GIF-2209, an Oxindole Derivative, Accelerates Melanogenesis and Melanosome Secretion via the Modification of Lysosomes in B16F10 Mouse Melanoma Cells
Abstract
:1. Introduction
2. Results
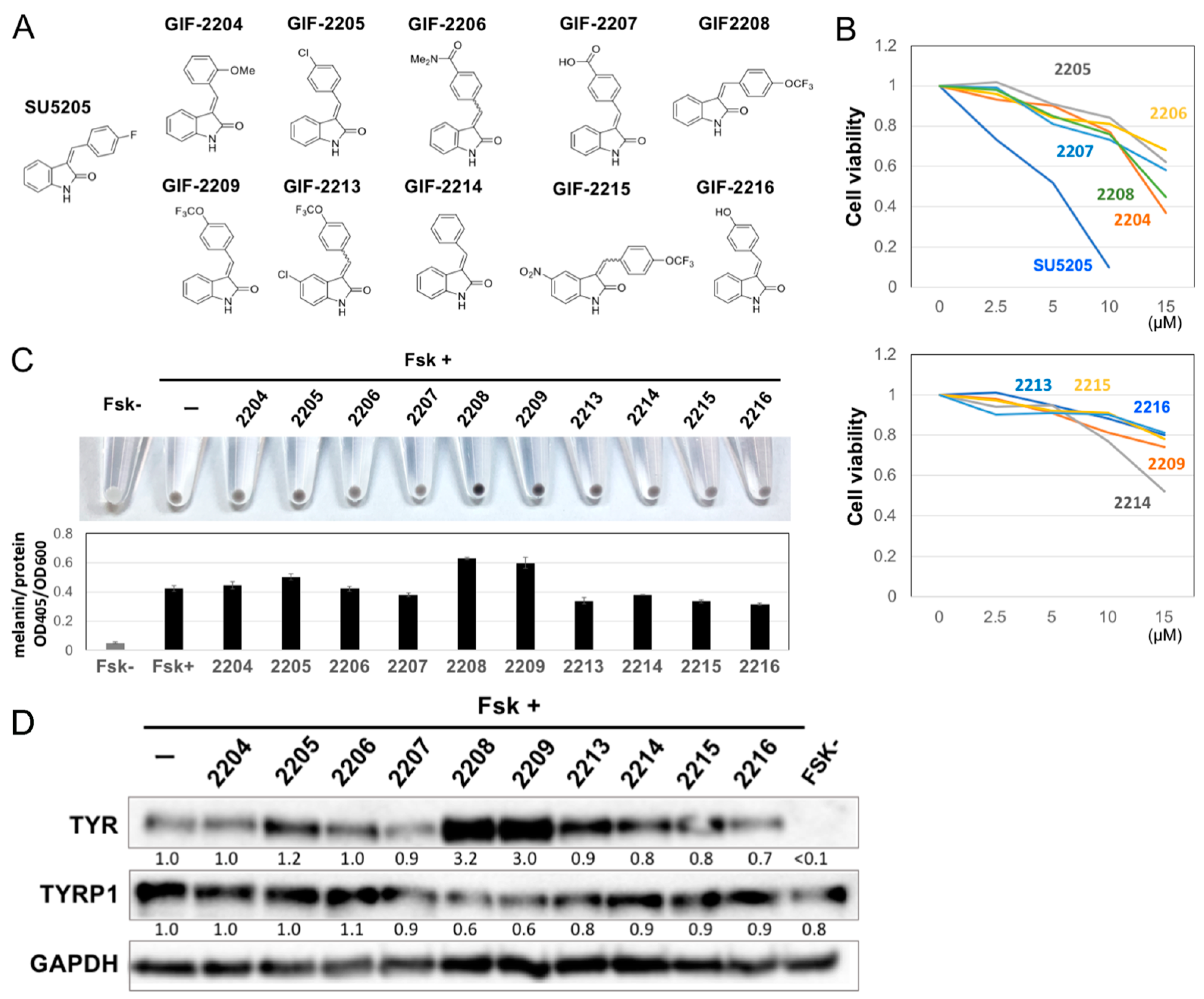
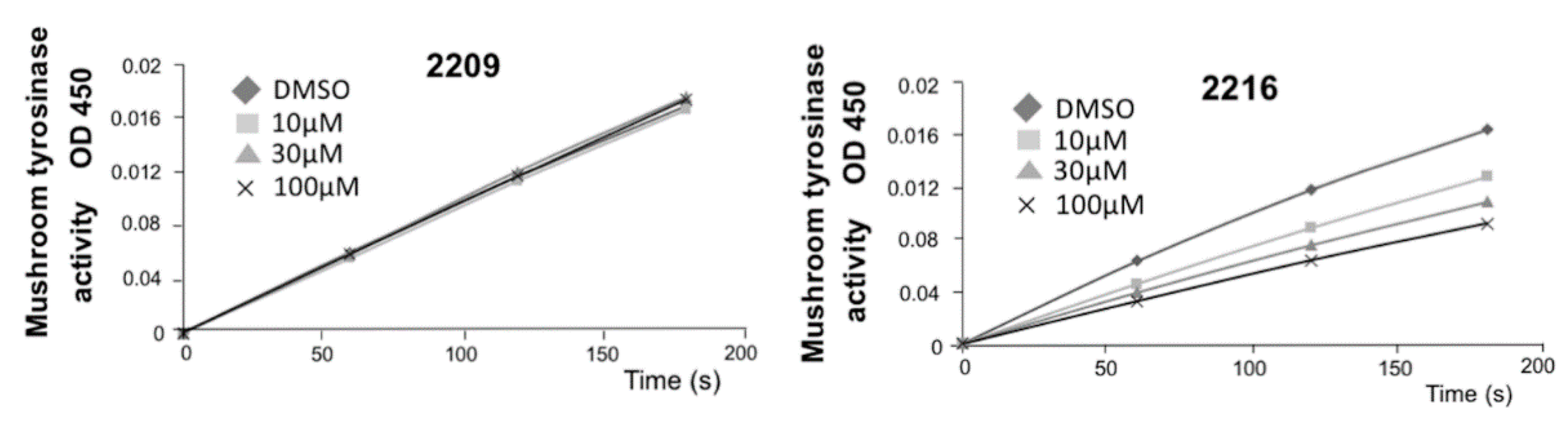
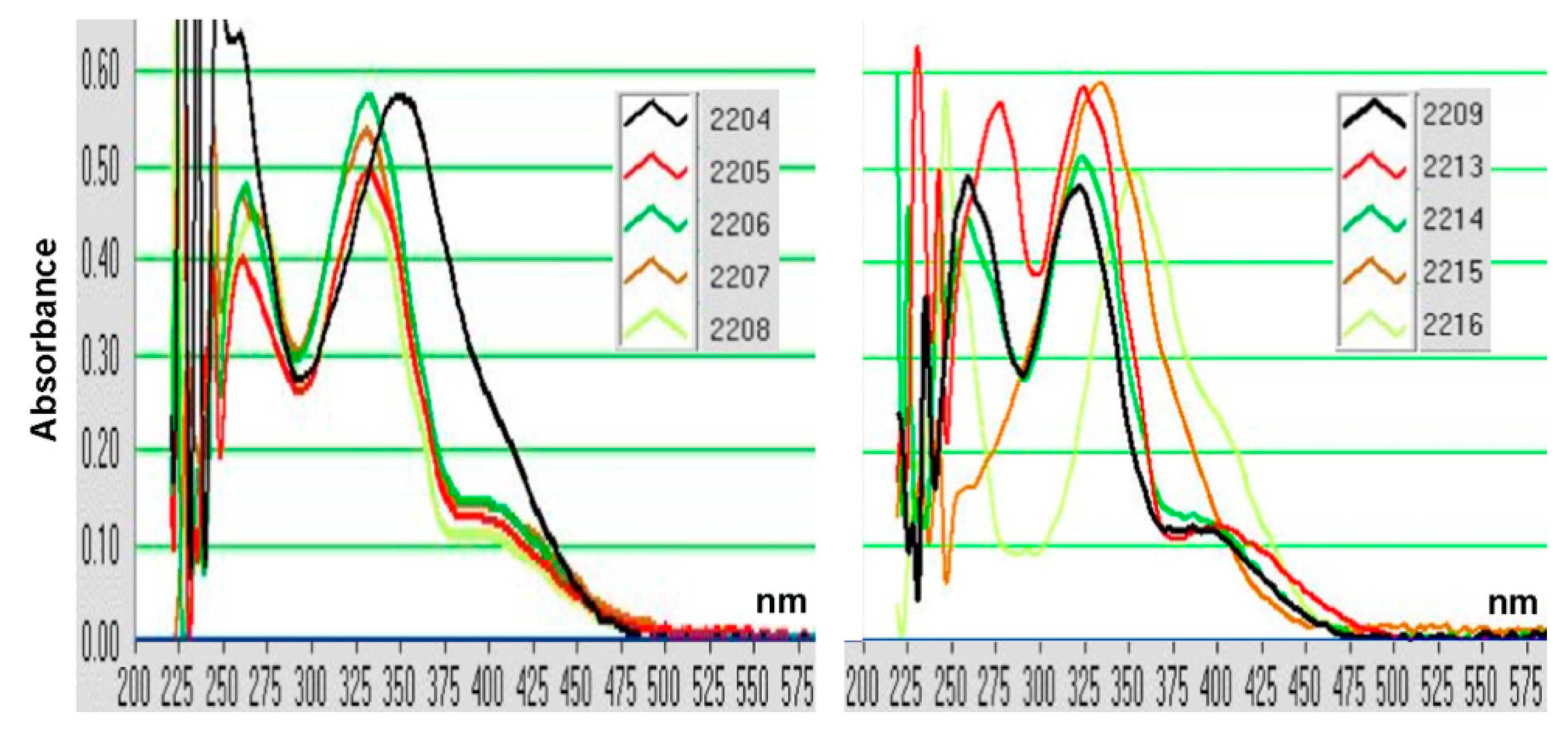
2.1. Structure–Activity Relationship of Oxindole Derivatives
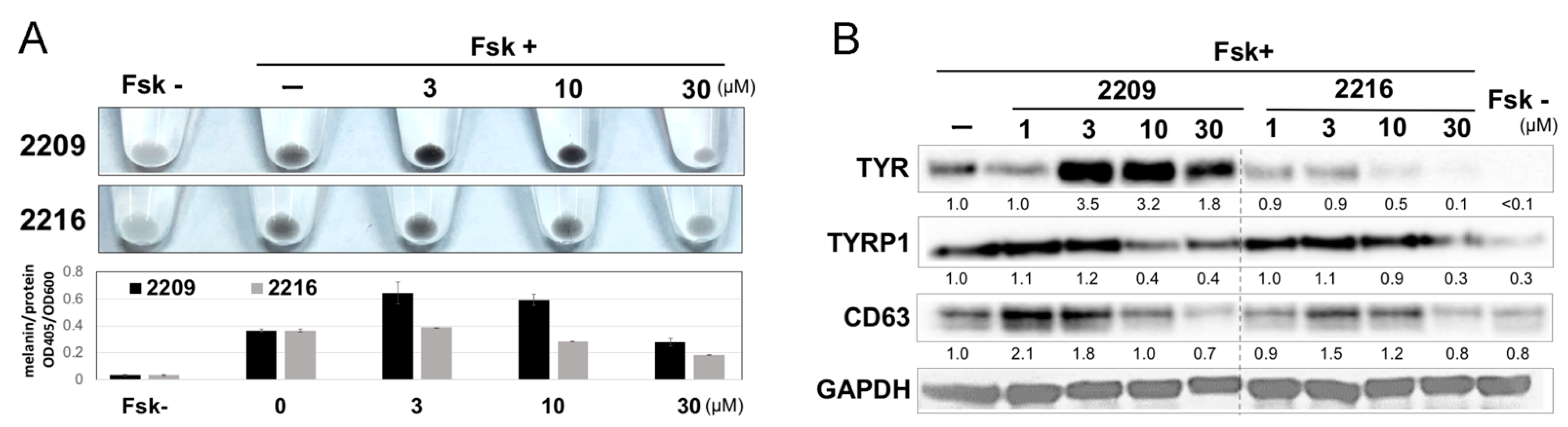
2.2. GIF-2209 Has Multiple Mechanisms of Action on Melanogenesis
2.3. GIF-2209 Upregulates TYR Transcription Independent of MITF Expression Level
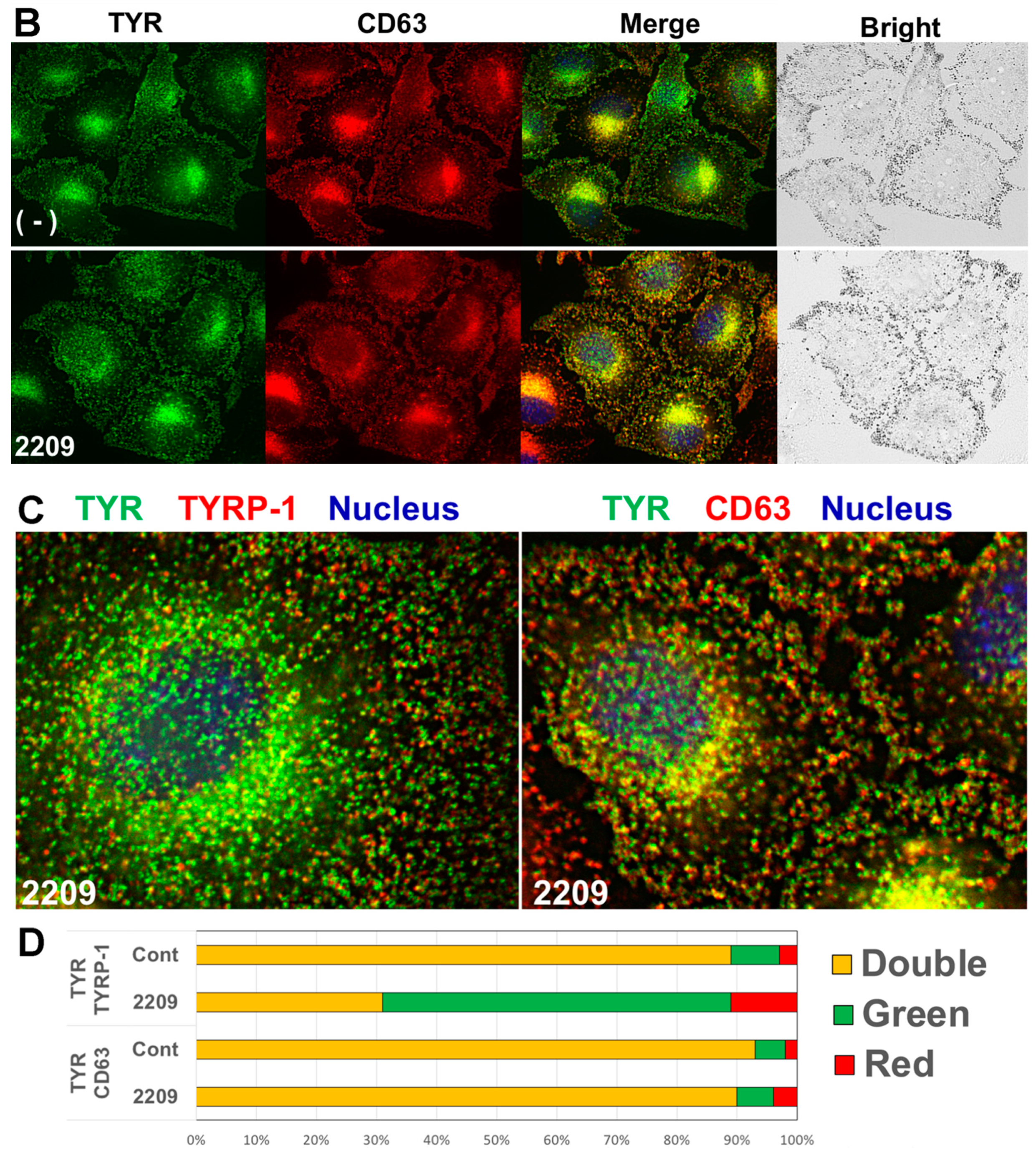
2.4. GIF-2209 Segregates TYR Containing Vesicles from Those of TYRP-1
2.5. GIF-2209 Stimulates Secretion of mELANOSOMes with High Amounts of TYR, TYRP-1, and CD63
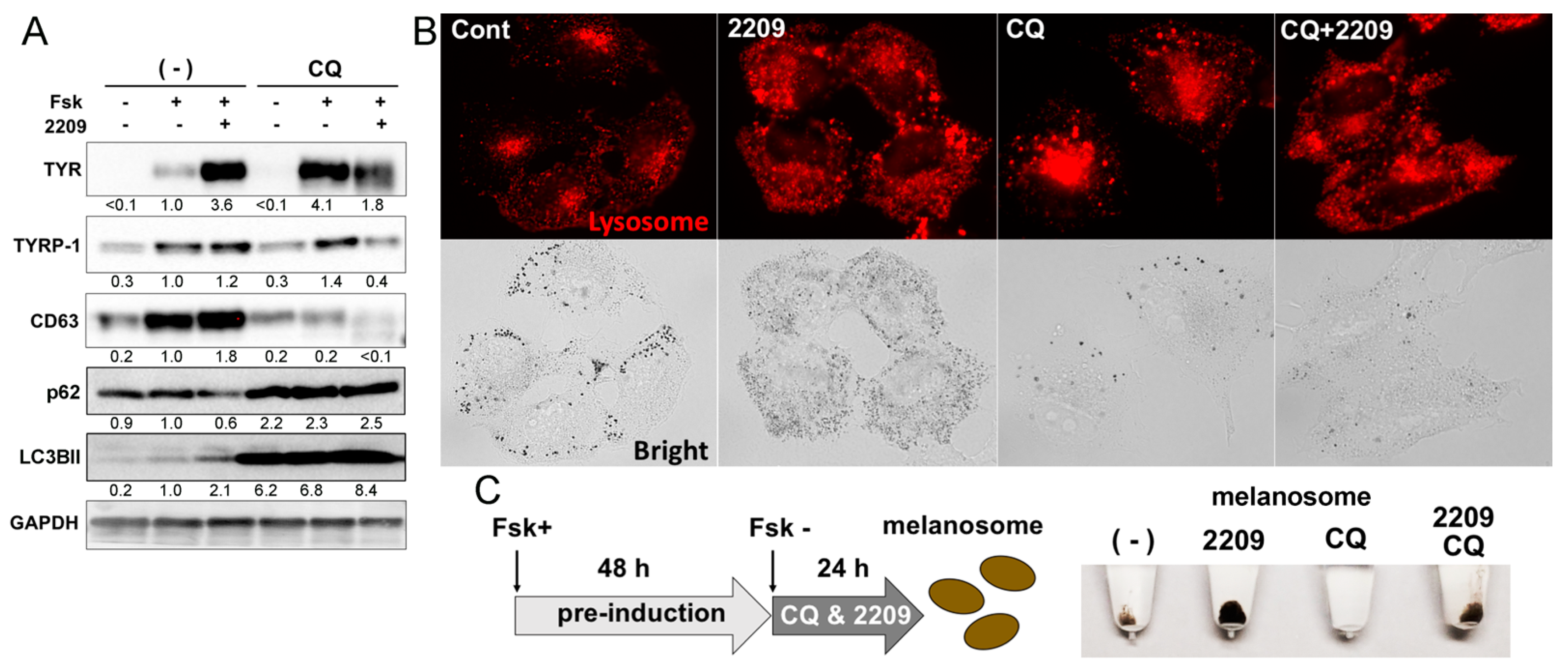
2.6. GIF-2209 Modulates the Function and Intracellular Distribution of Lysosomes
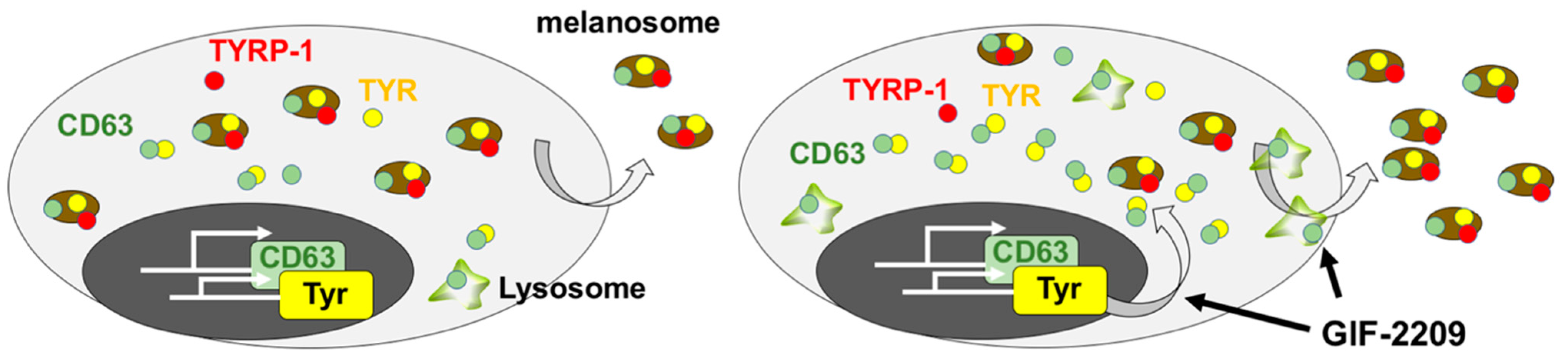
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Preparation of Extracellular Melanosomes
4.3. Western Blot
4.4. Quantitative PCR (qPCR)
4.5. Immunocytochemistry
4.6. Luciferase-Reporter Assay
4.7. Tyrosinase Activity
4.8. Colocalization Experiments
4.9. Statistical Analysis
4.10. GIF Compounds
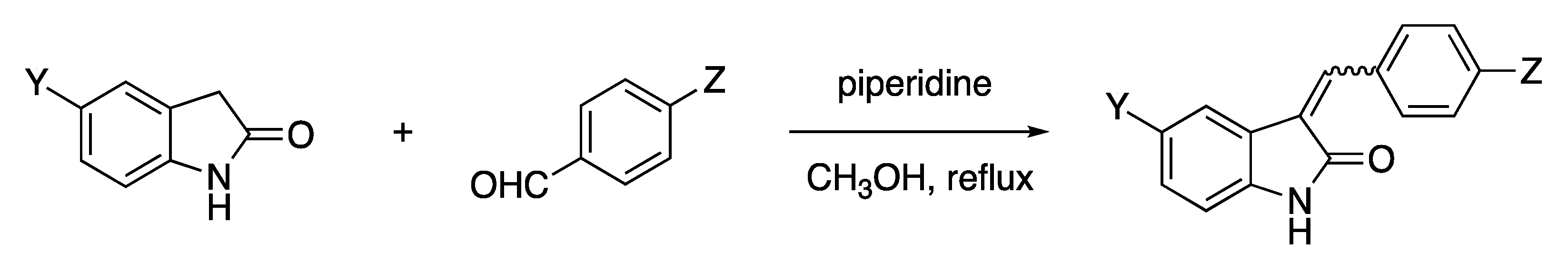
- GIF-2204 (E)-3-(2-methoxybenzylidene)indolin-2-one: 1H NMR (400 MHz, DMSO-d6) d = 3.86 (s, 3H, OCH3), 6.83 (t, J = 7.1 Hz, 1H, ArH), 6.86 (d, J = 7.7 Hz, 1H, ArH), 7.08 (t, J = 7.7 Hz, 1H, ArH), 7.16 (d, J = 8.3 Hz, 1H, ArH), 7.21 (t, J = 7.7 Hz, 1H, ArH), 7.40 (d, J = 7.7 Hz, 1H, ArH), 7.49 (t, J = 8.3 Hz, 1H, ArH), 7.65 (s, 1H, vinyl), 7.68 (d, J = 7.1 Hz, 1H, ArH), 10.6 (br s, 1H, NH).
- GIF-2205 (E)-3-(4-chlorobenzylidene)indolin-2-one: 1H NMR (400 MHz, DMSO-d6) d = 6.86 (t, J = 7.6 Hz, 1H, ArH), 6.88 (d, J = 7.8 Hz, 1H, ArH), 7.24 (t, J = 7.8 Hz, 1H, ArH), 7.48 (d, J = 7.8 Hz, 1H, ArH), 7.59 (d, J = 8.7 Hz, 2H, ArH), 7.59 (s, 1H, vinyl), 7.73 (d, J = 8.7 Hz, 2H, ArH, H-2′,6′), 10.6 (br s, 1H, NH).
- GIF-2206 (E)-N,N-dimethyl-4-((2-oxoindolin-3-ylidene)methyl)benzamide
- (Z)-N,N-dimethyl-4-((2-oxoindolin-3-ylidene)methyl)benzamide (E/Z mixture, 3.4/1): 1H NMR (400 MHz, DMSO-d6) d = 2.96 and 3.01 (s, 6H, NMe2), 6.83 (d, J = 7.3 Hz, 1H, ArH, Z-isomer) and 6.88 (d, J = 7.8 Hz, 1H, ArH, E-isomer), 6.86 (t, J = 7.8 Hz, 1H, ArH, E-isomer) and 7.00 (t, J = 7.3 Hz, 1H, ArH, Z-isomer), 7.23 (t, J = 7.3 Hz, 1H, ArH, Z-isomer) and 7.25 (t, J = 7.8 Hz, 1H, ArH, E-isomer), 7.48 (d, J = 8.3 Hz, 1H, ArH), 7.5–7.57 (complex, 3H, ArH), 7.63 (s, 1H, vinyl), 7.76 (d, J = 8.3 Hz, 2H, H-2′,6′, ArH, E-isomer), 8.40 (d, J = 8.3 Hz, 2H, ArH, H-2′,6′, Z-isomer), 10.64 and 10.66 (s, 1H, NH, E- and Z-isomers). 13C NMR (100 MHz, DMSO-d6) d = ~40 (buried under the DMSO signal), 109.97 and 110.77, 120.57 and 121.23, 121.69 and 121.78, 123.04, 127.27 and 127.91, 128.78 and 129.74, 130.97, 132.17, 135.41, 135.9, 137.91, 143.62, 169.06, 170.03.
- GIF-2207 (E)-4-((2-oxoindolin-3-ylidene)methyl)benzoic acid: 1H NMR (400 MHz, DMSO-d6) d = 6.85 (t, J = 7.3 Hz, 1H, ArH), 6.88 (d, J = 7.8 Hz, 1H, ArH), 7.25 (t, J = 7.8 Hz, 1H, ArH), 7.46 (d, J = 7.8 Hz, 1H, ArH), 7.65 (s, 1H, vinyl), 7.80 (d, J = 8.2 Hz, 2H, ArH), 8.06 (d, J = 8.2 Hz, 2H, ArH, H-2′,6′), 10.66 (br s, 1H, NH).
- GIF-2208 (Z)-3-(4-(trifluoromethoxy)benzylidene)indolin-2-one: 1H NMR (400 MHz, CDCl3) d = 6.85 (d, J = 7.8 Hz, 1H, ArH), 7.06 (t, J = 7.4 Hz, 1H, ArH), 7.25 (t, J = 7.8 Hz, 1H, ArH), 7.27 (2H, ArH), 7.50 (s, 1H, vinyl), 7.53 (d, J = 7.4 Hz 1H, ArH), 7.73 (br, 1H, NH), 8.32 (d, J = 9.2 Hz, 2H, ArH, H-2′,6′). 13C NMR (100 MHz, DMSO-d6) d = 110.00, 120.61, 120.95, 121.71, 125.13, 128.13, 129.89, 131.96, 133.67, 134.38, 135.34, 141.50, 149.65, 167.56.
- GIF-2209 (E)-3-(4-(trifluoromethoxy)benzylidene)indolin-2-one: 1H NMR (400 MHz, CDCl3) d = 6.90 (t, J = 7.6 Hz, 1H, ArH), 6.92 (d, J = 7.8 Hz, 1H, ArH), 7.24 (t, J = 7.6 Hz, 1H, ArH), 7.32 (d, J = 8.2 Hz, 2H, ArH), 7.57 (d, J = 7.8 Hz, 1H, ArH), 7.70 (d, J = 8.2 Hz, 2H, ArH, H-2′,6′), 7.77 (s, 1H, vinyl), 8.49 (br, 1H, NH). 13C NMR (100 MHz, DMSO-d6) d = 110.78, 121.11, 121.74, 121.81, 122.98, 128.92, 131.03, 131.96, 134.26, 134.58, 143.66, 149.27, 168.96.
- GIF-2213 (E)-5-chloro-3-(4-(trifluoromethoxy)benzylidene)indolin-2-one
- (Z)-5-chloro-3-(4-(trifluoromethoxy)benzylidene)indolin-2-one (E/Z mixture, 3.3/1): 1H NMR (400 MHz, DMSO-d6) d = 6.84 (d, J = 8.2 Hz, 1H, ArH, Z-isomer) and 6.90 (d, J = 9.2 Hz, 1H, ArH, E-isomer), 7.25–7.35 (complex, 2H, ArH), 7.47 (d, J = 8.7 Hz, 2H, ArH, Z-isomer) and 7.55 (d, J = 8.7 Hz, 2H, ArH, E-isomer), 7.72 (s, 1H, vinyl), 7.83 (d, J = 8.7 Hz, 2H, ArH, H-2′,6′, E-isomer) and 8.48 (d, J = 8.7 Hz, 2H, ArH, H-2′,6′, Z-isomer), 7.98 (s, 1H, vinyl), 10.8 (br, 1H, NH). 13C NMR (100 MHz, DMSO-d6) d = 111.38, 112.17, 119.23, 120.67, 120.99, 121.84, 122.40, 122.76, 125.54, 126.05, 127.06, 127.09, 128.18, 129.23, 130.49, 131.94, 133.40, 133.89, 134.67, 136.55, 137.33, 140.13, 142.43, 149.54, 149.97, 167.33, 168.61.
- GIF-2214 (E)-3-benzylideneindolin-2-one: 1H NMR (400 MHz, DMSO-d6) d = 6.85 (t, J = 8.4 Hz, 1H, ArH), 6.88 (d, J = 7.8 Hz, 1H, ArH), 7.23 (t, J = 7.8 Hz, 1H, ArH), 7.45–7.56 (complex, 4H, ArH), 7.63 (s, 1H, vinyl), 7.70 (d, J = 7.3 Hz, 2H, ArH, H-2′,6′), 10.61 (br s, 1H, NH).
- GIF-2215 (E)-5-nitro-3-(4-(trifluoromethoxy)benzylidene)indolin-2-one
- (Z)-5-nitro-3-(4-(trifluoromethoxy)benzylidene)indolin-2-one (E/Z mixture, 1/2.3): 1H NMR (400 MHz, DMSO-d6) d = 7.03 (d, J = 8.7 Hz, 1H, ArH, Z-isomer) and 7.08 (d, J = 9.2 Hz, 1H, ArH, E-isomer), 7.51 (d, J = 8.5 Hz, 2H, ArH, Z-isomer) and 7.58 (d, J = 7.8 Hz, 2H, ArH, E-isomer), 7.87 (s, 1H, vinyl, E-isomer), 7.91 (d, J = 8.7 Hz, 1H, ArH, Z-isomer), 8.17–8.24 (complex, 4H, ArH, Z-isomer), 8.28 (s, 1H, vinyl, E-isomer), 8.54 (d, J = 8.5 Hz, 2H, ArH, H-2′,6′, Z-isomer), 8.72 (s, 1H, ArH, Z-isomer), 11.39 (br s, 1H, NH). 13C NMR (100 MHz, DMSO-d6) d = 110.05, 110.78, 116.46, 118.05, 121.04, 121.92, 125.92, 126.17, 127.3, 132.14, 133.20, 134.97, 138.23, 139.30, 142.09, 142.66, 146.88, 149.2, 150.29, 167.80, 169.13.
- GIF-2216 (E)-3-(4-hydroxybenzylidene)indolin-2-one: 1H NMR (400 MHz, DMSO-d6) d = 6.87 (dt, J = 2.7 and 7.8 Hz, 1H, ArH), 6.90 (d, J = 8.5 Hz, 2H, ArH), 7.21 (t, J = 7.6 Hz, 1H, ArH), 7.53 (s, 1H, vinyl), 7.61 (d, J = 8.5 Hz, 2H, ArH, H-2′,6′), 7.69 (d, J = 7.8 Hz, 1H, ArH), 10.52 (br, 1H, NH).
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Serre, C.; Busuttil, V.; Botto, J.M. Intrinsic and extrinsic regulation of human skin melanogenesis and pigmentation. Int. J. Cosmet. Sci. 2018, 40, 328–347. [Google Scholar] [CrossRef] [Green Version]
- Tian, X.; Cui, Z.; Liu, S.; Zhou, J.; Cui, R. Melanosome transport and regulation in development and disease. Pharmacol. Ther. 2021, 219, 107707. [Google Scholar] [CrossRef] [PubMed]
- Bowman, S.L.; Bi-Karchin, J.; Le, L.; Marks, M.S. The road to lysosome-related organelles: Insights from Hermansky-Pudlak syndrome and other rare diseases. Traffic 2019, 20, 404–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumagai, A.; Horike, N.; Satoh, Y.; Uebi, T.; Sasaki, T.; Itoh, Y.; Hirata, Y.; Uchio-Yamada, K.; Kitagawa, K.; Uesato, S.; et al. A Potent Inhibitor of SIK2, 3, 3′, 7-Trihydroxy-4′-Methoxyflavon (4′-O-Methylfisetin), Promotes Melanogenesis in B16F10 Melanoma Cells. PLoS ONE 2011, 6, e26148. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.Y.; Won, K.J.; Kim, D.Y.; Kim, M.J.; Won, Y.R.; Kim, N.Y.; Lee, H.M. Wound Healing-Promoting and Melanogenesis-Inhibiting Activities of Angelica polymorpha Maxim. Flower Absolute In Vitro and Its Chemical Composition. Molecules 2021, 26, 6172. [Google Scholar] [CrossRef] [PubMed]
- Kuroi, A.; Sugimura, K.; Kumagai, A.; Kohara, A.; Nagaoka, Y.; Kawahara, H.; Yamahara, M.; Kawahara, N.; Takemori, H.; Fuchino, H. The Importance of 11α-OH, 15-oxo, and 16-en Moieties of 11α-Hydroxy-15-oxo-kaur-16-en-19-oic Acid in Its Inhibitory Activity on Melanogenesis. Skin Pharmacol. Physiol. 2017, 30, 205–215. [Google Scholar] [CrossRef] [Green Version]
- Alam, M.B.; Bajpai, V.K.; Lee, J.; Zhao, P.; Byeon, J.H.; Ra, J.S.; Majumder, R.; Lee, J.S.; Yoon, J.I.; Rather, I.A.; et al. Inhibition of melanogenesis by jineol from Scolopendra subspinipes mutilans via MAP-Kinase mediated MITF downregulation and the proteasomal degradation of tyrosinase. Sci. Rep. 2017, 7, 45858. [Google Scholar] [CrossRef]
- Isogawa, K.; Asano, M.; Hayazaki, M.; Koga, K.; Watanabe, M.; Suzuki, K.; Kobayashi, T.; Kawaguchi, K.; Ishizuka, A.; Kato, S.; et al. Thioxothiazolidin derivative, 4-OST, inhibits melanogenesis by enhancing the specific recruitment of tyrosinase-containing vesicles to lysosome. J. Cell Biochem. 2021, 122, 667–678. [Google Scholar] [CrossRef]
- Jeong, S.M.; Yoon, T.J. Development of Pigmentation-Regulating Agents by Drug Repositioning. Int. J. Mol. Sci. 2021, 22, 3894. [Google Scholar] [CrossRef]
- Niu, C.; Aisa, H.A. Upregulation of Melanogenesis and Tyrosinase Activity: Potential Agents for Vitiligo. Molecules 2017, 22, 1303. [Google Scholar] [CrossRef] [Green Version]
- van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell 2011, 21, 708–721. [Google Scholar] [CrossRef] [Green Version]
- Escola, J.M.; Kleijmeer, M.J.; Stoorvogel, W.; Griffith, J.M.; Yoshie, O.; Geuze, H.J. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 1998, 273, 20121–20127. [Google Scholar] [CrossRef] [Green Version]
- Pols, M.S.; Klumperman, J. Trafficking and function of the tetraspanin CD63. Exp. Cell Res. 2009, 315, 1584–1592. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Névo, N.; Jouve, M.; Valenzuela, J.I.; Maurin, M.; Verweij, F.J.; Palmulli, R.; Lankar, D.; Dingli, F.; Loew, D.; et al. Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nat. Commun. 2021, 12, 4389. [Google Scholar] [CrossRef] [PubMed]
- Ballabio, A.; Bonifacino, J.S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 101–118. [Google Scholar] [CrossRef]
- Ohbayashi, N.; Fukuda, M. Recent advances in understanding the molecular basis of melanogenesis in melanocytes. F1000Research 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Ni-Komatsu, L.; Tong, C.; Chen, G.; Brindzei, N.; Orlow, S.J. Identification of quinolines that inhibit melanogenesis by altering tyrosinase family trafficking. Mol. Pharmacol. 2008, 74, 1576–1586. [Google Scholar] [CrossRef] [Green Version]
- Okajima, S.; Hamamoto, A.; Asano, M.; Isogawa, K.; Ito, H.; Kato, S.; Hirata, Y.; Furuta, K.; Takemori, H. Azepine derivative T4FAT, a new copper chelator, inhibits tyrosinase. Biochem. Biophys. Res. Commun. 2019, 509, 209–215. [Google Scholar] [CrossRef]
- Muñoz, C.; Adasme, F.; Alzate-Morales, J.H.; Vergara-Jaque, A.; Kniess, T.; Caballero, J. Study of differences in the VEGFR2 inhibitory activities between semaxanib and SU5205 using 3D-QSAR, docking, and molecular dynamics simulations. J. Mol. Graph. Model. 2012, 32, 39–48. [Google Scholar] [CrossRef]
- Sato, T.; Sato, Y.; Iwai, K.; Kuge, S.; Teramae, N.; Nishizawa, S. Fluorescence Imaging of siRNA Delivery by Peptide Nucleic Acid-based Probe. Anal. Sci. 2015, 31, 315–320. [Google Scholar] [CrossRef] [Green Version]
- Slominski, A.; Tobin, D.J.; Shibahara, S.; Wortsman, J. Melanin pigmentation in mammalian skin and its hormonal regulation. Physiol. Rev. 2004, 84, 1155–1228. [Google Scholar] [CrossRef]
- Slominski, A.T.; Zmijewski, M.A.; Plonka, P.M.; Szaflarski, J.P.; Paus, R. How UV Light Touches the Brain and Endocrine System Through Skin, and Why. Endocrinology 2018, 159, 1992–2007. [Google Scholar] [CrossRef] [Green Version]
- Slominski, A.; Wortsman, J.; Plonka, P.M.; Schallreuter, K.U.; Paus, R.; Tobin, D.J. Hair follicle pigmentation. J. Investig. Dermatol. 2005, 124, 13–21. [Google Scholar] [CrossRef] [Green Version]
- Slominski, A.; Zmijewski, M.A.; Pawelek, J. L-tyrosine and L-dihydroxyphenylalanine as hormone-like regulators of melanocyte functions. Pigment. Cell Melanoma Res. 2012, 25, 14–27. [Google Scholar] [CrossRef] [Green Version]
- Bomirski, A.; Wrzolkowa, T.; Arendarczyk, M.; Bomirska, M.; Kuklinska, E.; Slominski, A.; Moellmann, G. Pathology and ultrastructural characteristics of a hypomelanotic variant of transplantable hamster melanoma with elevated tyrosinase activity. J. Investig. Dermatol. 1987, 89, 469–473. [Google Scholar] [CrossRef] [Green Version]
- White, B.H.; Kaczmarek, L.K. Identification of a vesicular pool of calcium channels in the bag cell neurons of Aplysia californica. J. Neurosci. 1997, 17, 1582–1595. [Google Scholar] [CrossRef]
- Wang, X.Y.; Guan, X.H.; Yu, Z.P.; Wu, J.; Huang, Q.M.; Deng, K.Y.; Xin, H.B. Human amniotic stem cells-derived exosmal miR-181a-5p and miR-199a inhibit melanogenesis and promote melanosome degradation in skin hyperpigmentation, respectively. Stem Cell Res. Ther. 2021, 12, 501. [Google Scholar] [CrossRef]
- Bentley, N.J.; Eisen, T.; Goding, C.R. Melanocyte-specific expression of the human tyrosinase promoter: Activation by the microphthalmia gene product and role of the initiator. Mol. Cell Biol. 1994, 14, 7996–8006. [Google Scholar] [PubMed] [Green Version]
- Yasumoto, K.; Yokoyama, K.; Shibata, K.; Tomita, Y.; Shibahara, S. Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Mol. Cell Biol. 1994, 14, 8058–8070. [Google Scholar] [PubMed]
- Maeda, M.; Suzuki, M.; Fuchino, H.; Tanaka, N.; Kobayashi, T.; Isogai, R.; Batubara, I.; Iswantini, D.; Matsuno, M.; Kawahara, N.; et al. Diversity of Adenostemma lavenia, multi-potential herbs, and its kaurenoic acid composition between Japan and Taiwan. J. Nat. Med. 2021. [Google Scholar] [CrossRef] [PubMed]
- Hseu, Y.C.; Vudhya Gowrisankar, Y.; Wang, L.W.; Zhang, Y.Z.; Chen, X.Z.; Huang, P.J.; Yen, H.R.; Yang, H.L. The in vitro and in vivo depigmenting activity of pterostilbene through induction of autophagy in melanocytes and inhibition of UVA-irradiated α-MSH in keratinocytes via Nrf2-mediated antioxidant pathways. Redox Biol. 2021, 44, 102007. [Google Scholar] [CrossRef]
- Perera, R.M.; Stoykova, S.; Nicolay, B.N.; Ross, K.N.; Fitamant, J.; Boukhali, M.; Lengrand, J.; Deshpande, V.; Selig, M.K.; Ferrone, C.R.; et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature 2015, 524, 361–365. [Google Scholar] [CrossRef]
- Zhu, J.W.; Ni, Y.J.; Tong, X.Y.; Guo, X.; Wu, X.P.; Lu, Z.F. Tranexamic Acid Inhibits Angiogenesis and Melanogenesis in Vitro by Targeting VEGF Receptors. Int. J. Med. Sci. 2020, 17, 903–911. [Google Scholar] [CrossRef] [Green Version]
- Brown, E.J.; Albers, M.W.; Shin, T.B.; Ichikawa, K.; Keith, C.T.; Lane, W.S.; Schreiber, S.L. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 1994, 369, 756–758. [Google Scholar] [CrossRef]
- Yun, W.J.; Kim, E.Y.; Park, J.E.; Jo, S.Y.; Bang, S.H.; Chang, E.J.; Chang, S.E. Microtubule-associated protein light chain 3 is involved in melanogenesis via regulation of MITF expression in melanocytes. Sci. Rep. 2016, 6, 19914. [Google Scholar] [CrossRef]
- Kim, J.; Choi, H.; Cho, E.G.; Lee, T.R. FoxO3a is an antimelanogenic factor that mediates antioxidant-induced depigmentation. J. Investig. Dermatol. 2014, 134, 1378–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pi, H.; Li, M.; Zou, L.; Yang, M.; Deng, P.; Fan, T.; Liu, M.; Tian, L.; Tu, M.; Xie, J.; et al. AKT inhibition-mediated dephosphorylation of TFE3 promotes overactive autophagy independent of MTORC1 in cadmium-exposed bone mesenchymal stem cells. Autophagy 2019, 15, 565–582. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, A.; Isogai, R.; Maeda, M.; Hayazaki, M.; Horiyama, E.; Takashima, S.; Koketsu, M.; Takemori, H. The High Content of Ent-11α-hydroxy-15-oxo-kaur- 16-en-19-oic Acid in Adenostemma lavenia (L.) O. Kuntze Leaf Extract: With Preliminary in Vivo Assays. Foods 2020, 9, 73. [Google Scholar] [CrossRef] [Green Version]
- Coda, A.C.; Invernizzi, A.G.; Righetti, P.P.; Tacconi, G.; Gatti, G. (Z)- and (E)-Arylidene-1,3-dihydroindol-2-ones: Configuration, conformation, and infrared carbonyl stretching frequencies. J. Chem. Soc. Perkin Trans. 1984, 2, 615–619. [Google Scholar] [CrossRef]
- Sun, L.; Tran, N.; Tang, F.; App, H.; Hirth, P.; McMahon, G.; Tang, C. Synthesis and biological evaluations of 3-substituted indolin-2-ones: A novel class of tyrosine kinase inhibitors that exhibit selectivity toward particular receptor tyrosine kinases. J. Med. Chem. 1998, 41, 2588–2603. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Go, M.-L. Functionalized 3-benzylidene-indolin-2-ones: Inducers of NAD(P)H-quinone oxidoreductase 1 (NQO1) with antiproliferative activity. Bioorg. Med. Chem. 2009, 17, 2077–2090. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watanabe, M.; Kawaguchi, K.; Nakamura, Y.; Furuta, K.; Takemori, H. GIF-2209, an Oxindole Derivative, Accelerates Melanogenesis and Melanosome Secretion via the Modification of Lysosomes in B16F10 Mouse Melanoma Cells. Molecules 2022, 27, 177. https://doi.org/10.3390/molecules27010177
Watanabe M, Kawaguchi K, Nakamura Y, Furuta K, Takemori H. GIF-2209, an Oxindole Derivative, Accelerates Melanogenesis and Melanosome Secretion via the Modification of Lysosomes in B16F10 Mouse Melanoma Cells. Molecules. 2022; 27(1):177. https://doi.org/10.3390/molecules27010177
Chicago/Turabian StyleWatanabe, Miyu, Kyoka Kawaguchi, Yusuke Nakamura, Kyoji Furuta, and Hiroshi Takemori. 2022. "GIF-2209, an Oxindole Derivative, Accelerates Melanogenesis and Melanosome Secretion via the Modification of Lysosomes in B16F10 Mouse Melanoma Cells" Molecules 27, no. 1: 177. https://doi.org/10.3390/molecules27010177
APA StyleWatanabe, M., Kawaguchi, K., Nakamura, Y., Furuta, K., & Takemori, H. (2022). GIF-2209, an Oxindole Derivative, Accelerates Melanogenesis and Melanosome Secretion via the Modification of Lysosomes in B16F10 Mouse Melanoma Cells. Molecules, 27(1), 177. https://doi.org/10.3390/molecules27010177





