DNA Interaction, DNA Photocleavage, Photocytotoxicity In Vitro, and Molecular Docking of Naphthyl-Appended Ruthenium Complexes
Abstract
:1. Introduction
2. Results and Discussion
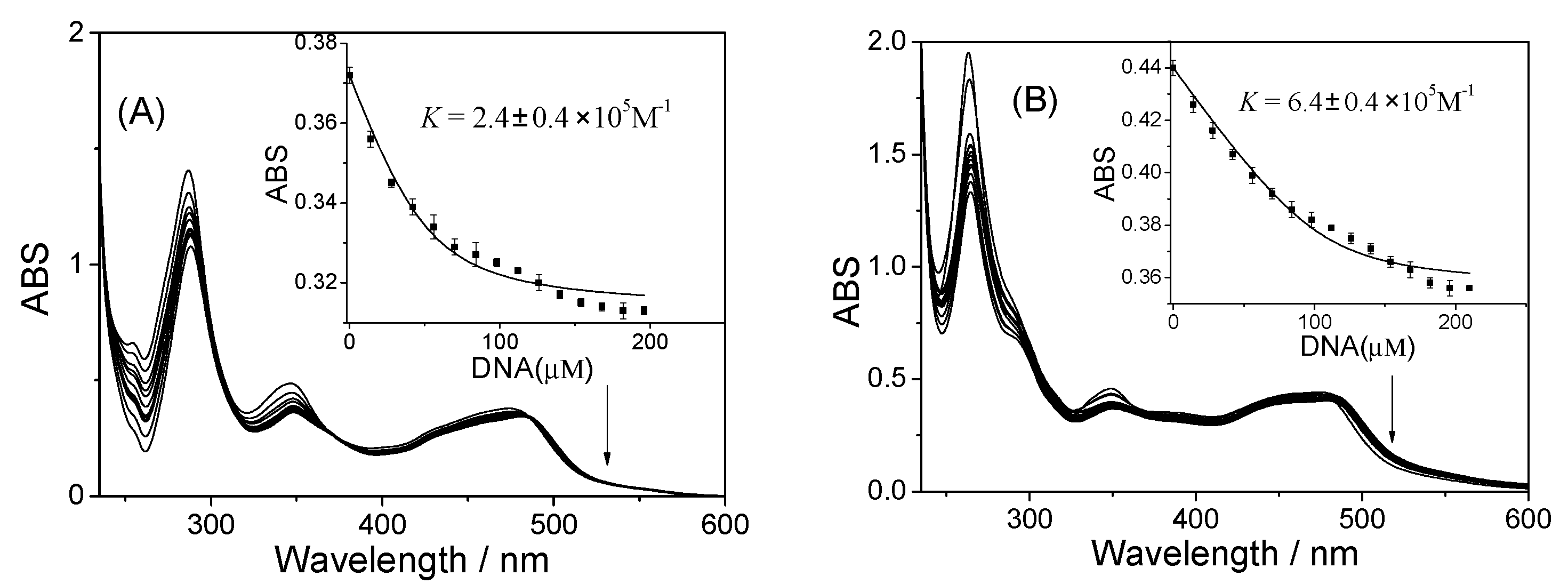
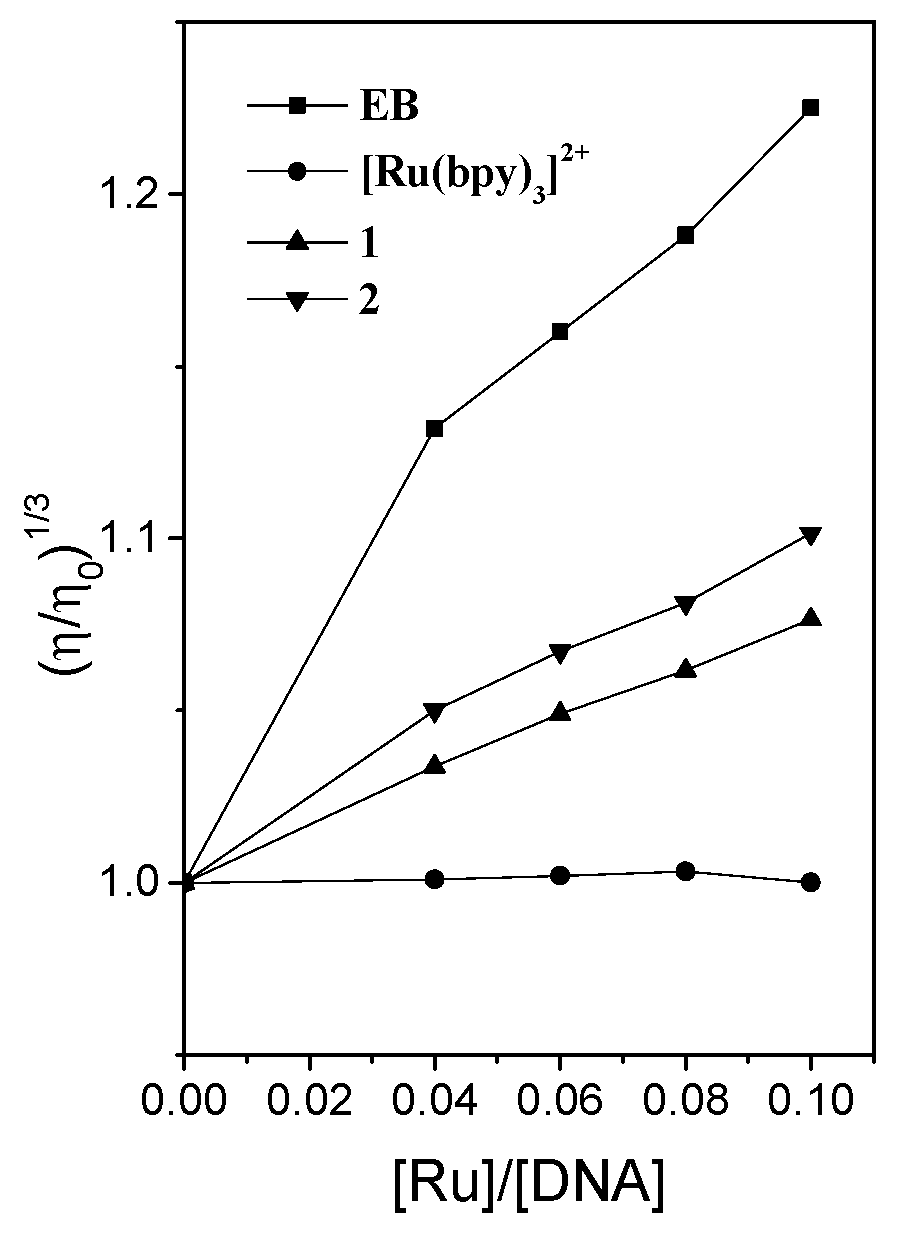
2.1. Studies on DNA Interaction
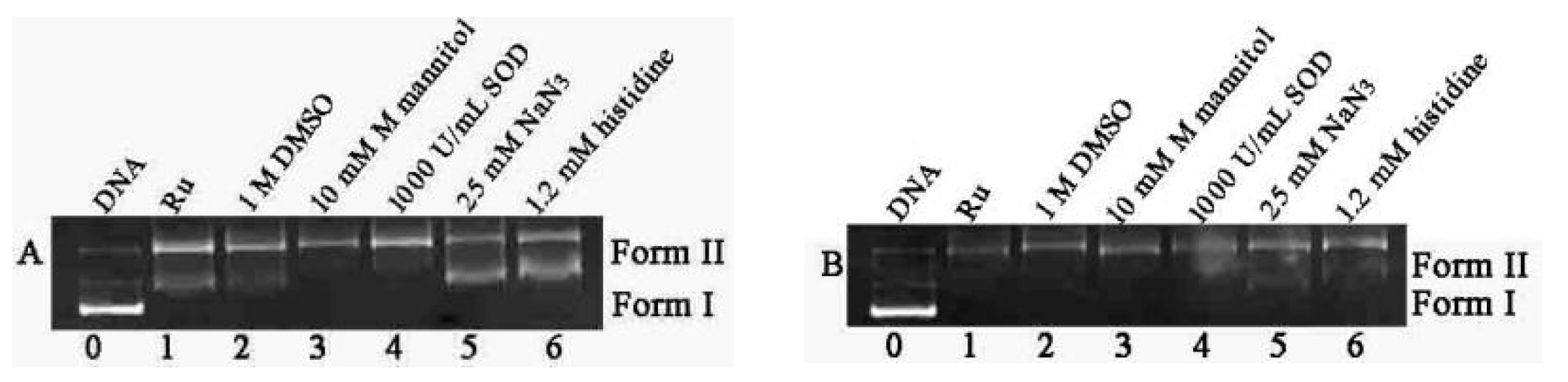
2.2. DNA Photocleavage Activities
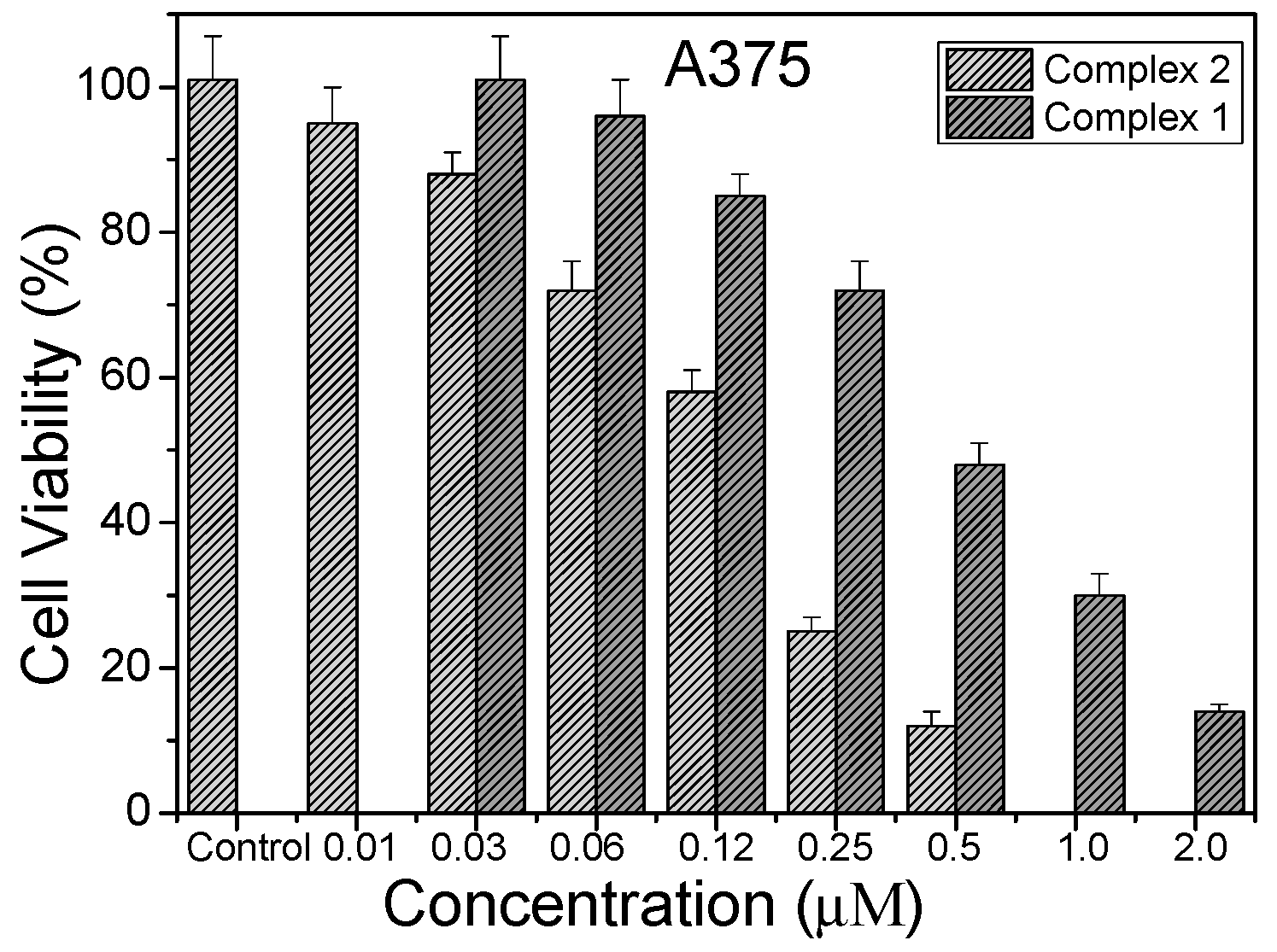
2.3. Photocytotoxic Activity In Vitro
3. Materials and Methods
3.1. Instrumentation
3.2. UV-Visible Titrations
3.3. Viscosity Measurement
3.4. Emission Measurements
3.5. DNA Photocleavage Experiments
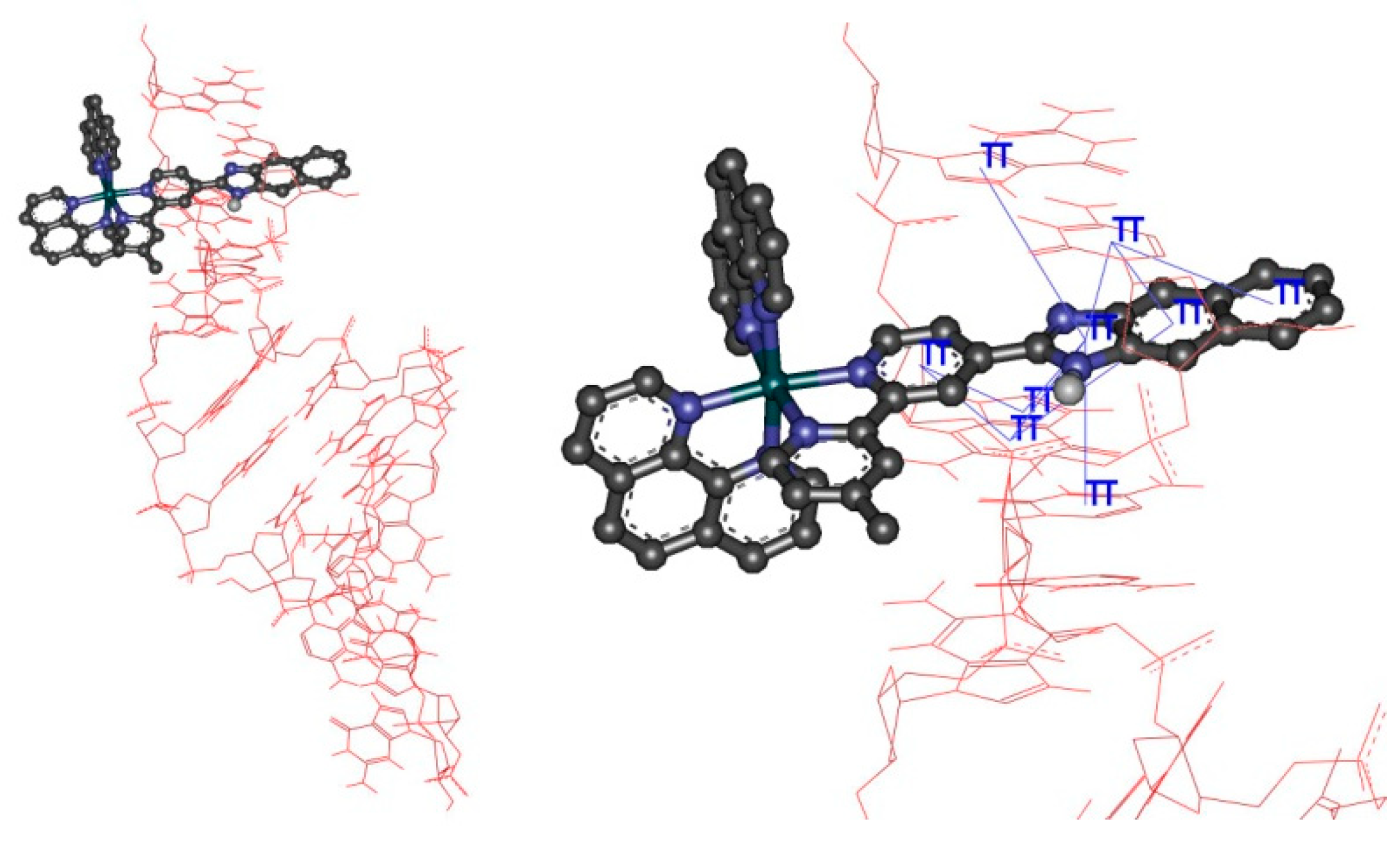
3.6. Molecular Docking
3.7. Photocytotoxicity
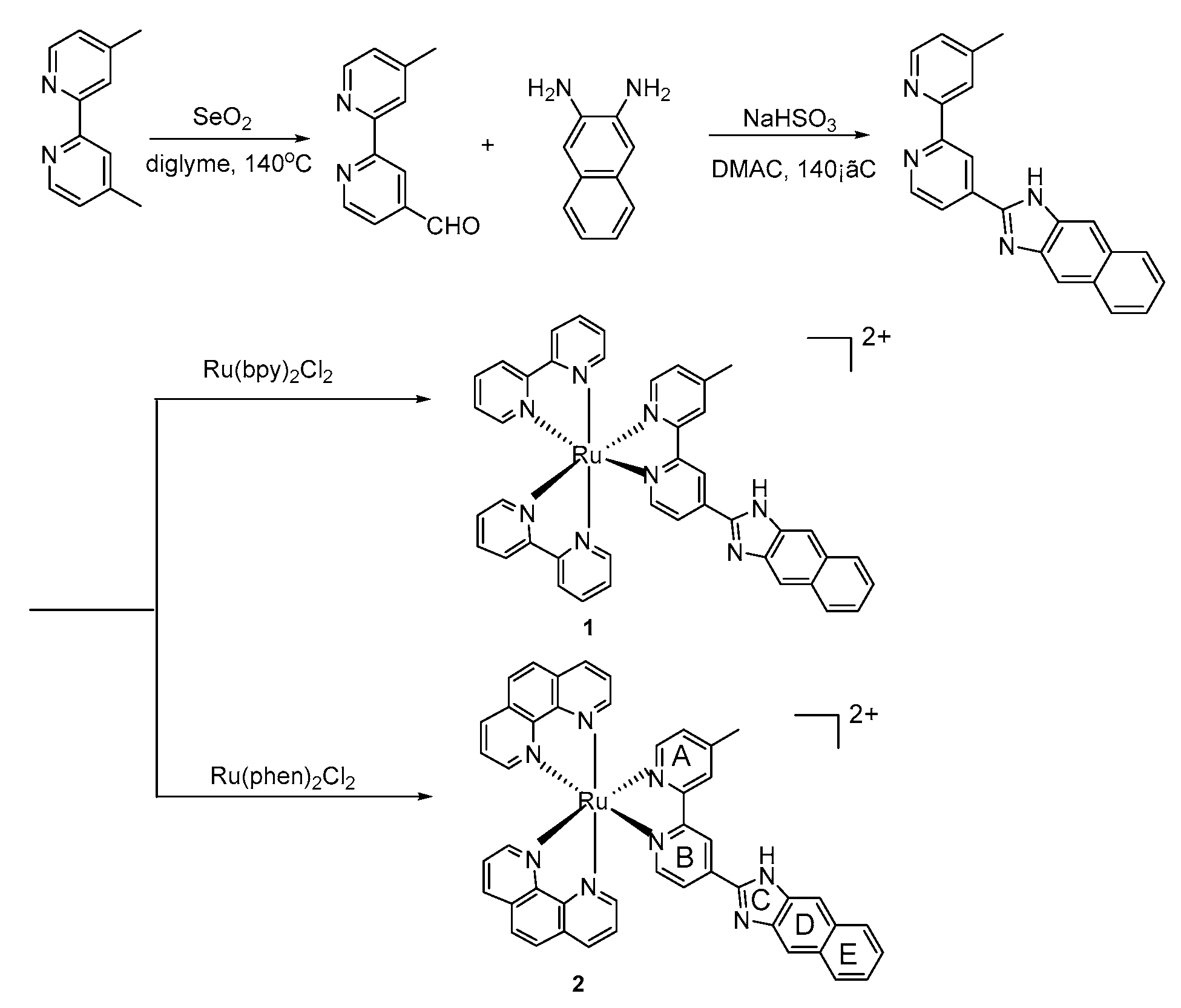
3.8. Synthesis
3.8.1. 2-(4′-Methyl-bipyridine-4-yl)-1H-imidazo[4,5-b]Naphthalene (Mbin)
3.8.2. Synthesis of Ruthenium Complexes
[Ru(bpy)2(mbin)](PF6)2(1)
[Ru(phen)2(mbin)](PF6)2 (2)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Wang, Y.Y.; Liu, Y.C.; Sun, H.; Guo, D.S. Type I photodynamic therapy by organic-inorganic hybrid materials: From strategies to applications. Coord. Chem. Rev. 2019, 395, 46–62. [Google Scholar] [CrossRef]
- Sztandera, K.; Gorzkiewicz, M.; Klajnert-Maculewicz, B. Nanocarriers in photodynamic therapy-in vitro and in vivo studies. Wires Nanomed. Nanobiotechnol. 2020, 12, e1509. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Shi, Z.L.; Wu, J.G.; Wu, J.L.; He, C.; Hao, X.R.; Duan, C.Y. Lighting Up Metallohelices: From DNA Binders to Chemotherapyand Photodynamic Therapy. Chem. Commun. 2020, 56, 7537–7548. [Google Scholar] [CrossRef]
- McKenzie, L.K.; Bryant, H.E.; Weinstein, J.A. Transition metal complexes as photosensitisers in one-and two-photon photodynamic therapy. Coord. Chem. Rev. 2019, 379, 2–29. [Google Scholar] [CrossRef] [Green Version]
- Shi, G.; Monro, S.; Hennigar, R.; Colpitts, J.; Fong, J.; Kasimova, K.; Yin, H.; DeCoste, R.; Spencer, C.; Chamberlain, L. Ru(II) dyads derived from α-oligothiophenes: A new class of potent and versatile photosensitizers for PDT. Coord. Chem. Rev. 2015, 282, 127–138. [Google Scholar] [CrossRef]
- Zeng, L.L.; Gupta, P.; Chen, Y.; Wang, E.; Ji, L.N.; Chao, H.; Chen, Z.S. The development of anticancer ruthenium (II) complexes: From single molecule compounds to nanomaterials. Chem. Soc. Rev. 2017, 46, 5771–5804. [Google Scholar] [CrossRef]
- Furrer, J.; Süss-Fink, G. Thiolato-bridged dinuclear arene ruthenium complexes and their potential as anticancer drugs. Coord. Chem. Rev. 2016, 309, 36–50. [Google Scholar] [CrossRef]
- Li., F.; Collins, J.G.; Keene, F.R. Ruthenium complexes as antimicrobial agents. Chem. Soc. Rev. 2015, 44, 2529–2542. [Google Scholar] [CrossRef] [Green Version]
- Martnez, R.; Chacn-Garca, L. The search of DNA-intercalators as. antitumoral drugs: What it worked and what did not work. Curr. Med. Chem. 2005, 12, 127–151. [Google Scholar] [CrossRef]
- Dougherty, T.J. Photodynamic therapy. Photochem. Photobiol. 1993, 58, 895–900. [Google Scholar] [CrossRef]
- Li, J.J.; Tian, Z.Z.; Xu, Z.S.; Zhang, S.M.; Feng, Y.Q.; Zhang, L.D.; Liu, Z. Highly potent half-sandwich iridium and ruthenium complexes as lysosome-targeted imaging and anticancer agents. Dalton Trans. 2018, 47, 15772–15782. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, A.; Shyamsivappan, S.; Kalagatur, N.K.; Suresh, T.; Maroli, N.; Bhuvanesh, N.; Kolandaivel, P.; Mohan, P.S. Application of real sample analysis and biosensing: Synthesis of new naphthyl derived chemosensor for detection of Al3+ ions. Spectrochim. Acta A 2020, 241, 118684. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Kundu, M.; Roy, S.; Roy, B.; Shah, S.S.; Nair, A.V.; Pal, B.; Mondal, M.; Singh, N.D.P. A two-photon responsive naphthyl tagged p-hydroxyphenacyl based drug delivery system: Uncaging of anti-cancer drug in the phototherapeutic window with real-time monitoring. Chem. Commun. 2020, 56, 9986–9989. [Google Scholar] [CrossRef] [PubMed]
- Goswami, T.K.; Gadadhar, S.; Roy, M.; Nethaji, M.; Karande, A.A.; Chakravarty, A.R. Ferrocene-Conjugated Copper(II) Complexes of L-Methionine andPhenanthroline Bases: Synthesis, Structure, and PhotocytotoxicActivity. Organometallics 2012, 31, 3010–3021. [Google Scholar] [CrossRef]
- Mondal, A.; Paira, P. Hypoxia efficient and glutathione-resistantcytoselective ruthenium(II)-p-cymenearylimidazophenanthroline complexes:biomolecular interaction and live cell imaging. Dalton Trans. 2020, 49, 12865–12878. [Google Scholar] [CrossRef]
- Ghosh, G.; Yin, H.M.; Monro, S.M.A.; Sainuddin, T.; Lapoot, L.; Greer, A.; McFarland, S.A. Synthesis and Characterization of Ru(II) Complexes with p-ExpansiveImidazophen Ligands for the Photokilling of Human Melanoma Cells. Photochem. Photobiol. 2020, 96, 349–357. [Google Scholar] [CrossRef]
- Cole, H.D.; Roque, J.A., III; Lifshits, L.M.; Hodges, R.; Barrett, P.C.; Havrylyuk, D.; Heidary, D.; Ramasamy, E.; Cameron, C.G.; Glazer, E.C.; et al. Fine-Feature Modifications to Strained Ruthenium Complexes RadicallyAlter Their Hypoxic Anticancer Activity. Photochem. Photobiol. 2021, 98, 73–84. [Google Scholar] [CrossRef]
- Abreu, F.D.; Paulo, T.F.; Gehlen, M.H.; Ando, R.A.; Lopes, L.G.F.; Gondim, A.C.S.; Vasconcelos, M.A.; Teixeira, E.H.; Sousa, E.H.S.; Carvalho, I.M.M. Aryl-Substituted Ruthenium(II) Complexes: A Strategy for EnhancedPhotocleavage and Efficient DNA Binding. Inorg. Chem. 2017, 56, 9084–9096. [Google Scholar] [CrossRef]
- Abreu, F.D.; Diógenes, I.C.N.; Lopes, L.G.d.; Sousa, E.H.S.; Carvalho, I.M.M. Ruthenium(II) Bipyridine Complexes with Pendant Anthracenyl and NaphthylMoieties: A Strategy for a ROS Generator with DNA binding Selectivity. Inorg. Chim. Acta 2016, 439, 92–99. [Google Scholar] [CrossRef]
- Zeng, L.L.; Kuang, S.; Li, G.Y.; Jin, C.Z.; Ji, L.N.; Ji, C.H. A GSH-activatable ruthenium(II)-azo photosensitizer for twophoton photodynamic therapy. Chem. Commun. 2017, 53, 1977–1980. [Google Scholar] [CrossRef]
- Chen, Y.J.; Lei, W.H.; Jiang, G.Y.; Zhou, Q.X.; Hou, Y.J.; Li, C.; Zhang, B.W.; Wang, X.S. A ruthenium(II) arene complex showing emissionenhancement and photocleavage activity towards DNAfrom singlet and triplet excited states respectively. Dalton Trans. 2013, 42, 5924–5931. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.G.; Ye, B.H.; Li, H.; Zhen, Q.X.; Ji, L.N.; Fu, R.H. Polypyridyl ruthenium(II) complexes containing intramolecular hydrogen-bond ligand: Syntheses, characterization, and DNA-binding properties. J. Inorg. Biochem. 1999, 76, 265–271. [Google Scholar] [CrossRef]
- McGhee, J.D.; Hippel von, P.H. Theoretical aspects of DNA-protein interactions: Co-operative and non-cooperative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974, 86, 469–489. [Google Scholar] [CrossRef]
- Ambroise, A.; Maiya, B.G. Ruthenium (II) complexes of 6; 7-dicyanodipyridoquinoxaline: Synthesis; luminescence studies and DNA interaction. Inorg. Chem. 2000, 39, 4256–4263. [Google Scholar] [CrossRef]
- Liu, J.G.; Ye, B.H.; Li, H.; Ji, L.N.; Li, R.H.; Zhou, J.Y. Synthesis, characterization and DNA-binding properties of novel dipyridophenazine complex of ruthenium (II):[Ru(IP)2(DPPZ)]2+. J. Inorg. Biochem. 1999, 73, 117–122. [Google Scholar] [CrossRef]
- Pyle, A.M.; Rehmann, J.P.; Meshoyrer, R.; Kumar, C.V.; Turro, N.J.; Barton, J.K. Mixed-Ligand Complexes of Ruthenium(11): Factors Governing Binding to DNA. J. Am. Chem. Soc. 1989, 111, 3051–3058. [Google Scholar] [CrossRef]
- Satyanarayana, S.; Dabroniak, J.C.; Chaires, J.B. NeitherDELTA- nor LAMBDA-tris (phenanthroline) ruthenium (II) binds to DNA by classical intercalation. Biochemistry 1992, 31, 9319–9324. [Google Scholar] [CrossRef]
- Satyanarayana, S.; Dabroniak, J.C.; Chaires, J.B. Tris(phenanthroline) ruthenium (II) enantiomer interactions with DNA: Mode and specificity of binding. Biochemistry 1993, 32, 2573–2584. [Google Scholar] [CrossRef]
- Cohen, G.; Eisenberg, H. Viscosity and sedimentation study of sonicated DNA–proflavine complexes. Biopolymers 1969, 8, 45–55. [Google Scholar] [CrossRef]
- Ji, L.N.; Zou, X.H.; Liu, J.G. Shape-and enantioselective interaction of Ru (II)/Co (III) polypyridyl complexes with DNA. Coord. Chem. Rev. 2001, 216, 513–536. [Google Scholar] [CrossRef]
- Barton, K.; Danishefsky, A.; Goldberg, J. Tris(phenanthroline)ruthenium(II): Stereoselectivity in binding to DNA. J. Am. Chem. Soc. 1984, 106, 2172–2176. [Google Scholar] [CrossRef]
- Burya, S.J.; Lutterman, D.A.; Turro, C. Absence of quenching by [Fe(CN)6]4− is not proof of DNA intercalation. Chem. Commun. 2011, 47, 1848–1850. [Google Scholar] [CrossRef]
- LePecq, J.B.; Paoletti, C. A fluorescent complex between ethidium bromide and nucleic acids: Physical-Chemical characterization. J. Mol. Biol. 1967, 27, 87–106. [Google Scholar] [CrossRef]
- Kumar, C.V.; Turro, N.J.; Barton, J.K. Photophysics of ruthenium complexes bound to double helical DNA. J. Am. Chem. Soc. 1985, 107, 5518–5523. [Google Scholar] [CrossRef]
- Waring, M.J. Complex formation between ethidium bromide and nucleic acids. J. Mol. Biol. 1965, 13, 269–282. [Google Scholar] [CrossRef]
- Mei, H.Y.; Barton, J.K. Tris (tetramethylphenanthroline) ruthenium (II): A chiral probe that cleaves A-DNA conformations. Proc. Natl. Acad. Sci. USA 1988, 85, 1339–1343. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, F.R.; Merkel, P.B.; Kearns, D.R. Unambiguous evidence for the participation of singlet oxygen (1δ) in photodynamic oxidation of amino acids. Photochem. Photobiol. 1972, 16, 117–124. [Google Scholar] [CrossRef]
- Yu, H.J.; Huang, S.M.; Li, L.Y.; Ji, H.N.; Chao, H.; Mao, Z.W.; Liu, J.Z.; Ji, L.N. Synthesis, DNA-binding and photocleavage studies of ruthenium complexes [Ru (bpy)2(mitatp)]2+ and [Ru (bpy)2(nitatp)]2+. J. Inorg. Biochem. 2009, 103, 881–890. [Google Scholar] [CrossRef]
- Gao, F.; Chao, H.; Zhou, F.; Yuan, Y.X.; Peng, B.; Ji, L.N. DNA interactions of a functionalized ruthenium(II) mixed-polypyridyl complex [Ru(bpy)2ppd]2+. J. Inorg. Biochem. 2006, 100, 1487–1494. [Google Scholar] [CrossRef]
- Abdel-Shafi, A.A.; Beer, P.D.; Mortimer, R.J.; Wilkinson, F. Photosensitized Generation of Singlet Oxygen from Vinyl Linked Benzo-Crown-Ether−Bipyridyl Ruthenium(II) Complexes. J. Phys. Chem. A 2000, 104, 192–202. [Google Scholar] [CrossRef]
- Stephenson, M.; Reichardt, C.; Pinto, M.; Wächtler, M.; Sainuddin, T.; Shi, G.; Yin, H.M.; Monro, S.; Sampson, E.; Dietzek, B.; et al. Ru(II) Dyads Derived from 2-(1-Pyrenyl)-1H-imidazo[4,5-f][1,10]phenanthroline: Versatile Photosensitizers for Photodynamic Applications. J. Phys. Chem. A 2014, 118, 10507–10521. [Google Scholar] [CrossRef] [PubMed]
- Albani, B.A.; Peña, B.; Leed, N.A.; de Paula, N.A.B.G.; Pavani, C.; Baptista, M.S.; Dunbar, K.R.; Turro, C. Marked Improvement in Photoinduced Cell Death bya New Tris-Heteroleptic Complex with Dual Action: Singlet Oxygen Sensitization and Ligand Dissociation. J. Am. Chem. Soc. 2014, 136, 17095–17101. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Ekka, M.K.; Maiti, S. Influence of ionic liquids on thermodynamics of small molecule–DNA interaction: The binding of ethidium bromide to calf thymus DNA. J. Phys. Chem. B 2016, 120, 2691–2700. [Google Scholar] [CrossRef]
- Huang, W.; Chen, Z.; Hou, L.; Feng, P.; Li, Y.; Chen, T. Adjusting the lipid–water distribution coefficient of iridium(iii) complexes to enhance the cellular penetration and treatment efficacy to antagonize cisplatin resistance in cervical cancer. Dalton Trans. 2020, 49, 11556–11564. [Google Scholar] [CrossRef]





| Complex | IC50 (μM) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| HeLa | A549 | A375 | |||||||
| Dark | Light | PI | Dark | Light | PI | Dark | Light | PI | |
| 1 | >100 | 0.95 ± 0.21 | 105 | >100 | 0.85 ± 0.26 | 117 | >100 | 0.48 ± 0.16 | 208 |
| 2 | >100 | 0.67 ± 0.15 | 149 | 92.67 ± 3.28 | 0.58 ± 0.06 | 160 | 90.34 ± 4.87 | 0.12 ± 0.01 | 753 |
| [Ru(bpy)3]2+ [20] | >300 | 161 ± 5.62 | 1.86 | >300 | 152 ± 4.34 | 1.97 | |||
| [Ru(bpy)2dppn]2+ [42] | 110 ± 28 | 0.39 ± 0.06 | 282 | ||||||
| Cisplatin | 45.75 ± 3.31 | 43.61 ± 5.52 | 1.05 | 38.27 ± 2.46 | 33.81 ± 5.17 | 1.13 | 33.86 ± 3.82 | 30.63 ± 5.47 | 1.11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, X.; Luo, Q.; Qin, Y.; Wu, Y.; Liu, X.-W. DNA Interaction, DNA Photocleavage, Photocytotoxicity In Vitro, and Molecular Docking of Naphthyl-Appended Ruthenium Complexes. Molecules 2022, 27, 3676. https://doi.org/10.3390/molecules27123676
Hu X, Luo Q, Qin Y, Wu Y, Liu X-W. DNA Interaction, DNA Photocleavage, Photocytotoxicity In Vitro, and Molecular Docking of Naphthyl-Appended Ruthenium Complexes. Molecules. 2022; 27(12):3676. https://doi.org/10.3390/molecules27123676
Chicago/Turabian StyleHu, Xia, Qian Luo, Yao Qin, Yao Wu, and Xue-Wen Liu. 2022. "DNA Interaction, DNA Photocleavage, Photocytotoxicity In Vitro, and Molecular Docking of Naphthyl-Appended Ruthenium Complexes" Molecules 27, no. 12: 3676. https://doi.org/10.3390/molecules27123676
APA StyleHu, X., Luo, Q., Qin, Y., Wu, Y., & Liu, X.-W. (2022). DNA Interaction, DNA Photocleavage, Photocytotoxicity In Vitro, and Molecular Docking of Naphthyl-Appended Ruthenium Complexes. Molecules, 27(12), 3676. https://doi.org/10.3390/molecules27123676






