The Effect of Dia2 Protein Deficiency on the Cell Cycle, Cell Size, and Recruitment of Ctf4 Protein in Saccharomyces cerevisiae
Abstract
1. Introduction
2. Results
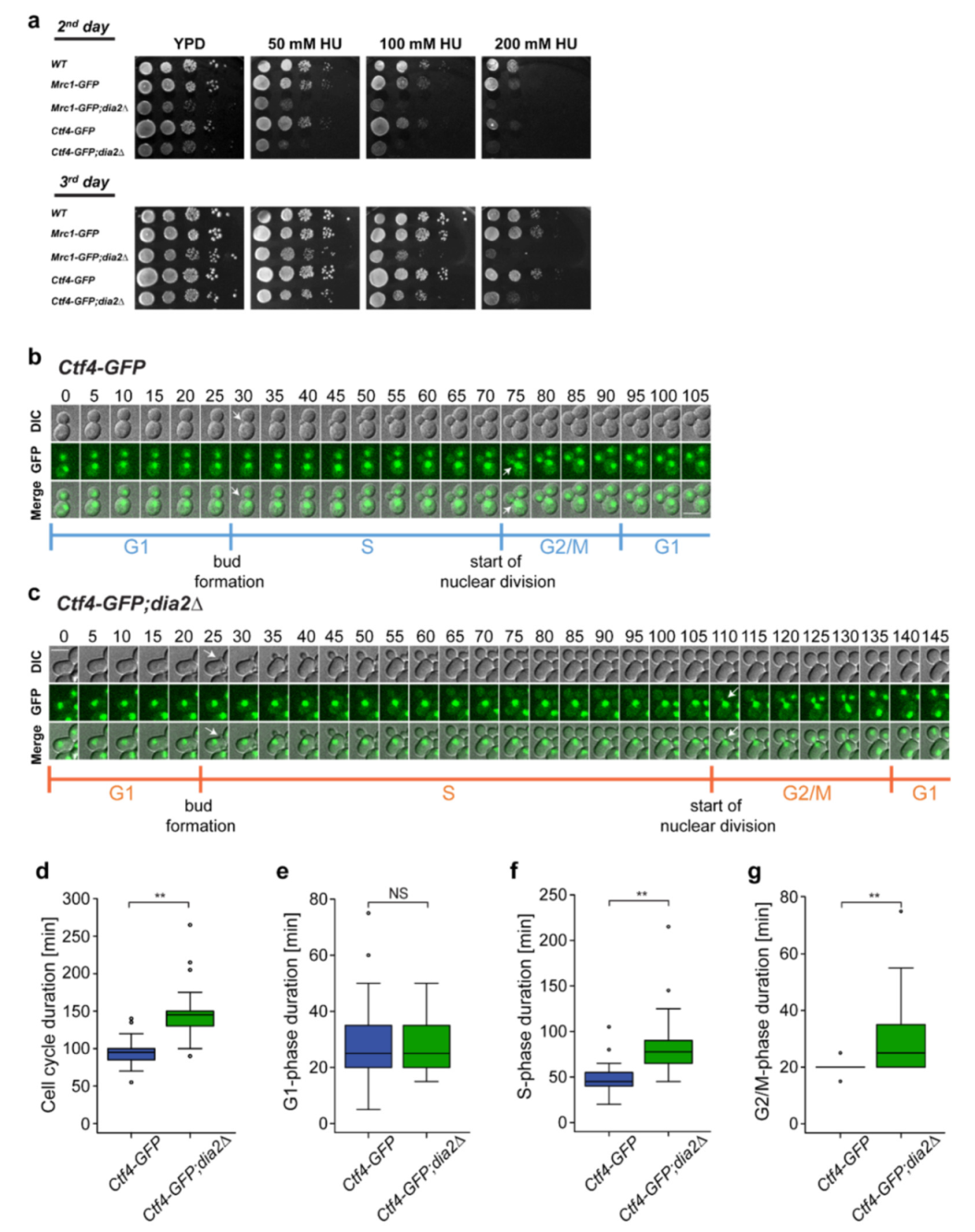
2.1. Loss of Dia2 Protein Extends Cell Cycle Duration
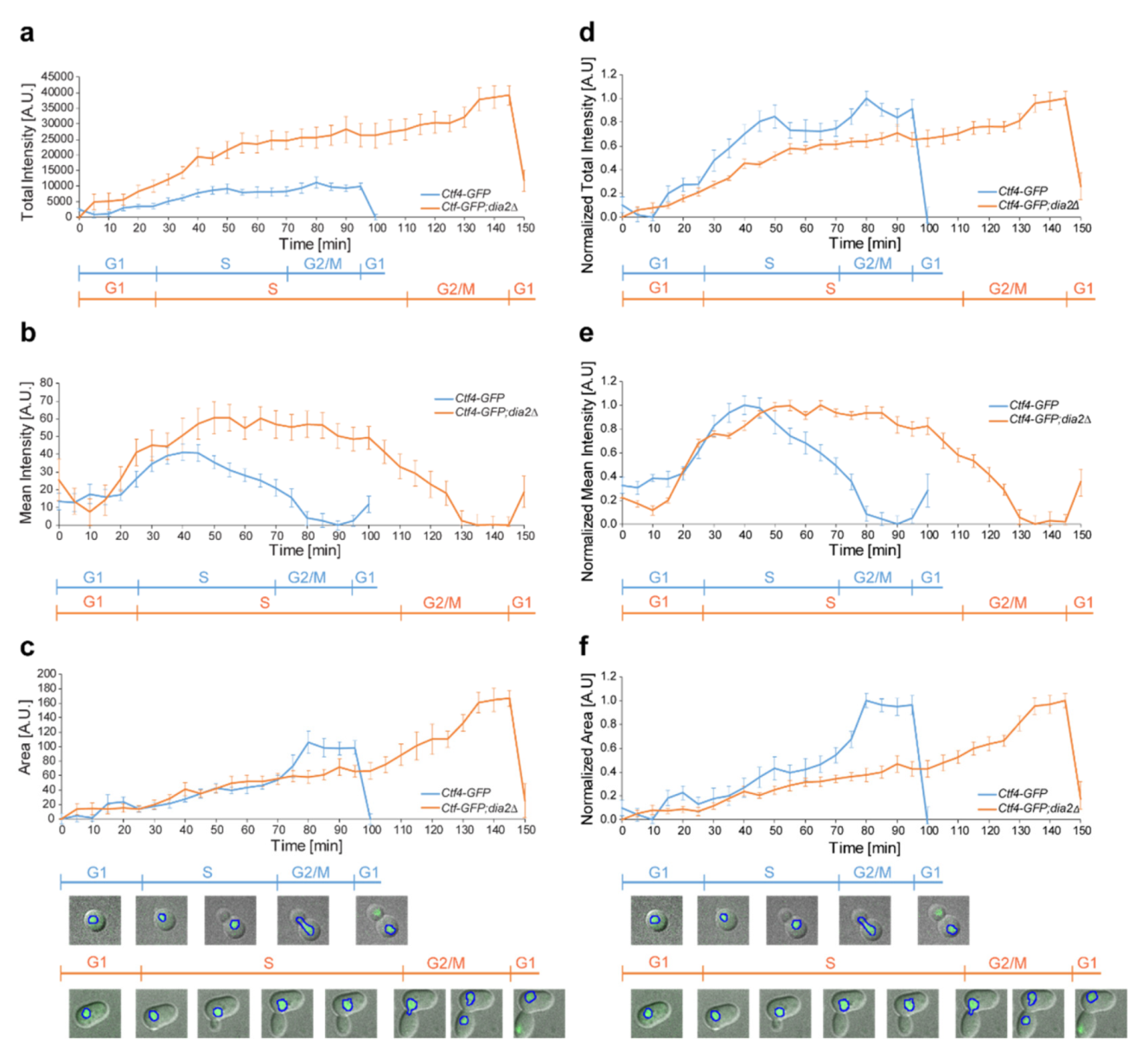
2.2. Deletion of DIA2 Leads to Increased Levels of Ctf4
2.3. Dia2 Deficiency Increases the Chromatin-Bound Fraction of Ctf4
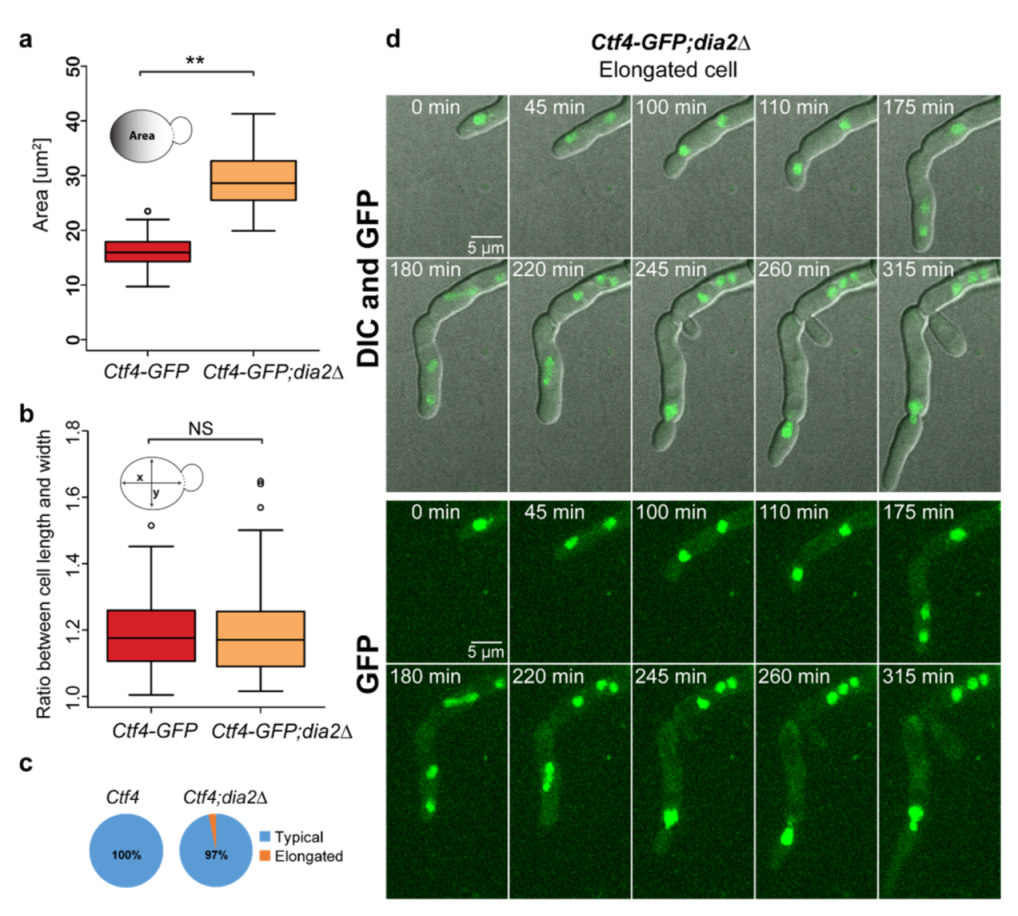
2.4. Lack of Dia2 Leads to Abnormal Cell Size
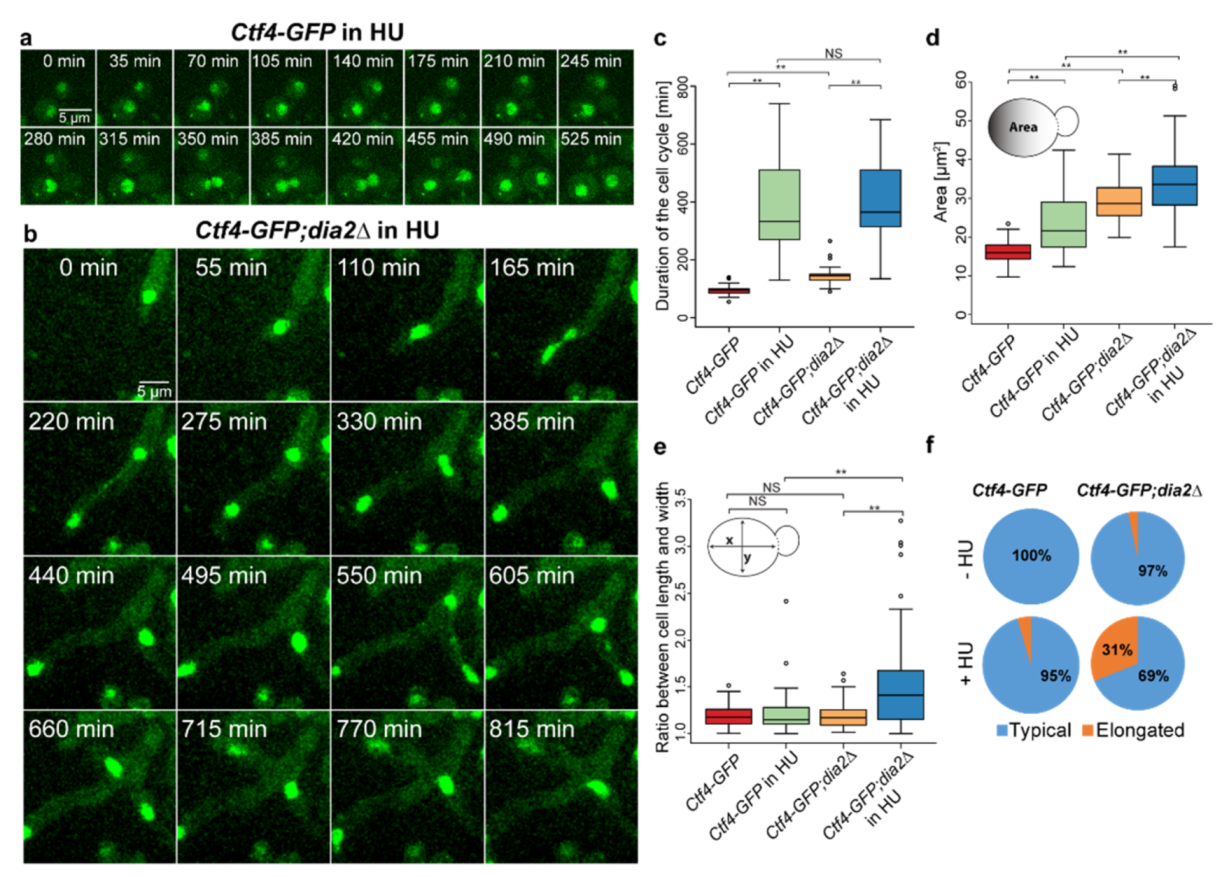
2.5. Inhibition of DNA Synthesis Enhances the Oversized Phenotype in dia2Δ Cells
3. Discussion
4. Materials and Methods
4.1. Strains
4.2. Viability Test
4.3. Time-Lapse Live-Cell Imaging of Yeast Cells
4.4. Bulk Chromatin Fractionation
4.5. Total Protein Extraction
4.6. Western Blotting
4.7. Flow Cytometry Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Jackson, A.P.; Laskey, R.A.; Coleman, N. Replication Proteins and Human Disease. Cold Spring Harb. Perspect. Biol. 2013, 6, a013060. [Google Scholar] [CrossRef] [PubMed]
- Kastan, M.B.; Bartek, J. Cell-cycle checkpoints and cancer. Nature 2004, 432, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.R.; McIntosh, J.R. Mcl1p is a polymerase α replication accessory factor important for S-phase DNA damage survival. Eukaryot. Cell 2005, 4, 166–177. [Google Scholar] [CrossRef][Green Version]
- Katayama, S.; Kitamura, K.; Lehmann, A.; Nikaido, O.; Toda, T. Fission yeast F-box protein Pof3 is required for genome integrity and telomere function. Mol. Biol. Cell 2002, 13, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Ukomadu, C.; Jha, S.; Senga, T.; Dhar, S.K.; Wohlschlegel, J.A.; Nutt, L.K.; Kornbluth, S.; Dutta, A. Mcm10 and And-1/CTF4 recruit DNA polymerase α to chromatin for initiation of DNA replication. Genes Dev. 2007, 21, 2288–2299. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, M.; Li, H. The eukaryotic replisome goes under the microscope. Curr. Biol. 2016, 26, R247. [Google Scholar] [CrossRef] [PubMed]
- Simon, A.C.; Zhou, J.C.; Perera, R.; Van Deursen, F.; Evrin, C.; Ivanova, M.E.; Kilkenny, M.L.; Renault, L.; Kjaer, S.; Matak-Vinković, D.; et al. A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature 2014, 510, 293–297. [Google Scholar] [CrossRef]
- Hanna, J.S.; Kroll, E.S.; Lundblad, V.; Spencer, F.A. Saccharomyces cerevisiae CTF18 and CTF4 Are Required for Sister Chromatid Cohesion. Mol. Cell. Biol. 2001, 21, 3144–3158. [Google Scholar] [CrossRef]
- Samora, C.P.; Saksouk, J.; Goswami, P.; Wade, B.O.; Singleton, M.; Bates, P.; Lengronne, A.; Costa, A.; Uhlmann, F. Ctf4 Links DNA Replication with Sister Chromatid Cohesion Establishment by Recruiting the Chl1 Helicase to the Replisome. Mol. Cell 2016, 63, 371–384. [Google Scholar] [CrossRef]
- Kouprina, N.; Kroll, E.; Bannikov, V.; Bliskovsky, V.; Gizatullin, R.; Kirillov, A.; Shestopalov, B.; Zakharyev, V.; Hieter, P.; Spencer, F. CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1992, 12, 5736. [Google Scholar] [PubMed]
- Tanaka, H.; Katou, Y.; Yagura, M.; Saitoh, K.; Itoh, T.; Araki, H.; Bando, M.; Shirahige, K. Ctf4 coordinates the progression of helicase and DNA polymerase α. Genes Cells 2009, 14, 807–820. [Google Scholar] [CrossRef] [PubMed]
- Villa, F.; Simon, A.C.; Bazan, M.A.O.; Kilkenny, M.L.; Wirthensohn, D.; Wightman, M.; Matak-Vinkovíc, D.; Pellegrini, L.; Labib, K. Ctf4 Is a Hub in the Eukaryotic Replisome that Links Multiple CIP-Box Proteins to the CMG Helicase. Mol. Cell 2016, 63, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Farina, A.; Bermudez, V.P.; Tappin, I.; Du, F.; Galal, W.C.; Hurwitz, J. Interaction between human Ctf4 and the Cdc45/Mcm2-7/GINS (CMG) replicative helicase. Proc. Natl. Acad. Sci. USA 2013, 110, 19760–19765. [Google Scholar] [CrossRef] [PubMed]
- Baretić, D.; Jenkyn-Bedford, M.; Aria, V.; Cannone, G.; Skehel, M.; Yeeles, J.T.P. Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork. Mol. Cell 2020, 78, 926–940.e13. [Google Scholar] [CrossRef] [PubMed]
- Byun, T.S.; Pacek, M.; Yee, M.C.; Walter, J.C.; Cimprich, K.A. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005, 19, 1040–1052. [Google Scholar] [CrossRef] [PubMed]
- Gambus, A.; Van Deursen, F.; Polychronopoulos, D.; Foltman, M.; Jones, R.C.; Edmondson, R.D.; Calzada, A.; Labib, K. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase α within the eukaryotic replisome. EMBO J. 2009, 28, 2992–3004. [Google Scholar] [CrossRef] [PubMed]
- Morohashi, H.; Maculins, T.; Labib, K. The Amino-Terminal TPR Domain of Dia2 Tethers SCFDia2 to the Replisome Progression Complex. Curr. Biol. 2009, 19, 1943–1949. [Google Scholar] [CrossRef] [PubMed]
- Willems, A.R.; Schwab, M.; Tyers, M. A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta Mol. Cell Res. 2004, 1695, 133–170. [Google Scholar] [CrossRef]
- Koepp, D.M. The replication stress response and the ubiquitin system: A new link in maintaining genomic integrity. Cell Div. 2010, 5, 2–5. [Google Scholar] [CrossRef] [PubMed]
- Blake, D.; Luke, B.; Kanellis, P.; Jorgensen, P.; Goh, T.; Penfold, S.; Breitkreutz, B.-J.; Durocher, D.; Peter, M.; Tyers, M. The F-Box Protein Dia2 Overcomes Replication Impedance to Promote Genome Stability in Saccharomyces cerevisiae. Genetics 2006, 174, 1709–1727. [Google Scholar] [CrossRef] [PubMed]
- Koepp, D.M.; Kile, A.C.; Swaminathan, S.; Rodriguez-Rivera, V. The F-box protein Dia2 regulates DNA replication. Mol. Biol. Cell 2006, 17, 1540–1548. [Google Scholar] [CrossRef]
- Maculins, T.; Nkosi, P.J.; Nishikawa, H.; Labib, K. Tethering of SCF(Dia2) to the Replisome Promotes Efficient Ubiquitylation and Disassembly of the CMG Helicase. Curr. Biol. 2015, 25, 2254–2259. [Google Scholar] [CrossRef] [PubMed]
- Mimura, S.; Komata, M.; Kishi, T.; Shirahige, K.; Kamura, T. SCF(Dia2) regulates DNA replication forks during S-phase in budding yeast. EMBO J. 2009, 28, 3693–3705. [Google Scholar] [CrossRef] [PubMed]
- Fong, C.M.; Arumugam, A.; Koepp, D.M. The Saccharomyces cerevisiae F-box protein Dia2 is a mediator of S-phase checkpoint recovery from DNA damage. Genetics 2013, 193, 483–499. [Google Scholar] [CrossRef] [PubMed]
- Deegan, T.D.; Mukherjee, P.P.; Fujisawa, R.; Rivera, C.P.; Labib, K. Cmg helicase disassembly is controlled by replication fork dna, replisome components and a ubiquitin threshold. eLife 2020, 9, e60371. [Google Scholar] [CrossRef] [PubMed]
- Palecek, S.P.; Parikh, A.S.; Kron, S.J. Genetic analysis reveals that FLO11 upregulation and cell polarization independently regulate invasive growth in Saccharomyces cerevisiae. Genetics 2000, 156, 1005–1023. [Google Scholar] [CrossRef]
- Cullen, P.J.; Sprague, G.F. The regulation of filamentous growth in yeast. Genetics 2012, 190, 23–49. [Google Scholar] [CrossRef] [PubMed]
- Uzunova, S.D.; Zarkov, A.S.; Ivanova, A.M.; Stoynov, S.S.; Nedelcheva-Veleva, M.N. The subunits of the S-phase checkpoint complex Mrc1/Tof1/Csm3: Dynamics and interdependence. Cell Div. 2014, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Calvert, M.E.K.; Lannigan, J.A.; Pemberton, L.F.; Signaling, C. Optimization of Yeast Cell Cycle Analysis and. Cytometry 2009, 73, 825–833. [Google Scholar]
- Neumann, F.; Taddei, A.; Gasser, S. Tracking individual chromosomes with integrated arrays of Lac op sites and GFP-Lac i repressor (PROT15) Procedure Preparations. Integr. Vlsi. J. 2007, 1–8. [Google Scholar]
- Garmendia-Torres, C.; Tassy, O.; Matifas, A.; Molina, N.; Charvin, G. Multiple inputs ensure yeast cell size homeostasis during cell cycle progression. eLife 2018, 7, e34025. [Google Scholar] [CrossRef] [PubMed]
- Alvino, G.M.; Collingwood, D.; Murphy, J.M.; Delrow, J.; Brewer, B.J.; Raghuraman, M.K. Replication in Hydroxyurea: It’s a Matter of Time. Mol. Cell. Biol. 2007, 27, 6396–6406. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-J.; Schwartz, M.F.; Duong, J.K.; Stern, D.F. Rad53 Phosphorylation Site Clusters Are Important for Rad53 Regulation and Signaling. Mol. Cell. Biol. 2003, 23, 6300–6314. [Google Scholar] [CrossRef] [PubMed]
- Cortez, D. Preventing replication fork collapse to maintain genome integrity. DNA Repair. 2015, 32, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Johnston, G.C.; Pringle, J.R.; Hartwell, L.H. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 1977, 105, 79–98. [Google Scholar] [CrossRef]
- Liu, H.; Styles, C.A.; Fink, G.R. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics 1996, 144, 967–978. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Rose, M.D. Stable Pseudohyphal Growth in Budding Yeast Induced by Synergism between Septin Defects and Altered MAP-kinase Signaling. PLoS Genet. 2015, 11, e1005684. [Google Scholar] [CrossRef]
- Tripathi, K.; Matmati, N.; Zheng, W.J.; Hannun, Y.A.; Mohanty, B.K. Cellular morphogenesis under stress is influenced by the sphingolipid pathway gene ISC1 and DNA integrity checkpoint genes in saccharomyces cerevisiae. Genetics 2011, 189, 533–547. [Google Scholar] [CrossRef] [PubMed]
- Kile, A.C.; Koepp, D.M. Activation of the S-Phase Checkpoint Inhibits Degradation of the F-Box Protein Dia2. Mol. Cell. Biol. 2010, 30, 160–171. [Google Scholar] [CrossRef]
- Labib, K.; de Piccoli, G. Surviving chromosome replication: The many roles of the S-phase checkpoint pathway. Philos. Trans. R Soc. L. B Biol. Sci. 2011, 366, 3554–3561. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, V. Mechanisms of replication fork protection: A safeguard for genome stability AU—Errico, Alessia. Crit. Rev. Biochem. Mol. Biol. 2012, 47, 222–235. [Google Scholar]
- Bando, M.; Katou, Y.; Komata, M.; Tanaka, H.; Itoh, T.; Sutani, T.; Shirahige, K. Csm3, Tof1, and Mrc1 Form a Heterotrimeric Mediator Complex That Associates with DNA Replication Forks. J. Biol. Chem. 2009, 284, 34355–34365. [Google Scholar] [CrossRef]
- Branzei, D.; Foiani, M. The checkpoint response to replication stress. DNA Repair 2009, 8, 1038–1046. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Ye, P.; Yuan, D.S.; Wang, X.; Bader, J.S.; Boeke, J.D. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 2006, 124, 1069–1081. [Google Scholar] [CrossRef]
- Santocanale, C.; Diffley, J.F. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 1998, 395, 615–618. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.; Cotta-Ramusino, C.; Pellicioli, A.; Liberi, G.; Plevani, P.; Muzi-Falconi, M.; Newlon, C.S.; Foiani, M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 2001, 412, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Segurado, M.; Diffley, J.F. Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev. 2008, 22, 1816–1827. [Google Scholar] [CrossRef] [PubMed]
- Maric, M.; Maculins, T.; De Piccoli, G.; Labib, K. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science 2014, 346, 1253596. [Google Scholar] [CrossRef]
- Huh, W.-K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O′Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Gietz, D.; Jean, A.S.; Woods, R.A.; Schiestl, R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992, 20, 1425. [Google Scholar] [CrossRef]
- Lõoke, M.; Kristjuhan, K.; Kristjuhan, A. UKPMC Funders Group Ukpmc Funders Group Author Manuscript Extraction of genomic DNA from yeasts for Pcr. Biotechniques 2011, 50, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Gallina, I.; Eckert-Boulet, N.; Lisby, M. Live Cell Microscopy of DNA Damage Response in Saccharomyces Cerevisiae BT—DNA Repair Protocols; Bjergbæk, L., Ed.; Humana Press: Totowa, NJ, USA, 2012; pp. 433–443. [Google Scholar]
- Danovski, G.; Dyankova, T.; Stoynov, S. CellTool: An open source software combining bio-image analysis and mathematical modeling. bioRxiv 2018, 454256. [Google Scholar]
- Liang, C.; Stillman, B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997, 11, 3375–3386. [Google Scholar] [CrossRef] [PubMed]
- Foiani, M.; Marini, F.; Gamba, D.; Lucchini, G.; Plevani, P. The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol. 1994, 14, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Poser, I.; Sarov, M.; A Hutchins, J.R.; Hériché, J.-K.; Toyoda, Y.; Pozniakovsky, A.; Weigl, D.; Nitzsche, A.; Hegemann, B.; Bird, A.; et al. BAC TransgeneOmics: A high-throughput method for exploration of protein function in mammals. Nat. Methods 2008, 5, 409–415. [Google Scholar] [CrossRef] [PubMed]

| Primer | Application | Sequence |
|---|---|---|
| Dia2-disr UP pUG6 | Gene disruption | TTCTCGAAAAATATTATAAATAGACATGCAAAATGATTAGCCATGCAGCTGAAGCTTCGTACGC |
| Dia2-disr DOWN pUG6 | Gene disruption | ATTTTCCGAAGGATACTGCATTATCATCAGTGATTTATTAATCTAGCATAGGCCACTAGTGGATCTG |
| Dia2-check 5′_UP | Diagnostic PCR | CGGCAATCTTCACACGGT |
| Dia2-check 3′_DOWN | Diagnostic PCR | GTTTGCCATCGGTGCATC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivanova, A.; Atemin, A.; Uzunova, S.; Danovski, G.; Aleksandrov, R.; Stoynov, S.; Nedelcheva-Veleva, M. The Effect of Dia2 Protein Deficiency on the Cell Cycle, Cell Size, and Recruitment of Ctf4 Protein in Saccharomyces cerevisiae. Molecules 2022, 27, 97. https://doi.org/10.3390/molecules27010097
Ivanova A, Atemin A, Uzunova S, Danovski G, Aleksandrov R, Stoynov S, Nedelcheva-Veleva M. The Effect of Dia2 Protein Deficiency on the Cell Cycle, Cell Size, and Recruitment of Ctf4 Protein in Saccharomyces cerevisiae. Molecules. 2022; 27(1):97. https://doi.org/10.3390/molecules27010097
Chicago/Turabian StyleIvanova, Aneliya, Aleksandar Atemin, Sonya Uzunova, Georgi Danovski, Radoslav Aleksandrov, Stoyno Stoynov, and Marina Nedelcheva-Veleva. 2022. "The Effect of Dia2 Protein Deficiency on the Cell Cycle, Cell Size, and Recruitment of Ctf4 Protein in Saccharomyces cerevisiae" Molecules 27, no. 1: 97. https://doi.org/10.3390/molecules27010097
APA StyleIvanova, A., Atemin, A., Uzunova, S., Danovski, G., Aleksandrov, R., Stoynov, S., & Nedelcheva-Veleva, M. (2022). The Effect of Dia2 Protein Deficiency on the Cell Cycle, Cell Size, and Recruitment of Ctf4 Protein in Saccharomyces cerevisiae. Molecules, 27(1), 97. https://doi.org/10.3390/molecules27010097





