The Chemical Constituents of Diaphragma Juglandis Fructus and Their Inhibitory Effect on α-Glucosidase Activity
Abstract
:1. Introduction
2. Results and Discussion
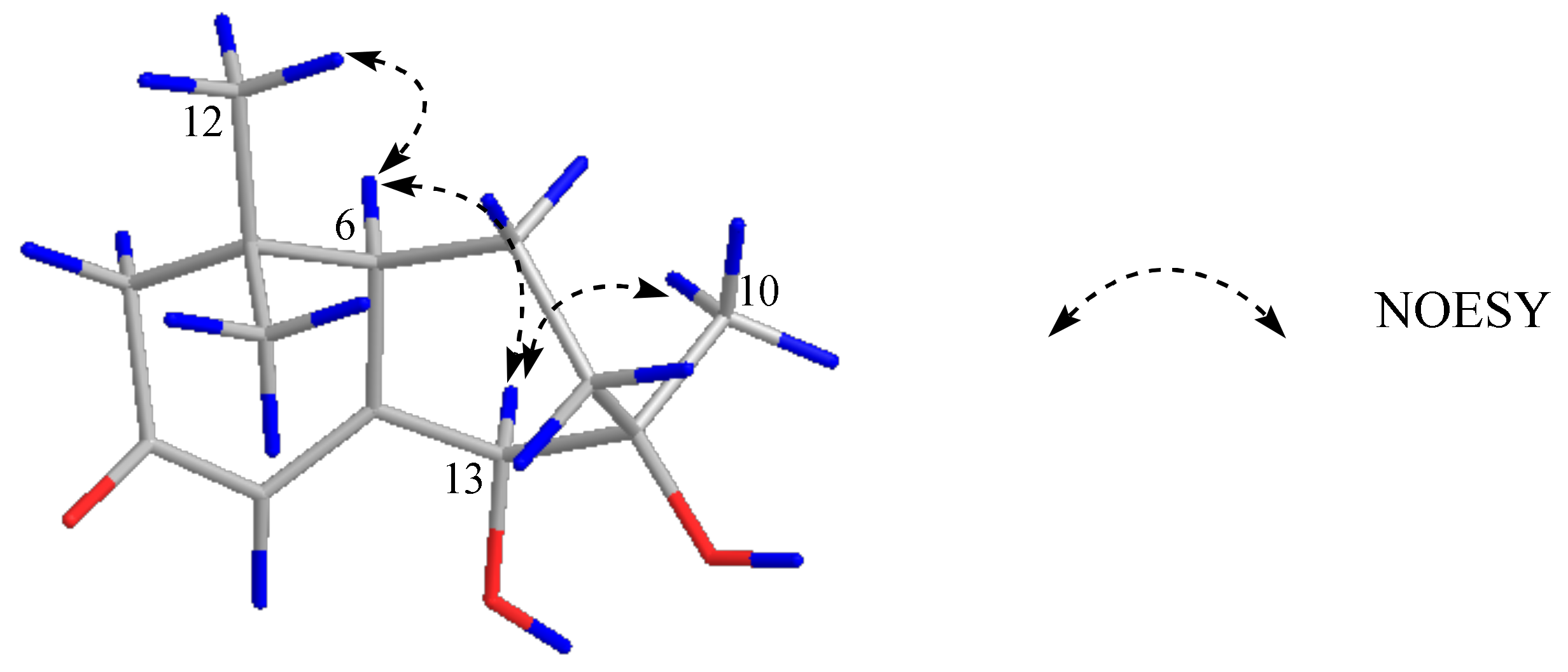
2.1. Structural Elucidation
2.2. Glucosidase Inhibitory Assay
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Characterization of Compounds 1–3, 12, 29
3.5. Acid Hydrolysis of Compounds 3, 12 and 29
3.6. α-Glucosidase Inhibitory Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Hu, Q.; Liu, J.; Li, J.; Liu, H.; Dong, N.; Geng, Y.Y.; Lu, Y.; Wang, Y. Phenolic composition and nutritional attributes of diaphragma juglandis fructus and shell of walnut (Juglans regia L.). Food Sci. Biotechnol. 2019, 29, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Zhao, Z.; Dai, S.; Che, X.; Liu, W. Identification and quantification of bioactive compounds in Diaphragma juglandis Fructus by UHPLC-Q-Orbitrap HRMS and UHPLC-MS/MS. J. Agric. Food Chem. 2019, 67, 3811–3825. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.G.; Kan, H.; Chen, S.X.; Thakur, K.; Wang, S.; Zhang, J.G.; Shang, Y.F.; Wei, Z.J. Comparison of phenolic compounds extracted from Diaphragma juglandis fructus, walnut pellicle, and flowers of Juglans regia using methanol, ultrasonic wave, and enzyme assisted-extraction. Food Chem. 2020, 321, 126672. [Google Scholar] [CrossRef]
- Zhao, H.; Bai, H.; Jing, Y.; Li, W.; Yin, S.; Zhou, H. A pair of taxifolin-3-O-arabinofuranoside isomers from Juglans regia L. Nat. Prod. Res. 2017, 31, 945–950. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Li, Y.; Xiao, T.; Zhang, L.; Xu, D. Antioxidant and antibacterial activities of polysaccharides isolated and purified from Diaphragma juglandis fructus. Int. J. Biol. Macromol. 2017, 105, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Wang, Y.; Chen, F.; Xiao, T.; Zhang, L. Polysaccharides from Diaphragma juglandis fructus: Extraction optimization, antitumor, and immune-enhancement effects. Int. J. Biol. Macromol. 2018, 115, 835–845. [Google Scholar] [CrossRef]
- Wang, D.; Mu, Y.; Dong, H.; Yan, H.; Hao, C.; Wang, X.; Zhang, L. Chemical constituents of the ethyl acetate extract from Diaphragma juglandis Fructus and their inhibitory activity on nitric oxide production in vitro. Molecules 2017, 23, 72. [Google Scholar] [CrossRef] [Green Version]
- Li, G.Y.; Cheng, Y.G.; Zeng, T.C.; Li, M.J.; Sun, R.R.; Li, H.F.; Kong, X.P.; Pei, M.R. Action mechanism of total flavonoids of Diaphragma Juglandis Fructus in treating type 2 diabetes mellitus based on network pharmacology and cellular experimental validation of AKT/FoxO1 signaling pathway. Drug Eval. Res. 2019, 42, 30–40. [Google Scholar]
- Muraoka, O.; Morikawa, T.; Zhang, Y.; Ninomiya, K.; Nakamura, S.; Matsuda, H.; Yoshikawa, M. Novel megastigmanes with lipid accumulation inhibitory and lipid metabolism-promoting activities in HepG2 cells from Sedum sarmentosum. Tetrahedron 2009, 65, 4142–4148. [Google Scholar] [CrossRef]
- Kanchanapoom, T.; Sirikatitham, A.; Otsuka, H.; Ruchirawat, S. A new megastigmane diglycoside from Erythroxylum cuneatum Blume. J. Asian Nat. Prod. Res. 2006, 8, 747–751. [Google Scholar] [CrossRef]
- Matsunami, K.; Otsuka, H.; Takeda, Y.; Miyase, T. Reinvestigation of the absolute stereochemistry of megastigmane glucoside, icariside B(5). Chem. Pharm. Bull. 2010, 58, 1399–1402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nomoto, Y.; Sugimoto, S.; Matsunami, K.; Otsuka, H. Hirtionosides A–C, gallates of megastigmane glucosides, 3-hydroxyoctanoic acid glucosides and a phenylpropanoid glucoside from the whole plants of Euphorbia hirta. J. Nat. Med. 2013, 67, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.K.; Shi, X.L.; Donkor, P.O.; Zhu, H.J.; Gao, X.M.; Ding, L.Q.; Qiu, F. Nine pairs of megastigmane enantiomers from the leaves of Eucommia ulmoides Oliver. J. Nat. Med. 2017, 71, 780–790. [Google Scholar] [CrossRef] [PubMed]
- Matsunami, K.; Ousuka, H.; Takeda, Y. Structural revisions of blumenol C glucoside and byzantionoside B. Chem. Pharm. Bull. 2010, 58, 438–441. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Jin, H.Z.; Li, X.F.; Zhang, Q.; Shen, Y.H.; Yan, S.K.; Zhang, W.D. A new chromone glycoside from Rhododendron spinuliferum. Arch. Pharm. Res. 2008, 31, 970–972. [Google Scholar] [CrossRef]
- Kim, H.J.; Woo, E.R.; Park, H. A novel lignan and flavonoids from Polygonum aviculare. J. Nat. Prod. 1994, 57, 581–586. [Google Scholar] [CrossRef]
- Madikizela, B.; Aderogba, M.A.; Finnie, J.F.; Van Staden, J. Isolation and characterization of antimicrobial compounds from Terminalia phanerophlebia Engl. & Diels leaf extracts. J. Ethnopharmacol. 2014, 156, 228–234. [Google Scholar]
- Shi, L.L.; Ma, G.X.; Yang, J.S.; Gulinar, S.; Jia, X.G. Chemical constituents from plant of Stelleropsis tianschanica. Chin. Tradit. Herb. Drugs 2016, 47, 223–226. [Google Scholar]
- Yang, J.Q.; He, W.J.; Tan, H.N.; Chu, H.B.; Zhang, Y.M.; Mei, W.L.; Dai, H.F. Chemical constituents of Pedicularis cephalantha Franch and P. siphonantha Don. Nat. Prod. Res. Dev. 2009, 21, 600–603. [Google Scholar]
- Yamamoto, M.; Akita, T.; Koyama, Y.; Sueyoshi, E.; Matsunami, K.; Otsuka, H.; Shinzato, T.; Takashima, A.; Aramoto, M.; Takeda, Y. Euodionosides A-G: Megastigmane glucosides from leaves of Euodia meliaefolia. Phytochemistry 2008, 69, 1586–1596. [Google Scholar] [CrossRef]
- Sueyoshi, E.; Liu, H.; Matsunami, K.; Otsuka, H.; Shinzato, T.; Aramoto, M.; Takeda, Y. Bridelionosides A–F: Megastigmane glucosides from Bridelia glauca f. balansae. Phytochemistry 2006, 67, 2483–2493. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, H.; Zhong, X.N.; Hirata, E.; Shinzato, T.; Takeda, Y. Myrsinionosides A-E: Megastigmane glycosides from the leaves of Myrsine seguinii Lev. Chem. Pharm. Bull. 2001, 49, 1093–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, Y.; Zhang, H.J.; Masuda, T.; Honda, G.; Otsuka, H.; Sezik, E.; Yesilada, E.; Sun, H.D. Megastigmane glucosides from Stachys byzantine. Phytochemistry 1997, 44, 1335–1337. [Google Scholar] [CrossRef]
- Bermúdez, J.; Rodríguez, M.; Hasegawa, M.; González-Mujica, F.; Duque, S.; Ito, Y. (6R,9S)-6”-(4”-hydroxybenzoyl)-roseoside, a new megastigmane derivative from Ouratea polyantha and its effect on hepatic glucose-6-phosphatase. Nat. Prod. Commun. 2012, 7, 973–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, P.K.; Agarwal, S.K.; Rastogi, R.P.; Osterdahal, B.G. Dihydroflavanonols from Cedrus deodara, A (13)C NMR study. Planta Medica 1981, 43, 82–85. [Google Scholar] [CrossRef]
- Yao, F.Y.; Huang, D.D.; Huang, G.H.; Xue, D.; Sun, L.N. Chemical constituents of Illicium brevistylum. Chin. Tradit. Herb. Drugs 2014, 45, 1536–1539. [Google Scholar]
- Liu, Y.; Rong, X.H.; Tan, J.Y.; Pan, J.; Guan, W.; Kuang, H.X.; Yang, B.Y. Chemical constituents of flavonoids and alkaloids from leaves of Datura metel. Chin. Tradit. Herb. Drugs 2021, 52, 4141–4152. [Google Scholar]
- Zhao, J.; Zhou, X.W.; Chen, X.B.; Wang, Q.X. α-Glucosidase inhibitory constituents from Toona sinensis. Chem. Nat. Compd. 2009, 45, 244–246. [Google Scholar] [CrossRef]
- Hong, S.S.; Suh, H.J.; Oh, J.S. Phenolic chemical constituents of the stem barks of Robinia pseudoacacia. Chem. Nat. Compd. 2017, 53, 359–361. [Google Scholar] [CrossRef]
- Qi, J.J.; Yan, Y.M.; Wang, C.X.; Cheng, Y.X. Compounds from Lycium ruthenicum. Nat. Prod. Res. Dev. 2018, 30, 345–353. [Google Scholar]
- Pan, J.Y.; Zhou, Y.; Zou, K.; Wu, J.; Li, Q.X.; Zhang, S. Chemical constituents of Balanophora involucrate. Chin. Tradit. Herb. Drugs 2008, 39, 327–331. [Google Scholar]
- Li, Y.L.; Li, K.M.; Su, M.X.; Leung, K.T.; Cheng, Y.W.; Zhang, Y.W. Studies on antiviral constituents in stems and leaves of Pithecellibium clypearia. China J. Chin. Mater. Med. 2006, 31, 397–400. [Google Scholar]
- Mu, L.H.; Zhang, D.M. Studies on chemical constituents of Cercis chinensis. China J. Chin. Mater. Med. 2006, 31, 1795–1797. [Google Scholar]
- Chen, G.; Jin, H.Z.; Li, X.F.; Zhang, Q.; Shen, Y.H.; Yan, S.K.; Zhang, W.D. Chemical constituents from Rhododendron spinu-liferum. Chem. Nat. Compd. 2009, 45, 725–727. [Google Scholar] [CrossRef]
- Park, B.J.; Matsuta, T.; Kanazawa, T.; Park, C.H.; Chang, K.J.; Onjo, M. Phenolic compounds from the leaves of Psidium guajava Ⅱ. Quercetin and its glycosides. Chem. Nat. Compd. 2012, 48, 477–479. [Google Scholar] [CrossRef]
- Masuda, T.; Iritani, K.; Yonemori, S.; Oyama, Y.; Takeda, Y. Isolation and antioxidant activity of galloyl flavonol glycosides from the seashore plant, Pemphis acidula. Biosci. Biotechnol. Biochem. 2001, 65, 1302–1309. [Google Scholar] [CrossRef]
- Wang, Q.; Xu, D.R.; Shi, X.H.; Qin, M.J. Flavones from Potentilla discolor Bunge. Chin. J. Nat. Med. 2009, 7, 361–364. [Google Scholar] [CrossRef]
- Ma, X.Q.; Zheng, C.J.; Zhang, Y.; Hu, C.L.; Lin, B.; Fu, X.Y.; Han, L.Y.; Xu, L.S.; Rahman, K.; Qin, L.P. Antiosteoporotic flavonoids from Podocarpium poducarpum. Phytochem. Lett. 2013, 6, 118–122. [Google Scholar] [CrossRef]
- Huang, X.X.; Zhou, C.C.; Li, L.Z.; Li, F.F.; Lou, L.L.; Li, D.M.; Ikejima, T.; Peng, Y.; Song, S.J. The cytotoxicity of 8-O-4′ neolignans from the seeds of Crataegus pinnatifida. Bioorg. Med. Chem. Lett. 2013, 23, 5599–5604. [Google Scholar] [CrossRef]
- Gao, L.; Tian, H.; Lv, P.J.; Wang, J.P.; Wang, Y.F. Chemical constituents of Sapium sebiferum leaves. China J. Chin. Mater. Med. 2015, 40, 1518–1522. [Google Scholar]
- Li, X.; Cao, W.; Shen, Y.; Li, N.; Dong, X.P.; Wang, K.J.; Cheng, Y.X. Antioxidant compounds from Rosa laevigata fruits. Food Chem. 2012, 130, 575–580. [Google Scholar] [CrossRef]
- Su, J.S.; Qin, F.Y.; Zhang, Y.; Cheng, Y.X. Compounds of Codonopsis pilosula. J. Chin. Med. Mater. 2018, 41, 863–867. [Google Scholar]
- Han, H.Y.; Liu, H.W.; Wang, N.L.; Yao, X.S. Sequilignans and dilignans from Campylotropis hirtella (Franch.) Schindl. Nat. Prod. Res. 2008, 22, 990–995. [Google Scholar] [CrossRef]
- Jiang, Z.H.; Tanaka, T.; Hirata, H.; Fukuoka, R.; Kouno, I. Three diarylheptanoids from Rhoiptelea chiliantha. Phytochemistry 1996, 43, 1049–1054. [Google Scholar] [CrossRef]
- Fang, J.M.; Lee, C.K.; Cheng, Y.S. Lignans from leaves of Juniperus chinensis. Phytochemistry 1992, 31, 3659–3661. [Google Scholar]
- Hu, G.L.; Peng, X.R.; Dong, D.; Nian, Y.; Gao, Y.; Wang, X.Y.; Hong, D.F.; Qiu, M.H. New ent-kaurane diterpenes from the roasted arabica coffee beans and molecular docking to α-glucosidase. Food Chem. 2021, 345, 128823. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.G.; Li, G.Y.; Tan, J.Y.; Liu, Y.; Li, H.F.; Yao, Y.J.; Zhang, J.Y.; Yang, B.Y.; Pei, M.R. Antioxidant of total favonoids from Diaphragma Juglandis Fructus. and its inhibitory effects on α-glucosidase and α-amylase activity in vitro. J. Liaoning Univ. TCM 2018, 20, 22–25. [Google Scholar]
- Tadera, K.; Minami, Y.; Takamatsu, K.; Matsuoka, T. Inhibition of α-glucosidase and α-amylase by flavonoids. J. Nutr. Sci. Vitaminol. 2006, 52, 149–153. [Google Scholar] [CrossRef] [Green Version]
- Jani, N.A.; Sirat, H.M.; Ahmad, F.; Aminudin, N.I. New sesquiterpene dilactone and β-carboline alkaloid and the α-glucosidase inhibitory activity of selected phytochemicals from Neolitsea cassia (L.) Kosterm. Nat. Prod. Res. 2021. Available online: https://www.tandfonline.com/doi/full/10.1080/14786419.2021.1961134 (accessed on 30 March 2022).
- Zhu, J.Z.; Chen, C.; Zhang, B.; Huang, Q. The inhibitory effects of flavonoids on α-amylase and α-glucosidase. Crit. Rev. Food Sci. Nutr. 2020, 60, 695–708. [Google Scholar] [CrossRef]
- Tan, J.Y.; Liu, Y.; Cheng, Y.G.; Sun, Y.P.; Pan, J.; Guan, W.; Li, X.M.; Huang, J.; Jiang, P.; Guo, S.; et al. New withanolides with anti-inflammatory activity from the leaves of Datura metel L. Bioorg. Chem. 2020, 95, 103541. [Google Scholar] [CrossRef]
- Peng, C.Y.; Lu, J.; Liu, J.Q.; Huang, H.L.; Zhu, Y.Y.; Shu, J.C. Three novel pterosin dimers form Pteris obtusiloba. Fitoterapia 2020, 146, 104713. [Google Scholar] [CrossRef] [PubMed]






| Position | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | |
| 1 | - | 35.5 | - | 43.1 | - | 37.2 |
| 2 | 2.33 (d, 15.6) 2.13 (d, 15.6) | 50.4 | 2.61 (d, 18.3) 2.20 (d, 18.3) | 50.8 | 2.34 (d, 17.3) 1.87 (d, 17.3) | 48.1 |
| 3 | - | 202.6 | - | 200.9 | - | 202.3 |
| 4 | 6.15 (br. s) | 121.0 | 6.10 (br. s) | 122.4 | 5.70 (br. s) | 125.3 |
| 5 | - | 168.2 | - | 173.9 | - | 169.9 |
| 6 | 2.16 (dd, 12.9, 4.5) | 49.2 | - | 78.8 | 1.81 (t, 5.2) | 52.2 |
| 7 | 1.94 (m) 1.39 (dd, 13.4, 4.0) | 25.3 | 1.98 (m) 1.78 (m) | 35.8 | 1.92 (m) 1.35 (m) | 26.8 |
| 8 | 1.86 (ddd, 13.4, 3.7, 3.0) 1.72 (td, 13.4, 4.3) | 39.1 | 1.67 (m) 1.43 (m) | 34.9 | 2.19 (m) 1.55 (m) | 37.6 |
| 9 | - | 76.8 | 3.67 (m) | 68.9 | 3.73 (m) | 76.5 |
| 10 | 1.03 (s) | 19.3 | 1.16 (d, 6.2) | 23.5 | 1.17 (d, 6.2) | 20.4 |
| 11 | 1.00 (s) | 25.8 | 1.09 (s) | 24.4 | 0.90 (s) | 28.9 |
| 12 | 1.05 (s) | 28.7 | 1.04 (s) | 23.5 | 0.96 (s) | 27.4 |
| 13 | 4.08 (d, 1.4) | 80.2 | 4.39 (t, 1.9) | 63.1 | 1.90 (d, 1.1) | 24.8 |
| 1′ | 4.34 (d, 7.7) | 102.8 | ||||
| 2′ | 3.17 (m) | 75.2 | ||||
| 3′ | 3.39 (m) | 78.0 | ||||
| 4′ | 3.39 (m) | 72.0 | ||||
| 5′ | 3.58 (m) | 75.2 | ||||
| 6′ | 4.56 (dd, 11.8, 2.3) 4.42 (dd, 11.8, 6.4) | 64.9 | ||||
| 1″ | - | 122.3 | ||||
| 2″, 6″ | 7.89 (d, 8.8) | 132.9 | ||||
| 3″, 5″ | 6.83 (d, 8.8) | 116.2 | ||||
| 4″ | - | 163.7 | ||||
| 7″ | - | 167.9 | ||||
| Position | δH | δC | Position | δH | δC |
|---|---|---|---|---|---|
| 2 | 8.09 (s) | 148.7 | 9 | - | 159.3 |
| 3 | - | 140.0 | 10 | 1.03 (s) | 106.4 |
| 4 | - | 179.0 | 1′ | 5.48 (s) | 109.5 |
| 5 | - | 163.4 | 2′ | 4.31 (dd, 3.2, 1.1) | 83.3 |
| 6 | 6.20 (dd, 2.1) | 100.0 | 3′ | 3.94 (dd, 5.9, 3.2) | 78.7 |
| 7 | - | 166.1 | 4′ | 4.13 (m) | 87.2 |
| 8 | 6.31 (d, 2.1) | 95.0 | 5′ | 3.78 (dd, 12.1, 3.4) 3.67 (dd, 12.1, 5.6) | 62.9 |
| Position | δH | δC | Position | δH | δC |
|---|---|---|---|---|---|
| 1 | 5.57 (d, 8.2) | 95.5 | 7′ | 7.64 (d, 15.9) | 146.9 |
| 2 | 3.39 (m) | 73.9 | 8′ | 6.37 (d, 15.9) | 114.9 |
| 3 | 3.48 (m) | 77.9 | 9′ | - | 169.1 |
| 4 | 3.42 (m) | 71.3 | 1″ | - | 127.4 |
| 5 | 3.67 (m) | 76.3 | 2″,6″ | 7.71 (d, 8.7) | 134.1 |
| 6 | 4.52 (dd, 12.0, 1.9) 4.32 (dd, 12.0, 5.5) | 64.4 | 3″,5″ | 6.75 (d, 8.7) | 115.9 |
| 1′ | - | 127.2 | 4″ | - | 160.4 |
| 2′,6′ | 7.45 (d, 8.6) | 131.2 | 7″ | 6.95 (d, 12.8) | 147.0 |
| 3′,5′ | 6.79 (d, 8.6) | 116.8 | 8″ | 5.83 (d, 12.8) | 115.4 |
| 4′ | - | 161.3 | 9″ | - | 166.5 |
| Compound | IC50 (µM) | Compound | IC50 (µM) |
|---|---|---|---|
| 1–11 | >100 | 22 | 67.74 ± 6.41 |
| 12 | 92.35 ± 7.24 | 23–25,27 | >100 |
| 13 | 40.39 ± 4.14 | 26 | 77.15 ± 12.36 |
| 14 | 95.78 ± 12.62 | 28 | 35.41 ± 3.87 |
| 15, 17–20 | >100 | 29–34,36,37 | >100 |
| 16 | 54.82 ± 7.47 | 35 | 87.74 ± 13.41 |
| 21 | 29.47 ± 2.95 | Acarbose | 60.01 ± 4.82 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, J.; Cheng, Y.; Wang, S.; Li, J.; Ren, H.; Qiao, Y.; Li, Q.; Wang, Y. The Chemical Constituents of Diaphragma Juglandis Fructus and Their Inhibitory Effect on α-Glucosidase Activity. Molecules 2022, 27, 3045. https://doi.org/10.3390/molecules27103045
Tan J, Cheng Y, Wang S, Li J, Ren H, Qiao Y, Li Q, Wang Y. The Chemical Constituents of Diaphragma Juglandis Fructus and Their Inhibitory Effect on α-Glucosidase Activity. Molecules. 2022; 27(10):3045. https://doi.org/10.3390/molecules27103045
Chicago/Turabian StyleTan, Jinyan, Yangang Cheng, Shihui Wang, Jianli Li, Haiqin Ren, Yuanbiao Qiao, Qingshan Li, and Yingli Wang. 2022. "The Chemical Constituents of Diaphragma Juglandis Fructus and Their Inhibitory Effect on α-Glucosidase Activity" Molecules 27, no. 10: 3045. https://doi.org/10.3390/molecules27103045
APA StyleTan, J., Cheng, Y., Wang, S., Li, J., Ren, H., Qiao, Y., Li, Q., & Wang, Y. (2022). The Chemical Constituents of Diaphragma Juglandis Fructus and Their Inhibitory Effect on α-Glucosidase Activity. Molecules, 27(10), 3045. https://doi.org/10.3390/molecules27103045







