with Candida saitoana and 2-Deoxy-d-glucose. Biol. Control. 2001, 20, 96–101. [Google Scholar] [CrossRef]
- Kulkarni, S. Commercialisation of Microbial Biopesticides for the Management of Pests and Diseases, Recent Advances in the Diagnosis and Management of Plant Diseases; Springer: New Delhi, India, 2015. [Google Scholar]
- Spadaro, D.; Droby, S. Development of biocontrol products for postharvest diseases of fruit: The importance of elucidating the mechanisms of action of yeast antagonists. Trends Food Sci. Technol. 2016, 47, 39–49. [Google Scholar] [CrossRef]
- Alvindia, D.G.; Natsuaki, K.T. Biocontrol activities of Bacillus amyloliquefaciens DGA14 isolated from banana fruit surface against banana crown rot-causing pathogens. Crop Prot. 2009, 28, 236–242. [Google Scholar] [CrossRef]
- Arguelles-Arias, A.; Ongena, M.; Halimi, B.; Lara, Y.; Brans, A.; Joris, B.; Fickers, P. Bacillus amyloliquefaciens GA1 as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogens. Microb. Cell Factories 2009, 8, 63. [Google Scholar] [CrossRef]
- Arrebola, E.; Jacobs, R.; Korsten, L. Iturin A is the principal inhibitor in the biocontrol activity of Bacillus amyloliquefaciens PPCB004 against postharvest fungal pathogens. J. Appl. Microbiol. 2010, 108, 386–395. [Google Scholar] [CrossRef]
- Fravel, D.R. Commercialization and implementation of biocontrol. Annu. Rev. Phytopathol. 2005, 43, 337–359. [Google Scholar] [CrossRef]
- Nabila, E.A.; Soufiyan, E.A. Microbial biocontrol of post-harvest fungal rot in apples: Current state of the science. Bull. Bot. Res. 2020, 2, 28. [Google Scholar]
- Cai, W.; Xia, B.; Xia, Y.; Yi, Y.; Wang, Y. Effect of Bacillus amyloliquefaciens fermentation liquid on different types of peppers for biological control of postharvest A. alternate. Trans. Chin. Soc. Agric. Eng. 2013, 29, 253–261. [Google Scholar]
- Tan, S.; Dong, Y.; Liao, H.; Huang, J.; Song, S.; Xu, Y.; Shen, Q. Antagonistic bacterium Bacillus amyloliquefaciens induces resistance and controls the bacterial wilt of tomato. Pest Manag. Sci. 2013, 69, 1245–1252. [Google Scholar] [CrossRef]
- Ye, F.; Yang, R.; Hua, X.; Shen, Q.; Zhao, W.; Zhang, W. Modification of stevioside using transglucosylation activity of Bacillus amyloliquefaciens alpha-amylase to reduce its bitter aftertaste. LWT Food Sci. Technol. 2013, 51, 524–530. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Li, B.Q.; Wang, Y.; Guo, Q.G.; Lu, X.Y.; Li, S.Z.; Ma, P. Lipopeptides, a novel protein, and volatile compounds contribute to the antifungal activity of the biocontrol agent Bacillus atrophaeus CAB-1. Appl. Microbiol. Biotechnol. 2013, 97, 9525–9534. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhang, X.; Huang, H.; Zhai, F.; An, T.C.; An, D.R. Control effect and mechanism of Bacillus amyloliquefaciens Ba-168 on wheat take-all. J. Plant Prot. 2013, 40, 475–476. [Google Scholar]
- Chao, J.J.; Yang, H.Y.; Xie, X.S.; Tian, J.; Wu, Y.F.; An, D.R. Control effect of Bacillus amyloliquefaciens on wheat yellow mosaic disease. Acta Phytophylacica Sin. 2016, 43, 588–593. [Google Scholar]
- Kesarwani, M.; Azam, M.; Natarajan, K.; Mehta, A.; Datta, A. Oxalate decarboxylase from collybia velutipes. J. Biol. Chem. 2000, 275, 7230–7238. [Google Scholar] [CrossRef]
- De, D.L.A. Plant-derived antifungal proteins and peptides. Can. J. Microbiol. 2005, 51, 1001–1014. [Google Scholar]
- Zhao, X.; Zhao, X.; Wei, Y.; Shang, Q.; Liu, Z. Isolation and identification of a novel antifungal protein from a rhizobacterium Bacillus subtilisstrain F3. J. Phytopathol. 2012, 161, 43–48. [Google Scholar] [CrossRef]
- Garciabustos, J.F.; Pezzi, N.; Mendez, E. Structure and mode of action of microcin 7, an antibacterial peptide produced by Escherichia coli. Antimicrob. Agents Chemother. 1985, 27, 791. [Google Scholar] [CrossRef]
- Sharma, A.K.; Kumar, S.K.H.; Dhakan, D.B.; Sharma, V.K. Prediction of peptidoglycan hydrolases- a new class of antibacterial proteins. BMC Genom. 2016, 17, 411. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Guo, Q.G.; Lu, X.Y.; Zhao, W.S.; Wang, P.P.; Li, S.Z.; Ma, P. Isolation and identification of an antifungal protein produced by Bacillus subtilis NCD-2. Acta Phytopathol. Sin. 2018, 48, 395–401. [Google Scholar]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.L.; Fang, Y.Q.; Sun, Y.; Luan, N.; Chen, X.; Chen, M.R.; Han, Y.J.; Yin, Y.Z.; Mwangi, J.; Niu, J.K.; et al. Antimicrobial peptide LL-37 forms complex with bacterial DNA to facilitate blood translocation of bacterial DNA and aggravate ulcerative colitis. Chin. Sci. Bull. 2018, 63, 1364–1375. [Google Scholar] [CrossRef]
- Muthu, S.; Gopal, V.B.; Soundararajan, S.; Nattarayan, K.; Narayan, K.S.; Lakshmikanthan, M.; Malairaj, S.; Perumal, P. Antibacterial serine protease from Wrightia tinctoria: Purification and characterization. Plant Physiol. Biochem. 2017, 112, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.J.; Park, S.M.; Ahn, B.Y. Isolation and characterization of an antimicrobial substance from Bacillus subtilis BY08 antagonistic to Bacillus cereus and Listeria monocytogenes. Food Sci. Biotechnol. 2013, 22, 433–440. [Google Scholar] [CrossRef]
- Li, B.L.; Wang, H.B.; Kong, F.Y.; Zhang, C.S.; Wang, J.; Li, X.K. Purification and partial characterization of antagonistic proteins from Bacillus Subtilis Tpb55. Chin. Tob. Sci. 2011, 11, 565. [Google Scholar]
- Antonio, G.; Maqueda, M.; Manuel, M.-B.; Lebbadi, M.; Vaidivia, E. Isolation and physico-chemical characterization of an antifungal and antibacterial peptide produced by Bacillus licheniformis A12. Appl. Microbiol. Biotechnol. 1993, 39, 438–442. [Google Scholar]
- Sandrin, C.; Peypoux, F.; Michel, G. Coproduction of surfactin and iturin A, lipopeptides with surfactant and antifungal properties, by Bacillus subtilis. Biotechnol. Appl. Biochem. 1990, 12, 370–375. [Google Scholar]
- Chen, X.H.; Koumoutsi, A.; Scholz, R.; Schneider, K.; Vater, J.; Süssmuth, R.; Piel, J.; Borriss, R.; Pühler, A.; Selbitschka, W. Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J. Biotechnol. 2008, 140, 27–37. [Google Scholar] [CrossRef]
- Chen, X.H.; Scholza, R.; Borrissb, M.; Jungeb, H.; Mögel, G.; Kunz, S.; Borrissa, R. Difficidin and bacilysin produced by plant-associated Bacillus amyloliquefaciens are efficient in controlling fire blight disease. J. Biotechnol. 2009, 140, 38–44. [Google Scholar] [CrossRef]
- Wu, L.M.; Wu, H.J.; Chen, L.N.; Yu, X.F.; Borriss, R.; Gao, X.W. Difficidin and bacilysin from Bacillus amyloliquefaciens FZB42 have antibacterial activity against Xanthomonas oryzae rice pathogens. Sci. Rep. 2015, 5, 12975. [Google Scholar] [CrossRef]
- Zheng, G.; Yan, L.Z.; Vederas, J.C.; Zuber, P. Genes of the sbo-alb locus of Bacillus subtilis are required for production of the antilisterial bacteriocin subtilosin. J. Bacteriol. 1999, 181, 7346–7355. [Google Scholar] [CrossRef]
- Chen, Z.; Huang, J.; Zhao, J.; Wang, C.B.; Liang, H. Research advances on antibacterial mechanism of Bacillus amyloliquefaciens. Biotechnol Bull. 2015, 17, 293–296. [Google Scholar]
- Abdullah, M.T.; Ali, N.Y.; Suleman, P. Biological control of Sclerotinia sclerotiorum (Lib.) de Bary with Trichoderma harzianum and Bacillus amyloliquefaciens. Crop Prot. 2008, 27, 1354–1359. [Google Scholar] [CrossRef]
- Caldeira, A.T.; Arteiro, J.M.S.; Coelho, A.V.; Roseiro, J.C. Combined use of LC–ESI-MS and antifungal tests for rapid identification of bioactive lipopeptides produced by Bacillus amyloliquefaciens CCMI 1051. Process Biochem. 2011, 46, 1738–1746. [Google Scholar] [CrossRef]
- Neupane, S.; Ma, Q.; Mathew, F.M.; Varenhorst, A.J.; Andersen, E.J.; Nepal, M.P. Evolutionary divergence of TNL disease-resistant proteins in soybean (glycine max) and common bean (phaseolus vulgaris). Biochem. Genet. 2018, 56, 397–422. [Google Scholar] [CrossRef] [PubMed]
- Seung, H.J.; Narayan, C.P.; Xin, D.J.; Young, S.K.; Bong-Sik, Y.; Seung, H.Y. Biocontrol activity of Bacillus amyloliquefaciens CNU114001 against fungal plant diseases. Mycobiology 2013, 41, 234–242. [Google Scholar]
- Bacon, J.S.; Gordon, A.H.; Jones, D.; Tayor, I.; Webley, D. The separation of beta-glucanases produced by Cytophaga johnsonii and their role in the lysis of yeast cell walls. Biochem. J. 1970, 120, 67. [Google Scholar] [CrossRef]
- Mehmood, M.; Hussain, K.; Latif, F.; Tabassum, M.; Gull, M.; Gill, S.; Anam, S.; Zahid, I. Synergistic action of the antifungal β-chitin binding protein cbp50 from Bacillus thuringiensis with bacterial chitinases. Curr. Proteom. 2014, 11, 23–26. [Google Scholar] [CrossRef]
- Kpb, V.D.B.; Rouge, P.P.; Coosemans, J.; Krouglova, T. Synergistic antifungal activity of two chitin-binding proteins from spindle tree (Euonymus europaeus L.). Planta 2004, 219, 221–232. [Google Scholar]
- He, P.; Hao, K.; Blom, J.; Rückert, C.; Vater, J.; Mao, Z.; Wu, Y.; Hou, M.; He, P.; He, Y. Genome sequence of the plant growth promoting strain Bacillus amyloliquefaciens subsp. plantarum B9601-Y2 and expression of mersacidin and other secondary metabolites. J. Biotechnol. 2013, 164, 281–291. [Google Scholar] [CrossRef]
- Eckhard, U.; Bandukwala, H.; Mansfield, M.J.; Marino, G.; Cheng, J.J.; Wallace, I.; Holyoak, T.; Charles, T.C.; Austin, J.; Overall, C.M.; et al. Discovery of a proteolytic flagellin family in diverse bacterial phyla that assembles enzymatically active flagella. Nat. Commun. 2017, 8, 521. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.I.; Lee, C.; Park, J.; Jeon, B.Y.; Hong, M. Structure-guided fusion-protein designs using Bacillus flagellin as a vaccine adjuvant. Acta Crystallogr. Sect. A Found. Adv. 2018, 74, a134. [Google Scholar] [CrossRef]
- Felix, G.; Duran, J.D.; Volko, S.; Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 2010, 18, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.J.; Shi, G.L.; Xiao, Q.W.; Liu, J.G.; Wang, Y.N. Identification and characterization of a novel Bacillus subtilis strain with potent antifungal activity of a flagellin-like protein. World J. Microbiol. Biotechnol. 2013, 29, 2343–2352. [Google Scholar] [CrossRef]
- Fang, Y.Y.; Li, L.L.; Zheng, Y.L.; Yuan, H.H.; Zhang, X.H.; Cheng, S.Y.; Cheng, H. Extraction, isolation and identification of antimicrobial substances from Bacillus amyloliquefaciens CMN1308. Not. Bot. Horti Agrobot. Cluj-Napoca 2017, 45, 308. [Google Scholar] [CrossRef][Green Version]
- Djordje, F.; Ivica, D.; Tanja, B.; Jelena, L.; Slaviša, S. Biological control of plant pathogens by Bacillus species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Wiggins, B.E.; Kinkel, L.L. Green manures and crop sequences influence potato diseases and pathogen inhibitory activity of indigenous streptomycetes. Phytopathology 2005, 95, 178–185. [Google Scholar] [CrossRef]
- Sadeghian, M.; Bonjar, G.H.S.; Sirchi, G.R.S. Post harvest biological control of apple bitter rot by soil-borne actinomycetes and molecular identification of the active antagonist. Postharvest Biol. Technol. 2016, 112, 46–54. [Google Scholar] [CrossRef]
- Laemmli, U.K. Maturation of the head of bacteriophage T4. I. DNA packaging events. J. Mol. Biol. 1973, 80, 575–599. [Google Scholar] [CrossRef]
- Zhao, X.; Zhou, Z.J.; Han, Y.; Wang, Z.Z.; Fan, J.; Xiao, H.Z. Isolation and identification of antifungal peptides from Bacillus BH072, a novel bacterium isolated from honey. Microbiol. Res. 2013, 168, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Kwok, S.; Higuchi, R. Avoiding false positives with PCR. Nature 1989, 339, 237–238. [Google Scholar] [CrossRef] [PubMed]





| Project | Protein Name | Protein ID(UniProt) | Gene Name | Molecular Weight (Da) | References |
|---|---|---|---|---|---|
| Antimicrobial protein | Oxalate decarboxylase | A0A0G3VD82 | oxdC | 43,574.56 | [16] |
| Chitin-binding proteins | A0A0D7XZQ5 | gbpA | 22,450.79 | [17] | |
| Flagellin | I2CAR1 | hag | 35,479.84 | [18] | |
| Carboxypeptidase | A0A0K6LSU1 | ykfA | 33,685.03 | [19,20] | |
| Aminopeptidase | I2C7H0 | yqjE | 39,509.56 | [21] | |
| Antimicrobial peptide | I2C153 | lci | 9770.11 | [22,23] | |
| Serine protease | A0A0D7XMJ4 | UZ38_16760 | 47,241.97 | [24] | |
| Subtilisin | A0A0K6L9A7 | apr | 39,099.63 | [25] | |
| Leucine dehydrogenase | A0A0G3VBQ2 | ldh | 39,688.26 | [26] | |
| β-glucanase | A0A0D7XPS0 | UZ38_11685 | 27,386.34 | [27] | |
| Proteins related to the formation of antimicrobial proteins | Surfactin synthase A | I2C187 | srfAA | 402,709.4 | [28] |
| Polyketide synthase | A0A1J0C866 | BAMY_11440 | 573,237.2 | [29] | |
| bacillomycin D synthetase C | H9TE62 | bmyC | 299,764.2 | NCBI | |
| Bacillomycin D synthetase A | H9TE64 | bmyA | 448,739.5 | NCBI | |
| bacilysin biosynthesis protein | A0A142F9I5 | bacA | 27,999.68 | [30,31] | |
| Antilisterial bacteriocin subtilosin biosynthesis protein | A0A0K6M2R3 | albE | 48,507.52 | [32] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, M.; Chen, Y.; Li, L.; Ma, Y.; Tong, Z.; Guo, D.; Sun, P.; An, D. Analysis and Evaluation of the Flagellin Activity of Bacillus amyloliquefaciens Ba168 Antimicrobial Proteins against Penicillium expansum. Molecules 2022, 27, 4259. https://doi.org/10.3390/molecules27134259
Lu M, Chen Y, Li L, Ma Y, Tong Z, Guo D, Sun P, An D. Analysis and Evaluation of the Flagellin Activity of Bacillus amyloliquefaciens Ba168 Antimicrobial Proteins against Penicillium expansum. Molecules. 2022; 27(13):4259. https://doi.org/10.3390/molecules27134259
Chicago/Turabian StyleLu, Meihuan, Yahan Chen, Lijun Li, Yinghui Ma, Zefang Tong, Dongsheng Guo, Pingping Sun, and Derong An. 2022. "Analysis and Evaluation of the Flagellin Activity of Bacillus amyloliquefaciens Ba168 Antimicrobial Proteins against Penicillium expansum" Molecules 27, no. 13: 4259. https://doi.org/10.3390/molecules27134259
APA StyleLu, M., Chen, Y., Li, L., Ma, Y., Tong, Z., Guo, D., Sun, P., & An, D. (2022). Analysis and Evaluation of the Flagellin Activity of Bacillus amyloliquefaciens Ba168 Antimicrobial Proteins against Penicillium expansum. Molecules, 27(13), 4259. https://doi.org/10.3390/molecules27134259





