Ratiometric Electrochemistry: Improving the Robustness, Reproducibility and Reliability of Biosensors
Abstract
1. Introduction
2. Secondary Redox-Active Labelling
2.1. Secondary Labelling of Solid-Supported DNA Structures
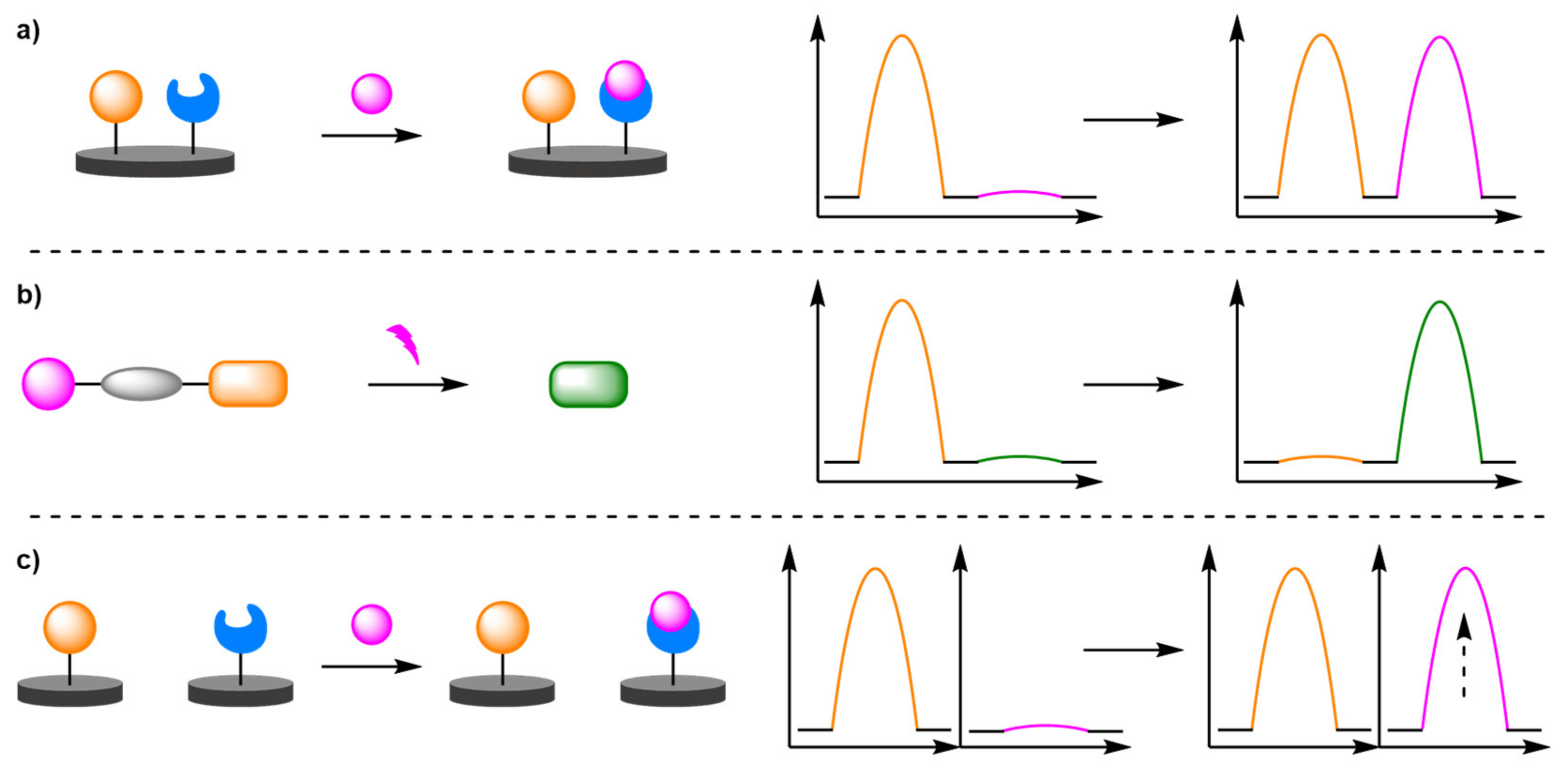
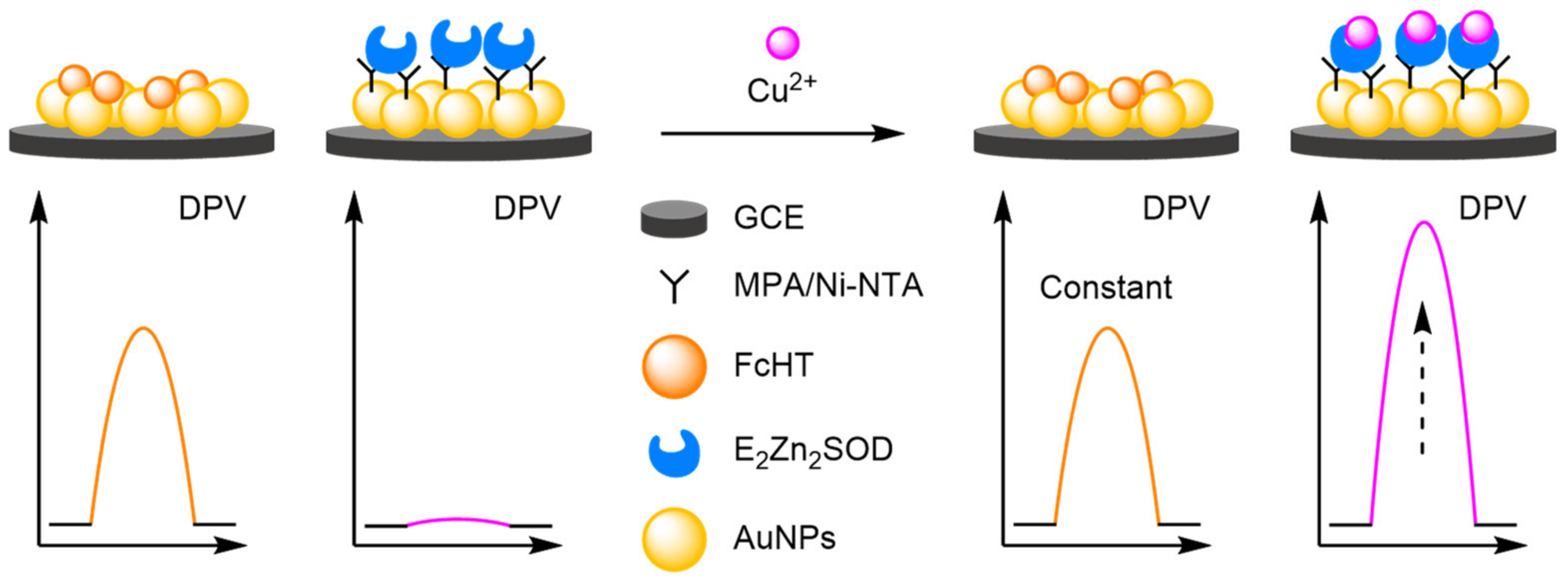
2.1.1. Binding Modes
- (a)
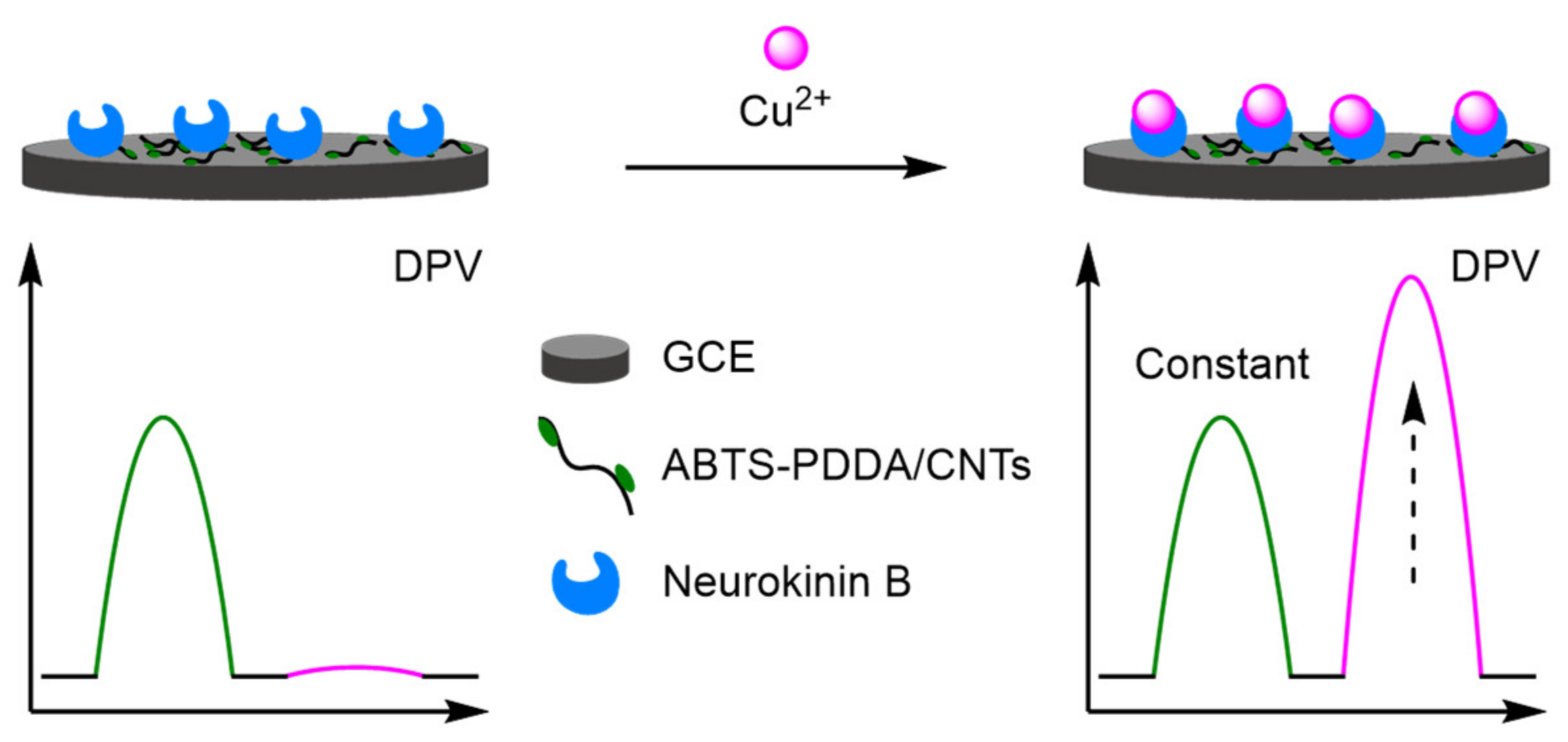
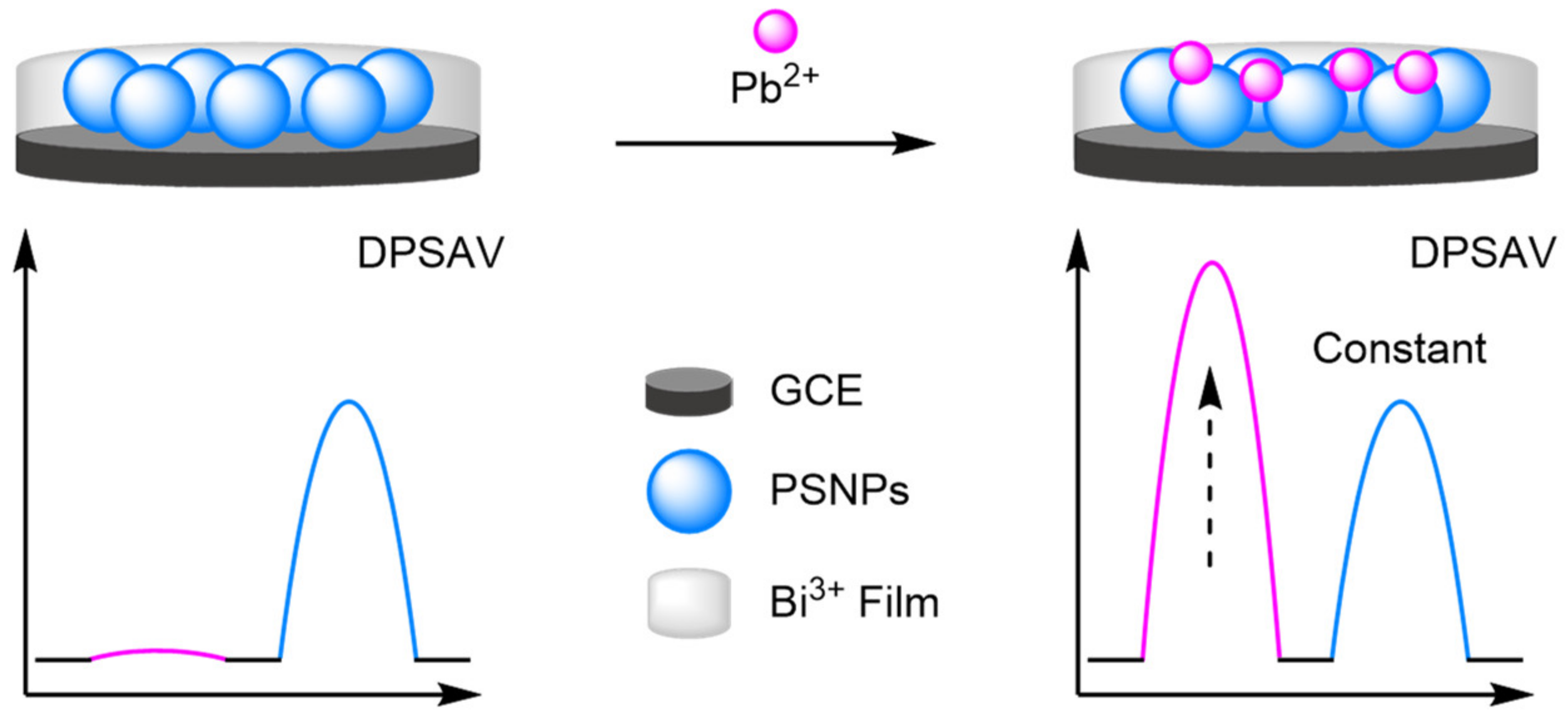
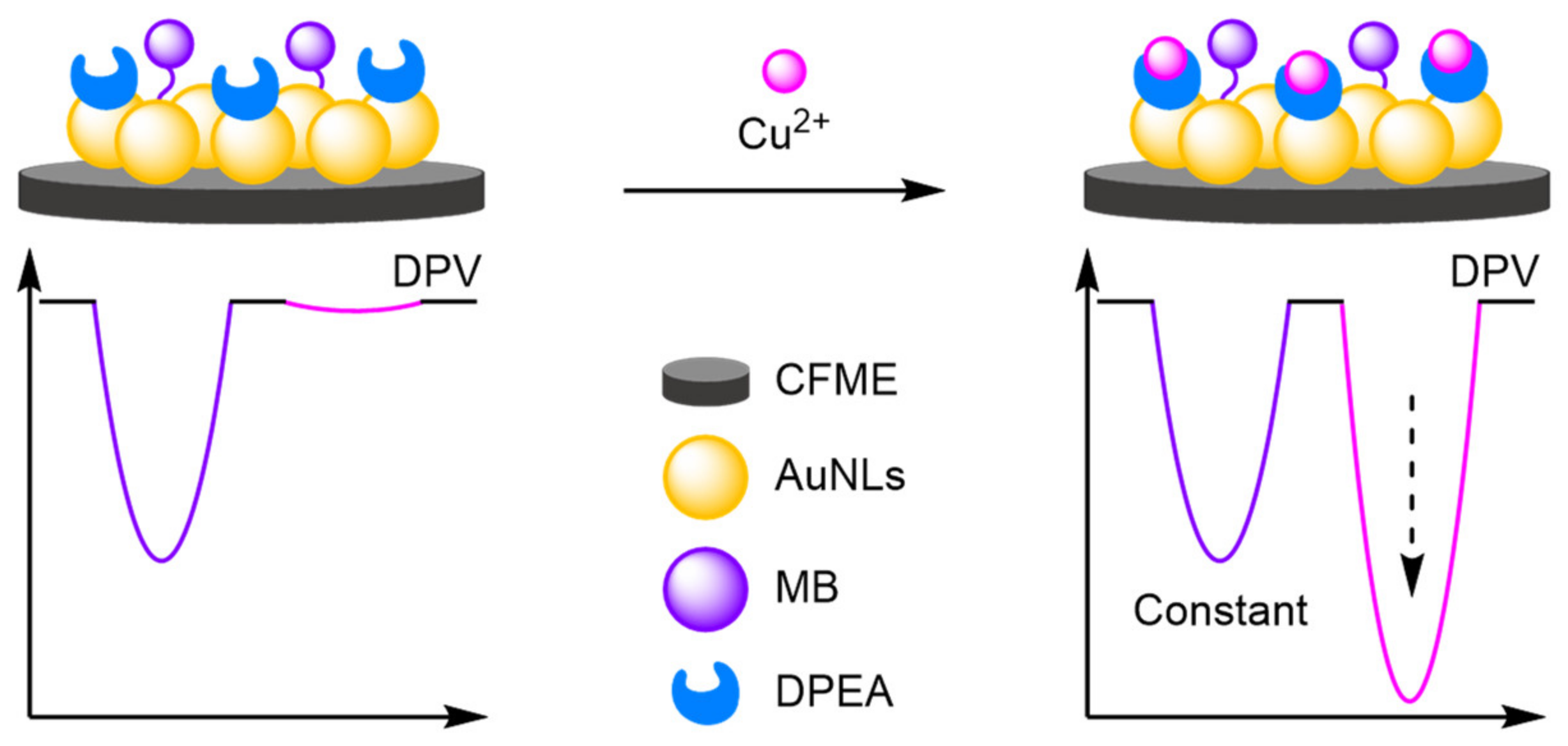
- Heavy Metals
- (b)
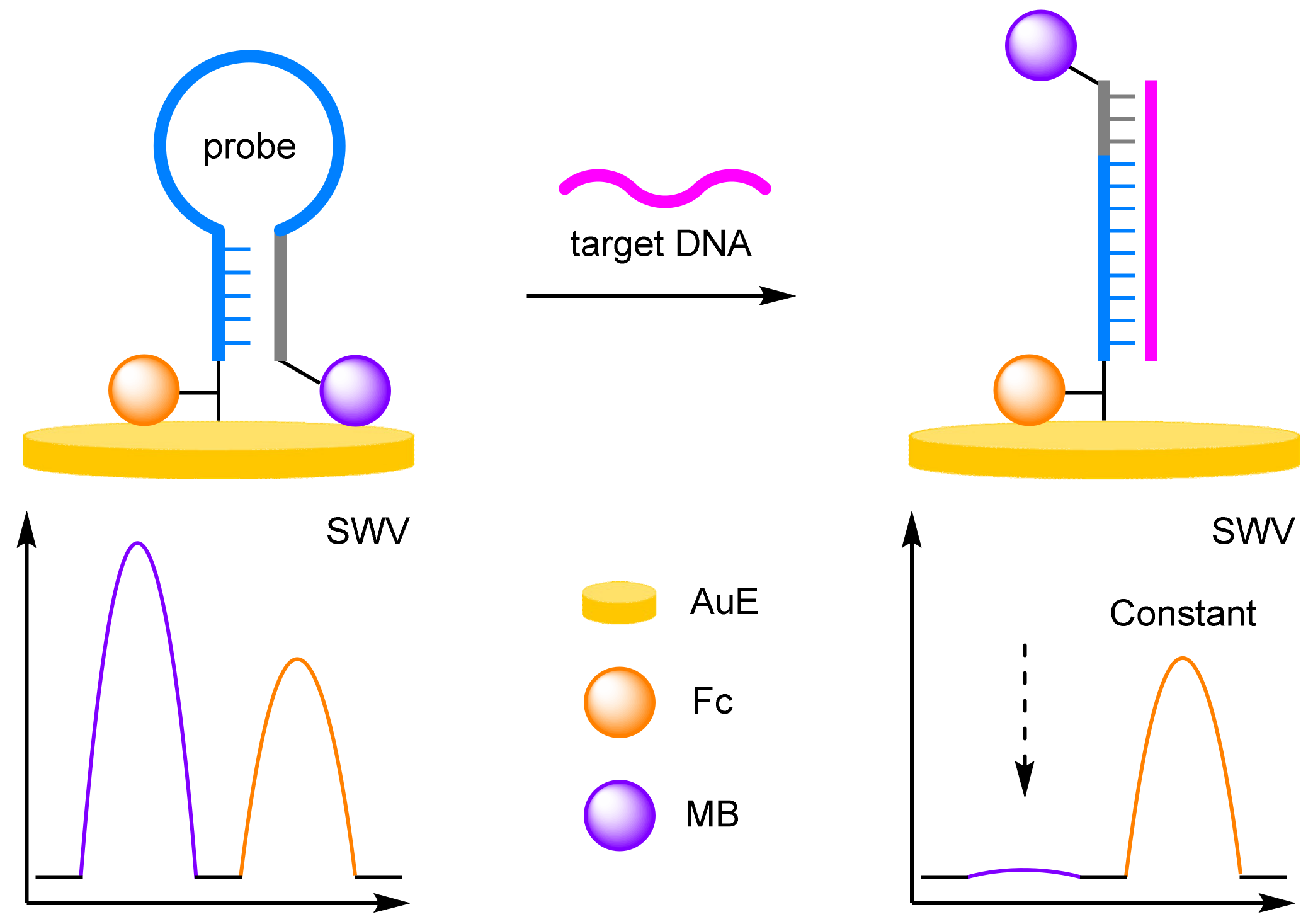
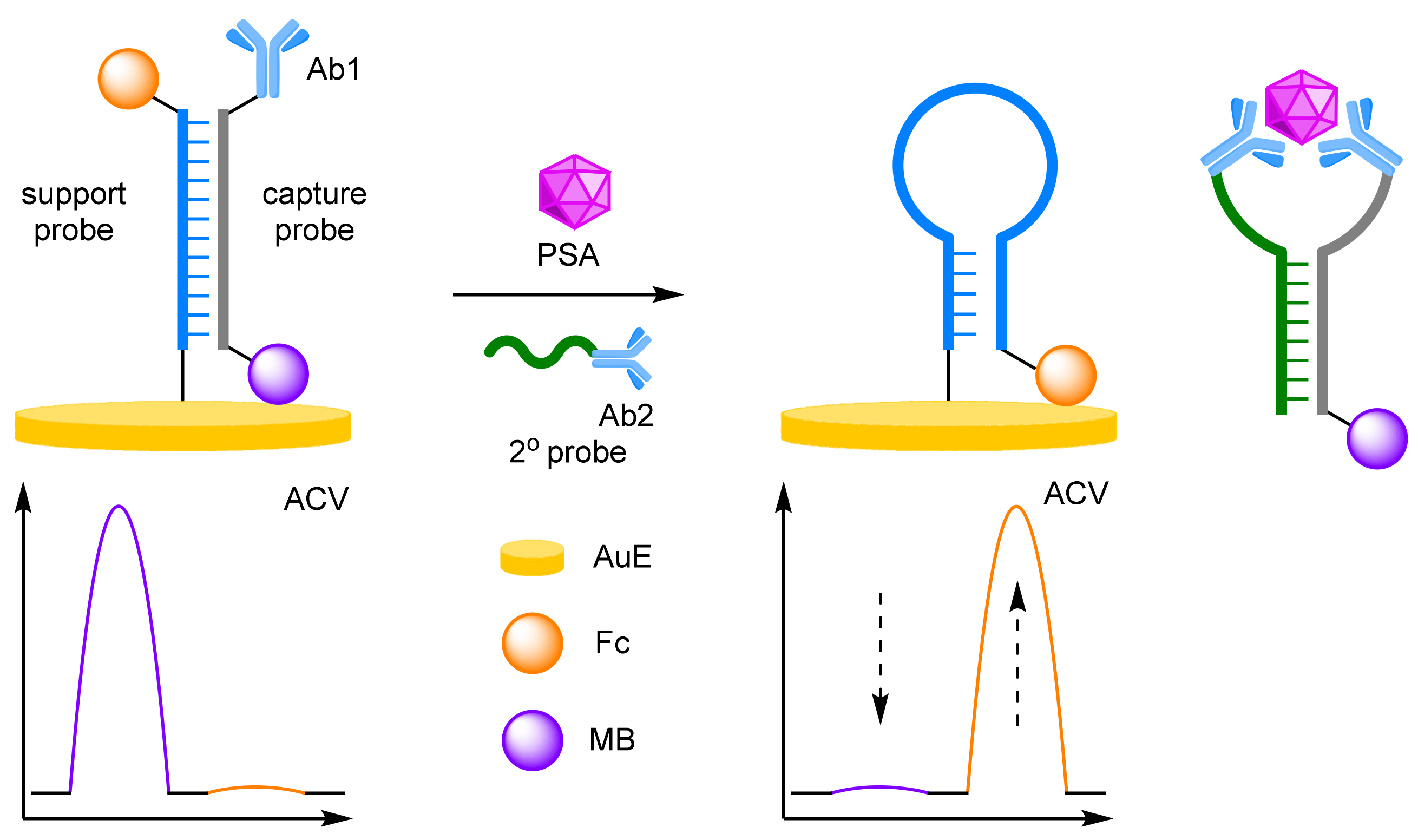
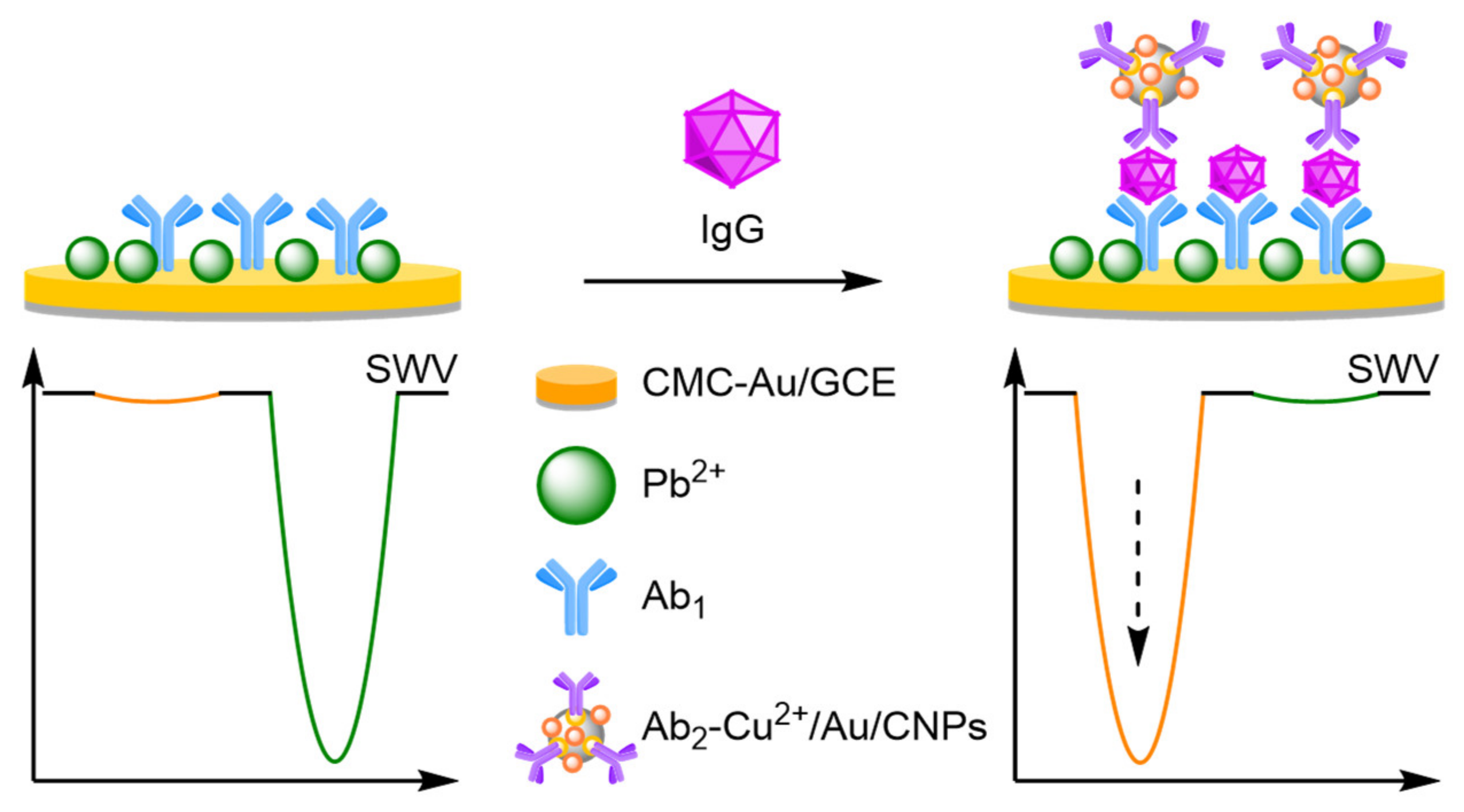
- Antibodies
- (c)
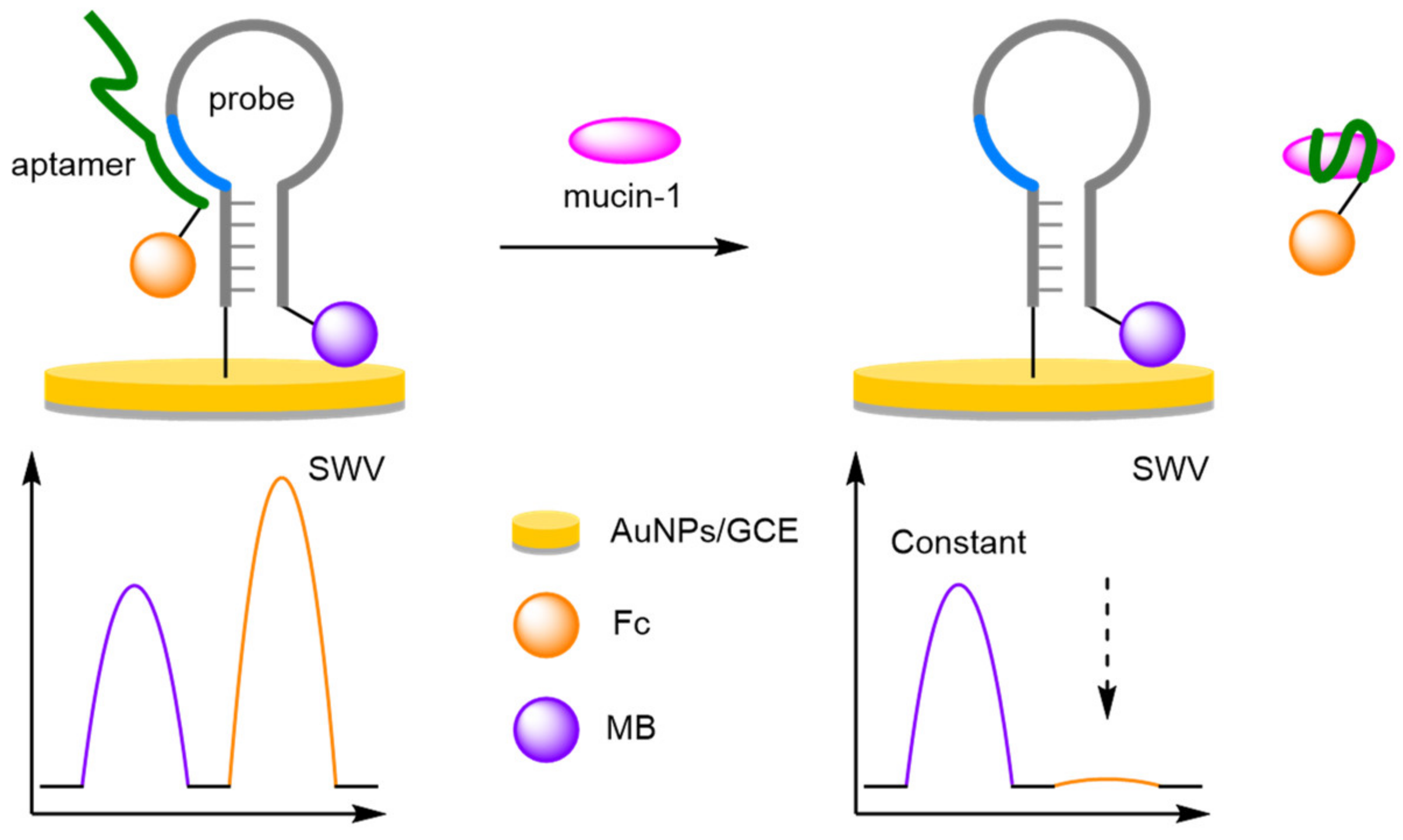
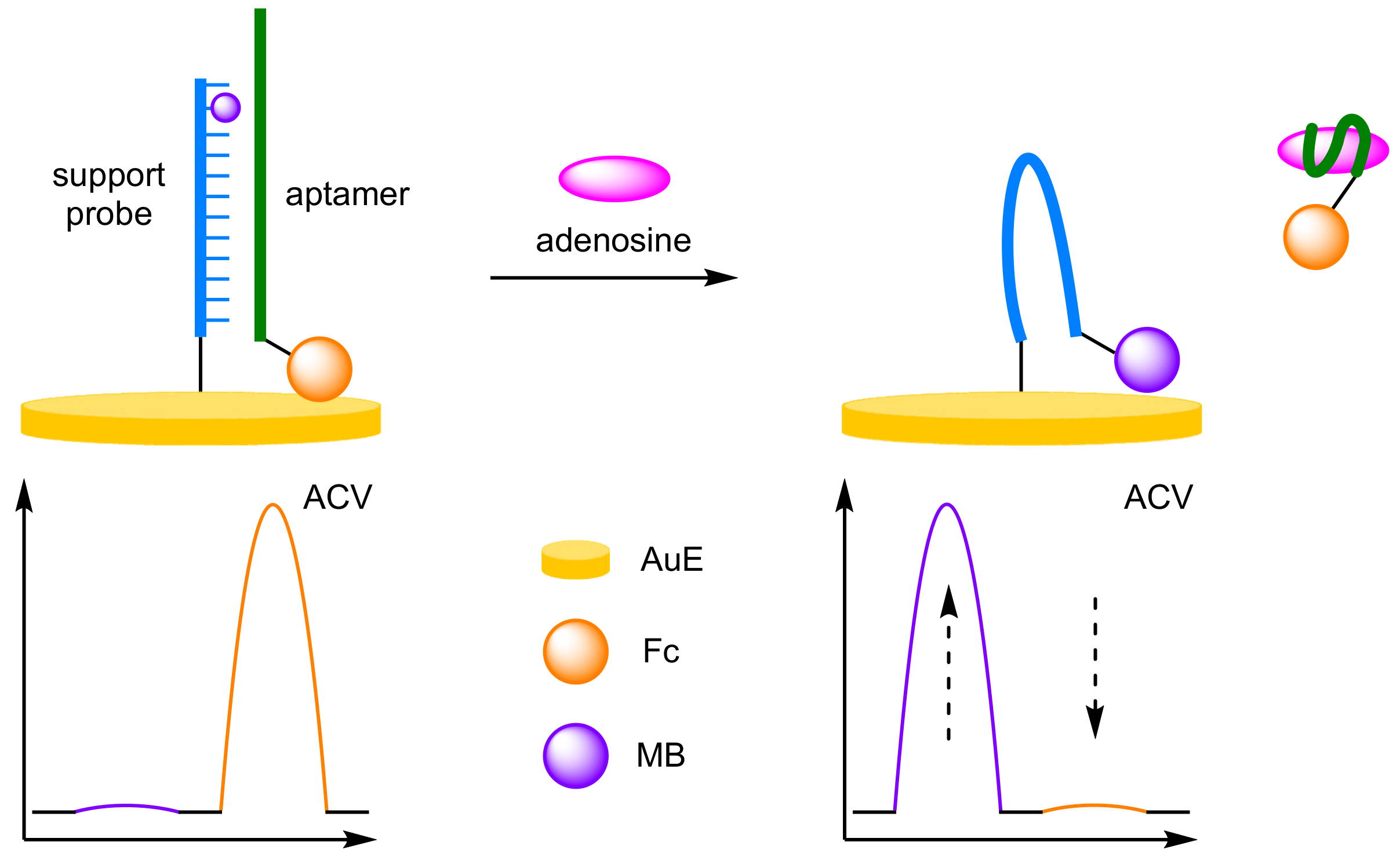
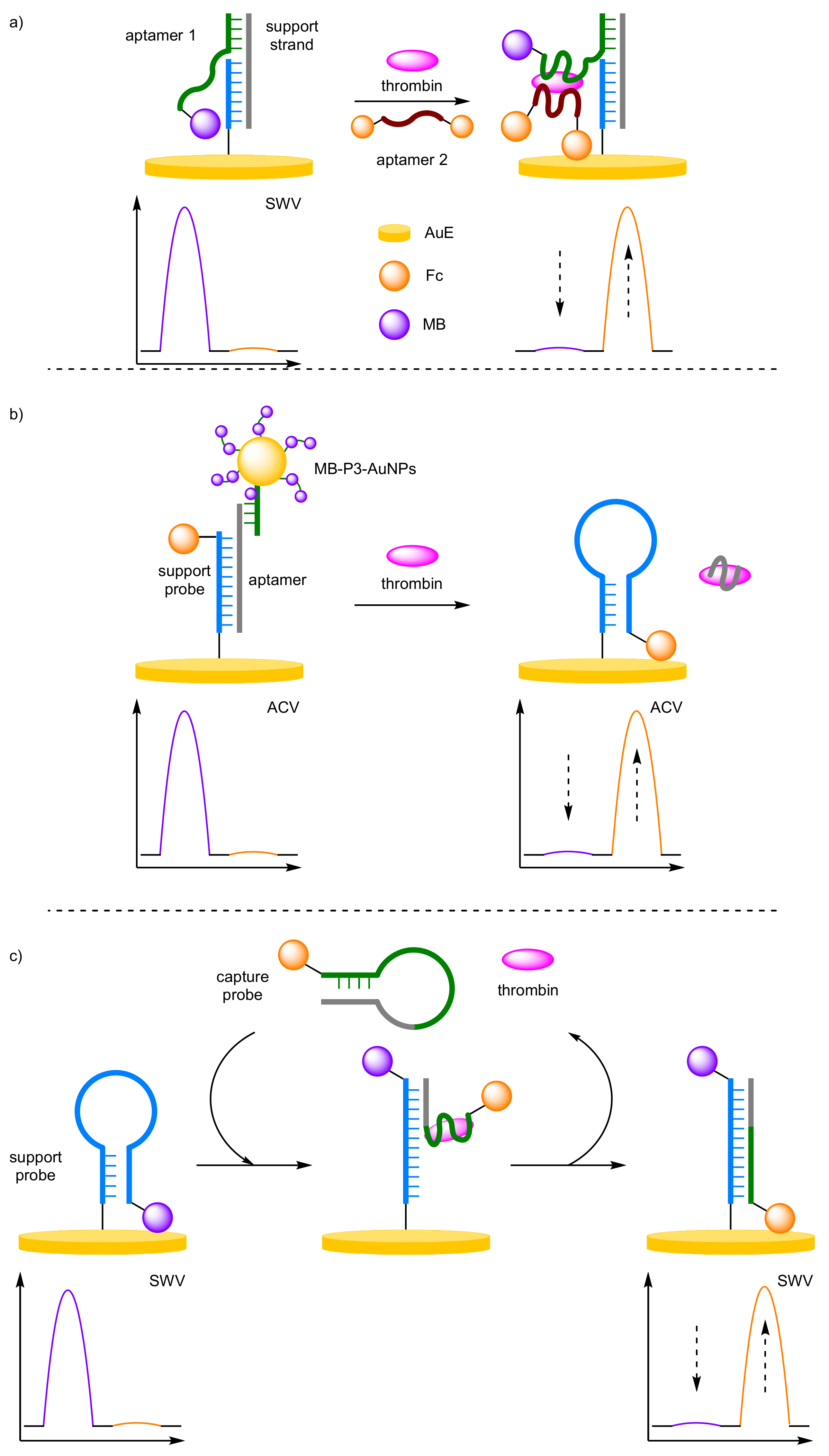
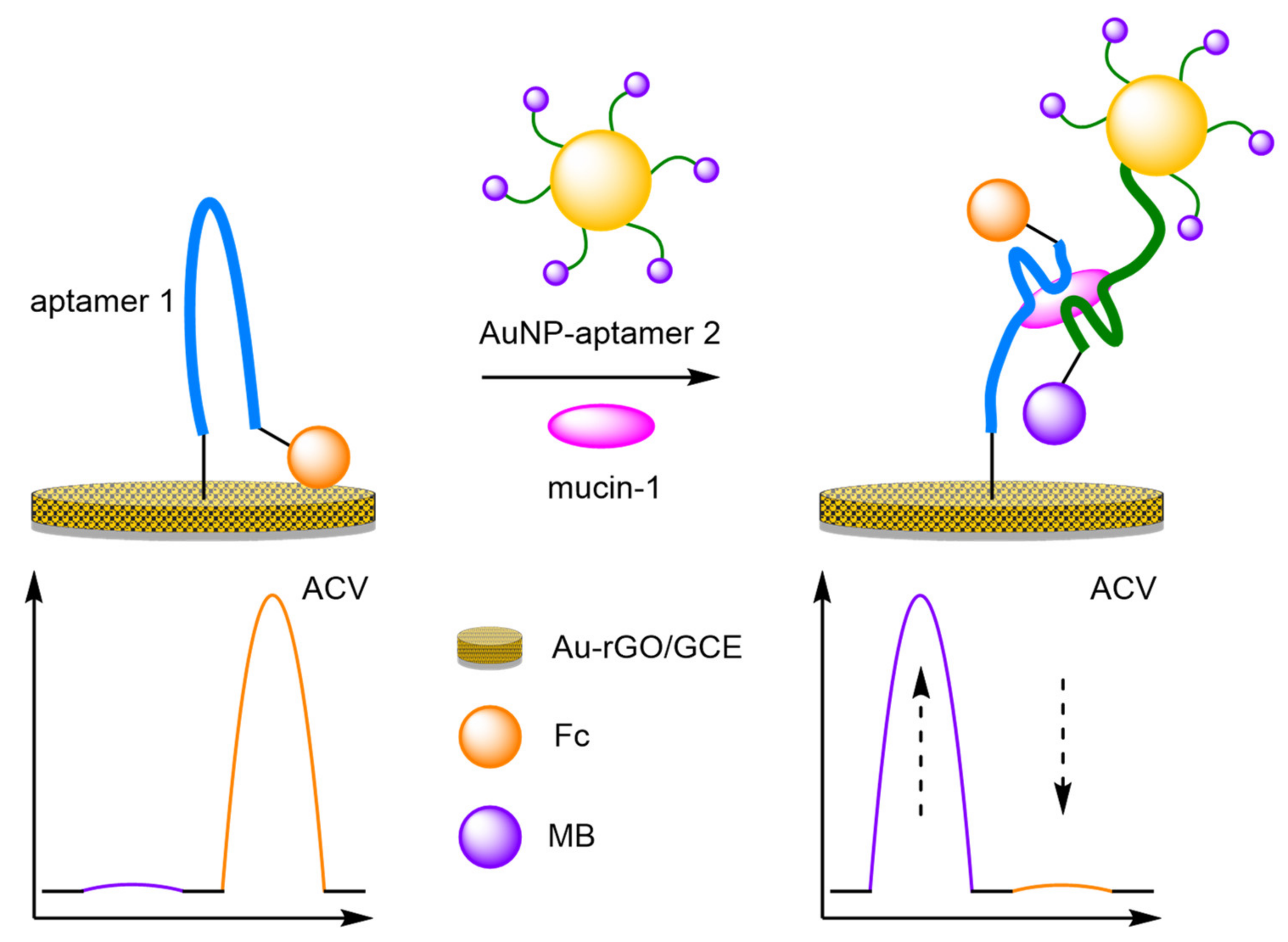
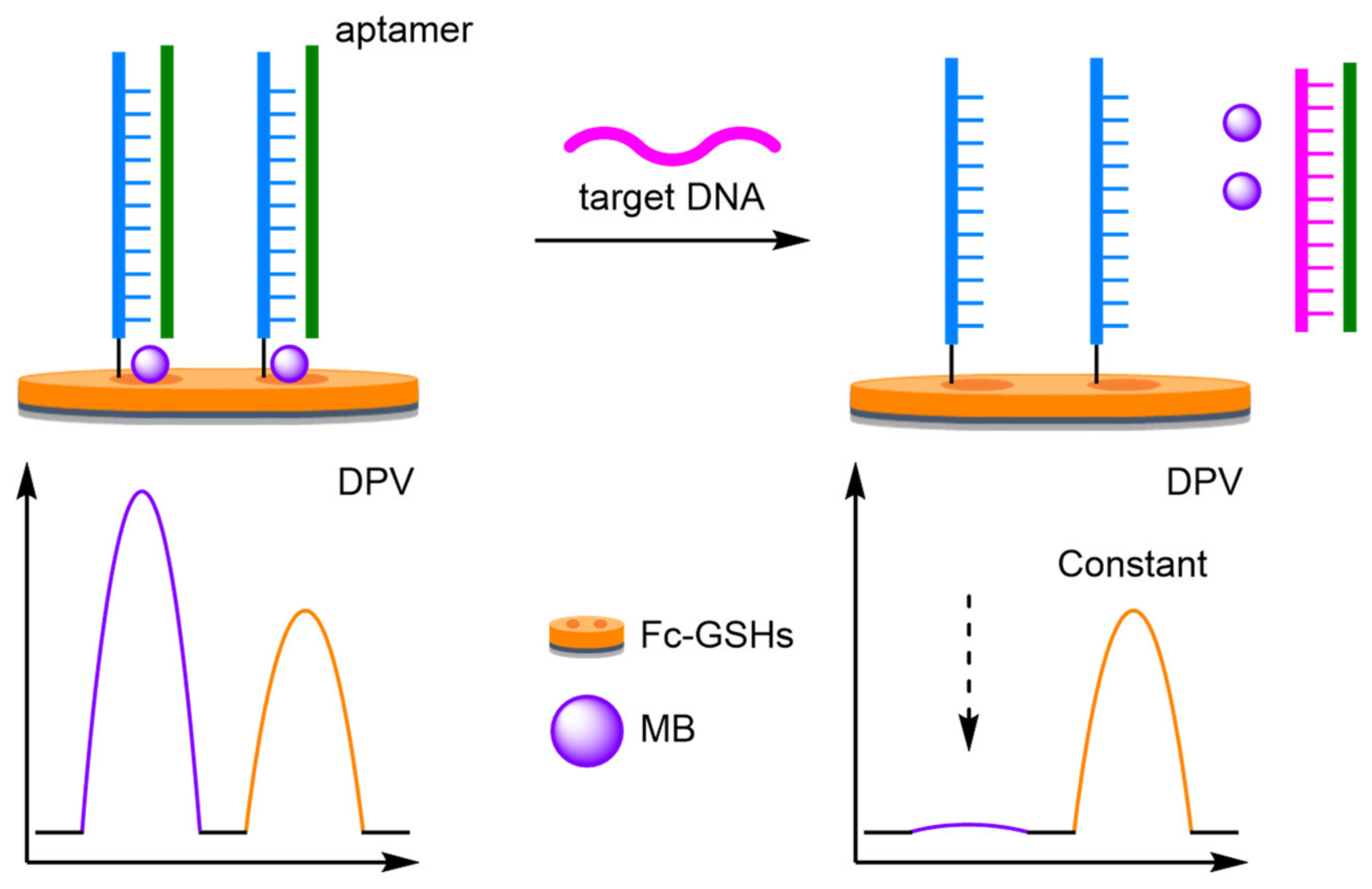
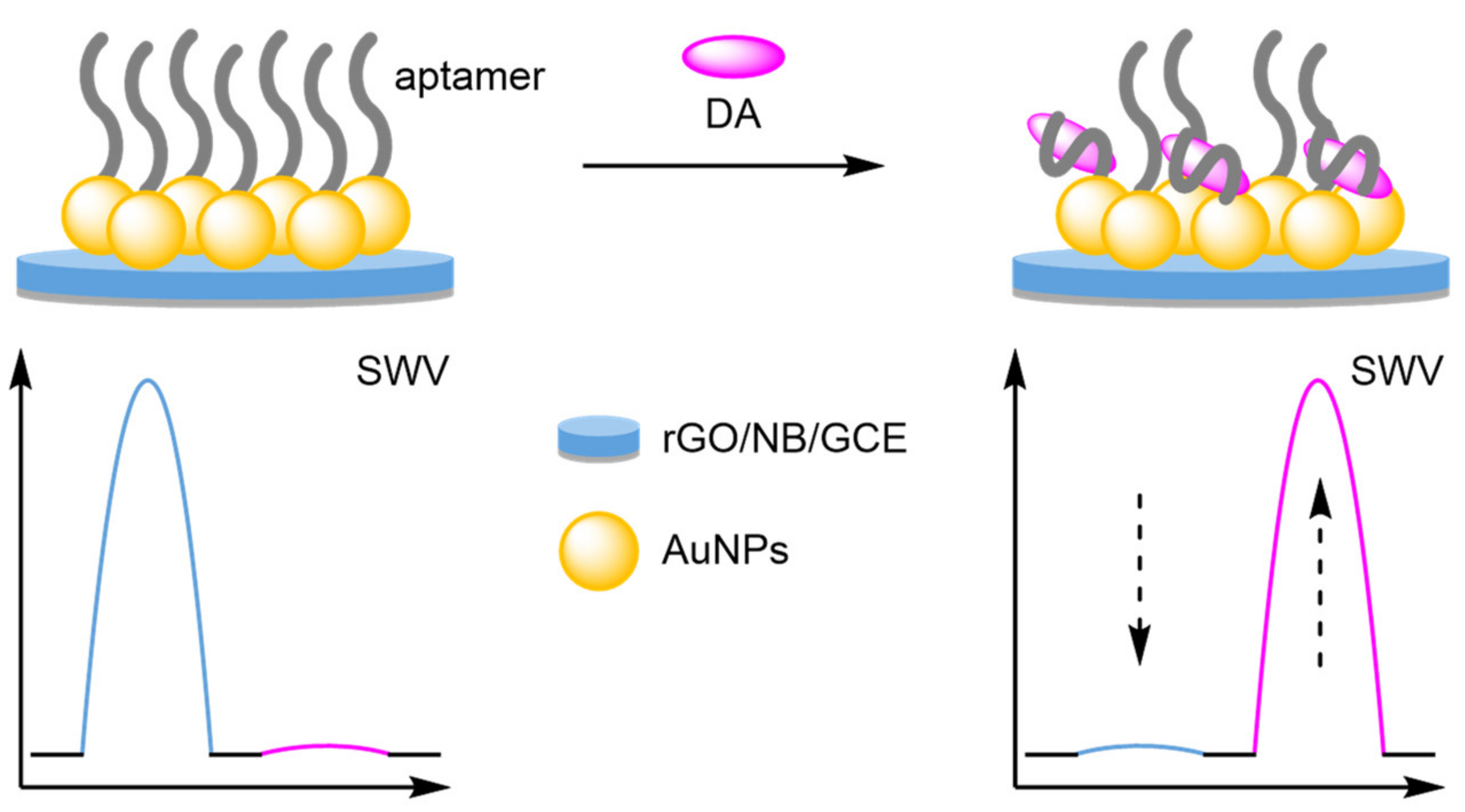
- Aptamers
- (d)
- Enzyme Detection
2.1.2. Selectivity Strategies
- (a)
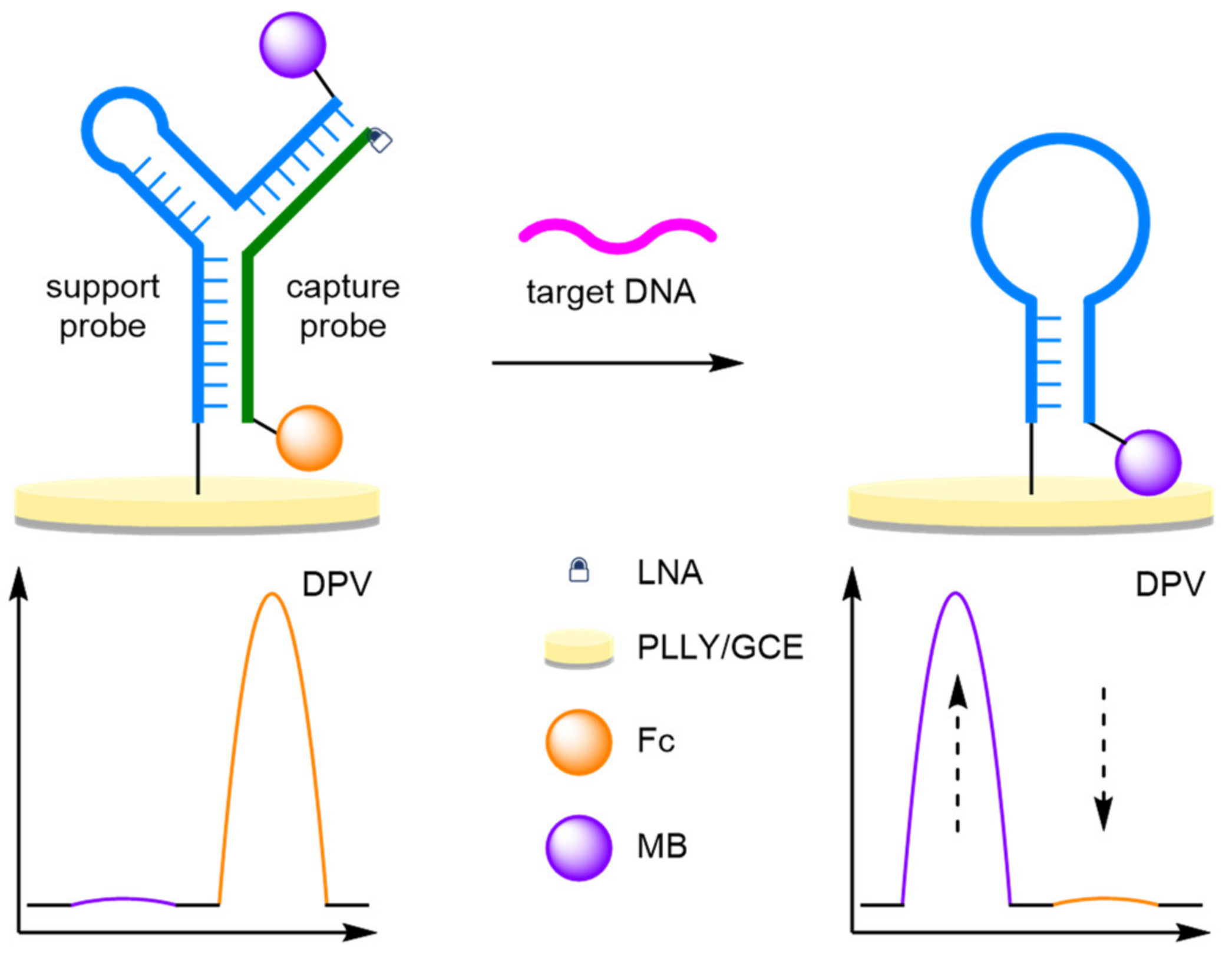
- Locked Nucleic Acids
- (b)
- Triple-Helix Molecular Beacons
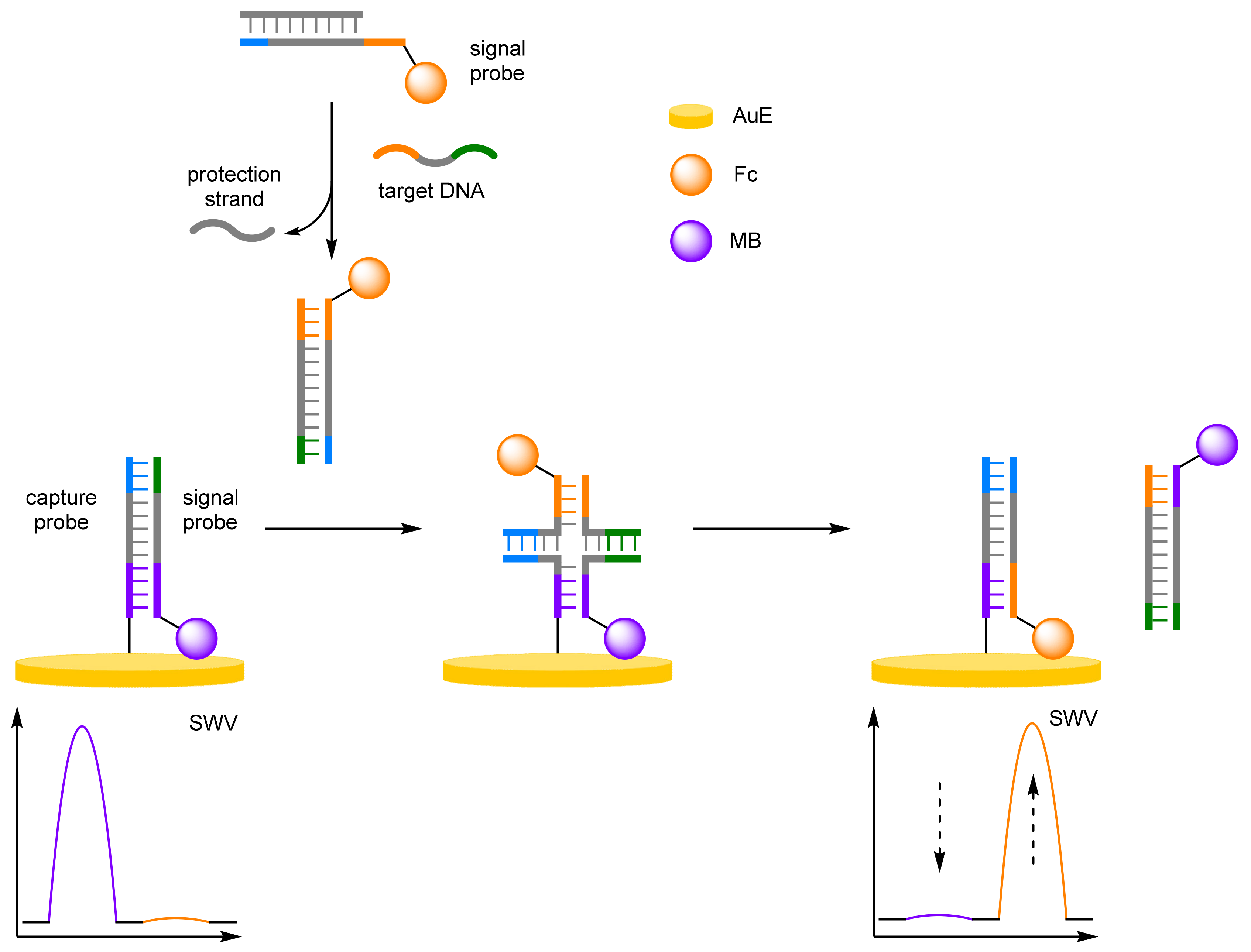
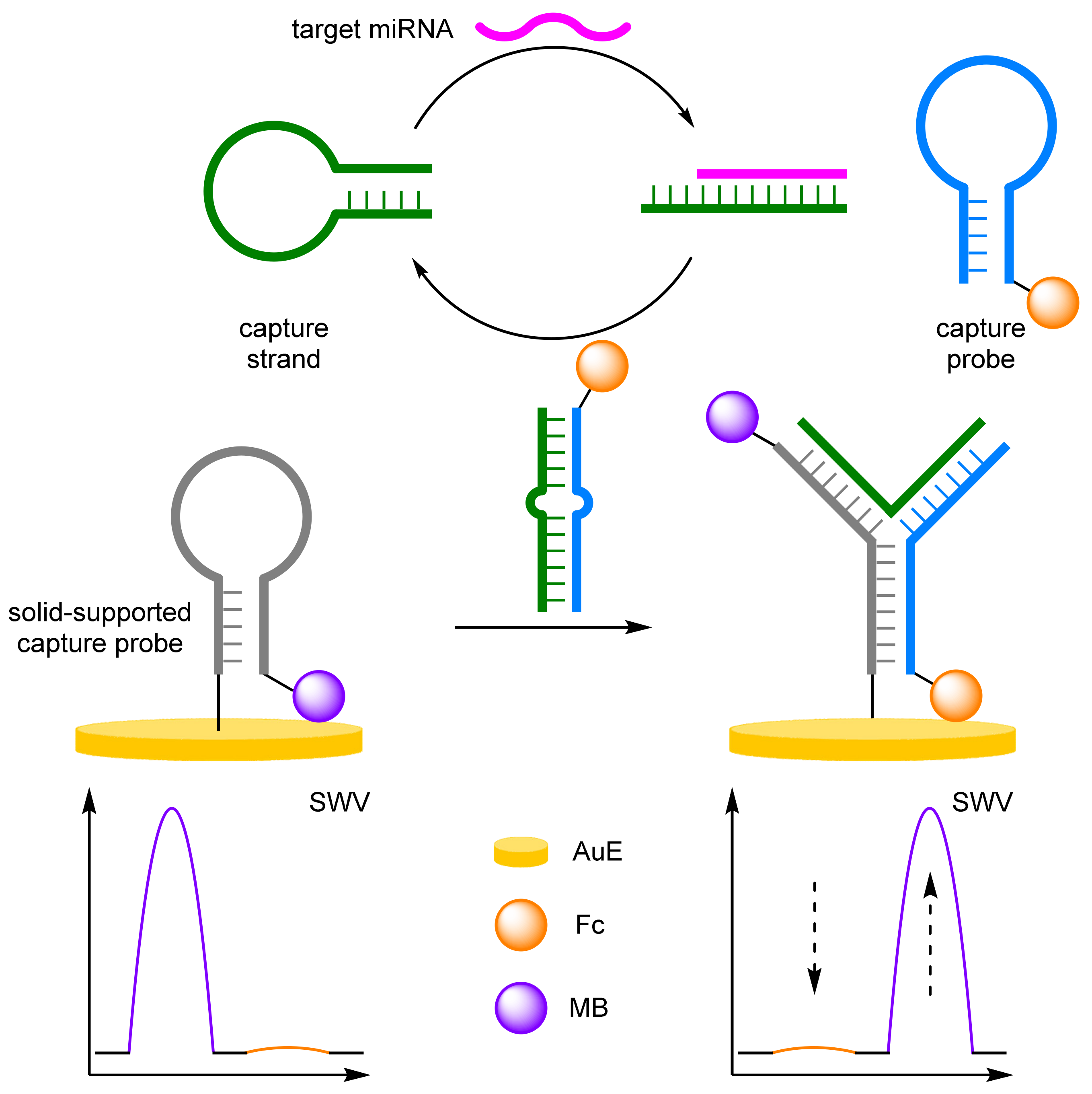
DNA Four-Way Junctions
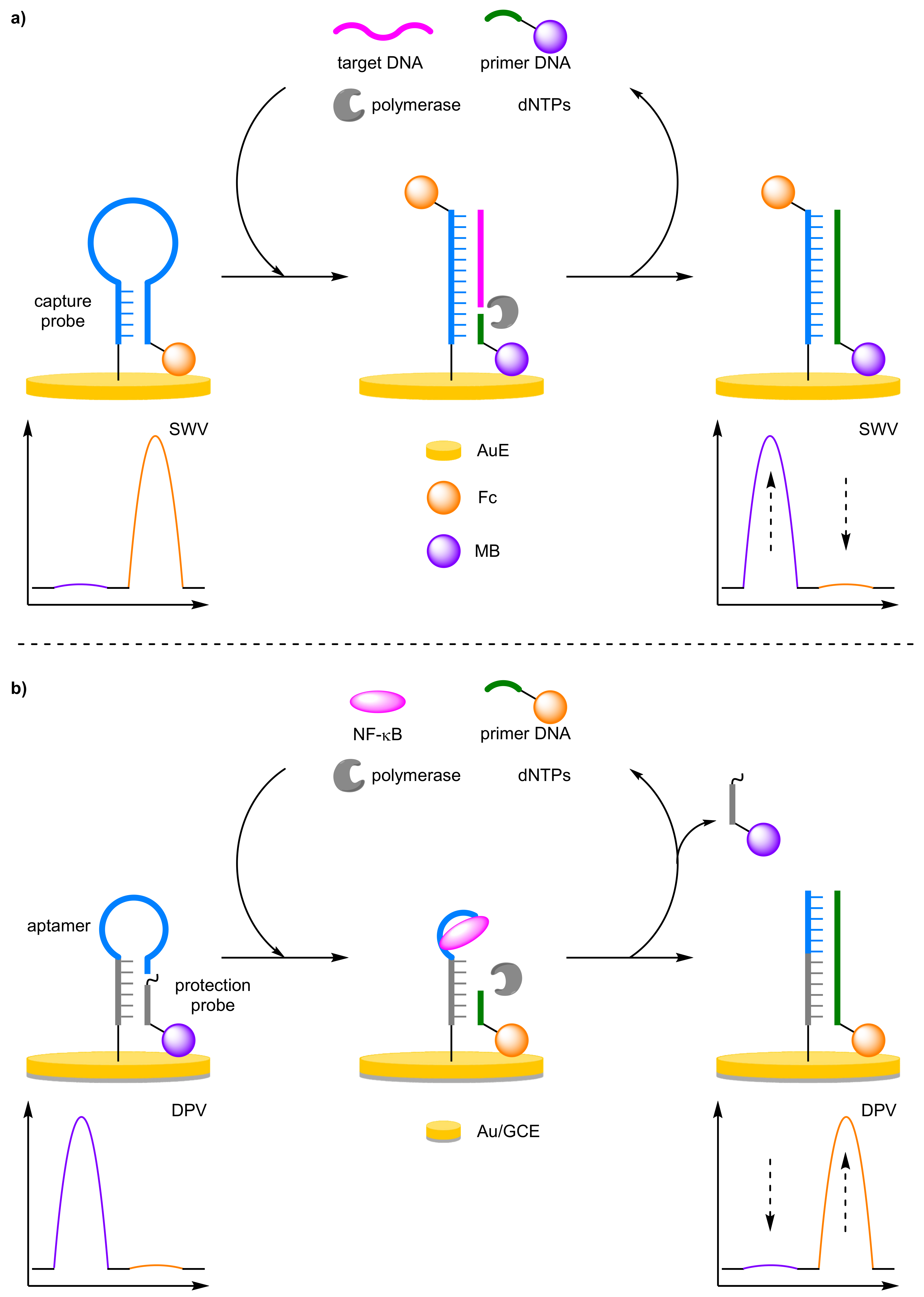
2.1.3. Amplification Strategies
- (a)
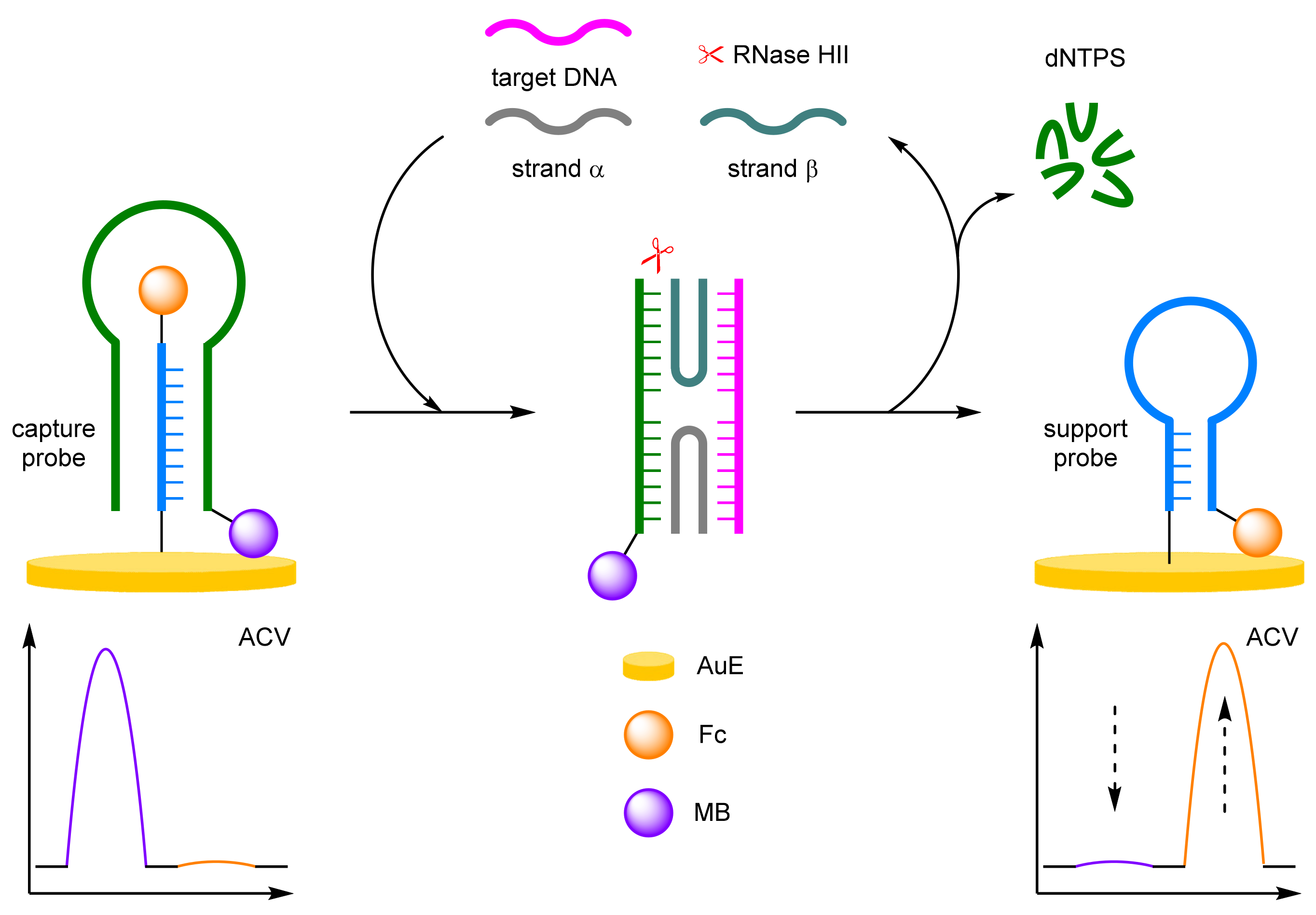
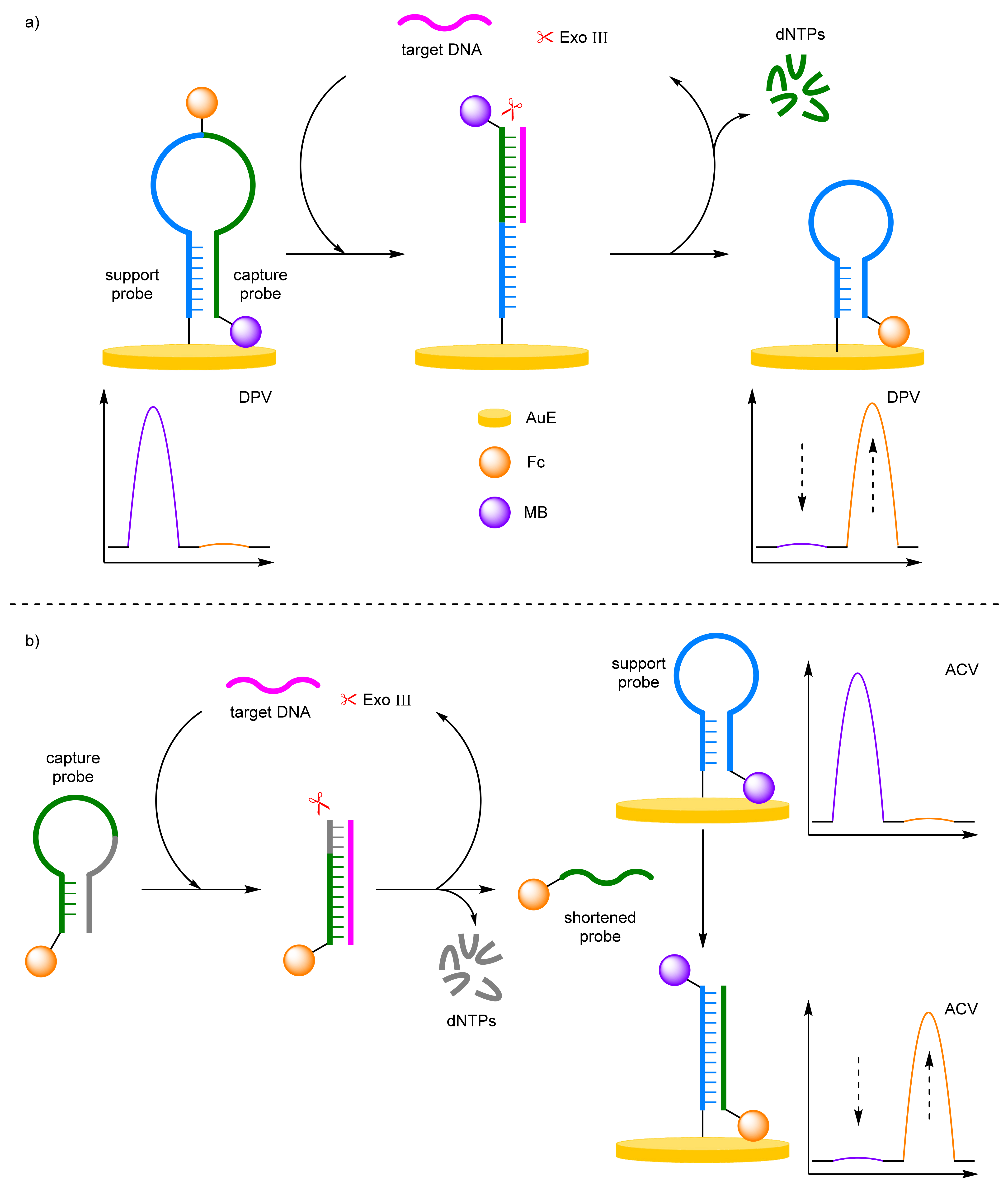
- Enzyme-Assisted Amplification
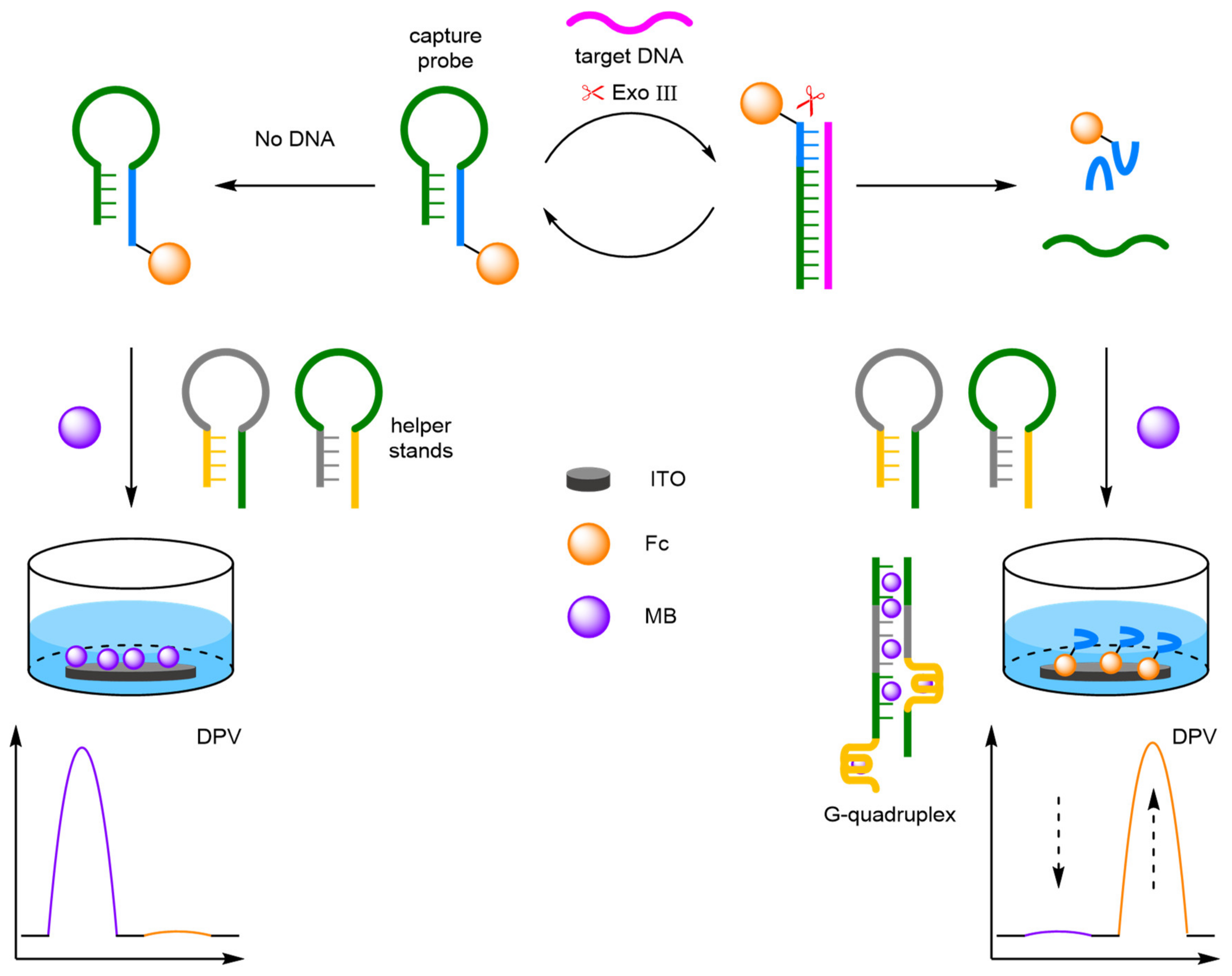
Exonuclease
Strand Displacement Polymerase Reaction
Rolling Circle Amplification
Catalytic Cascades
- (b)
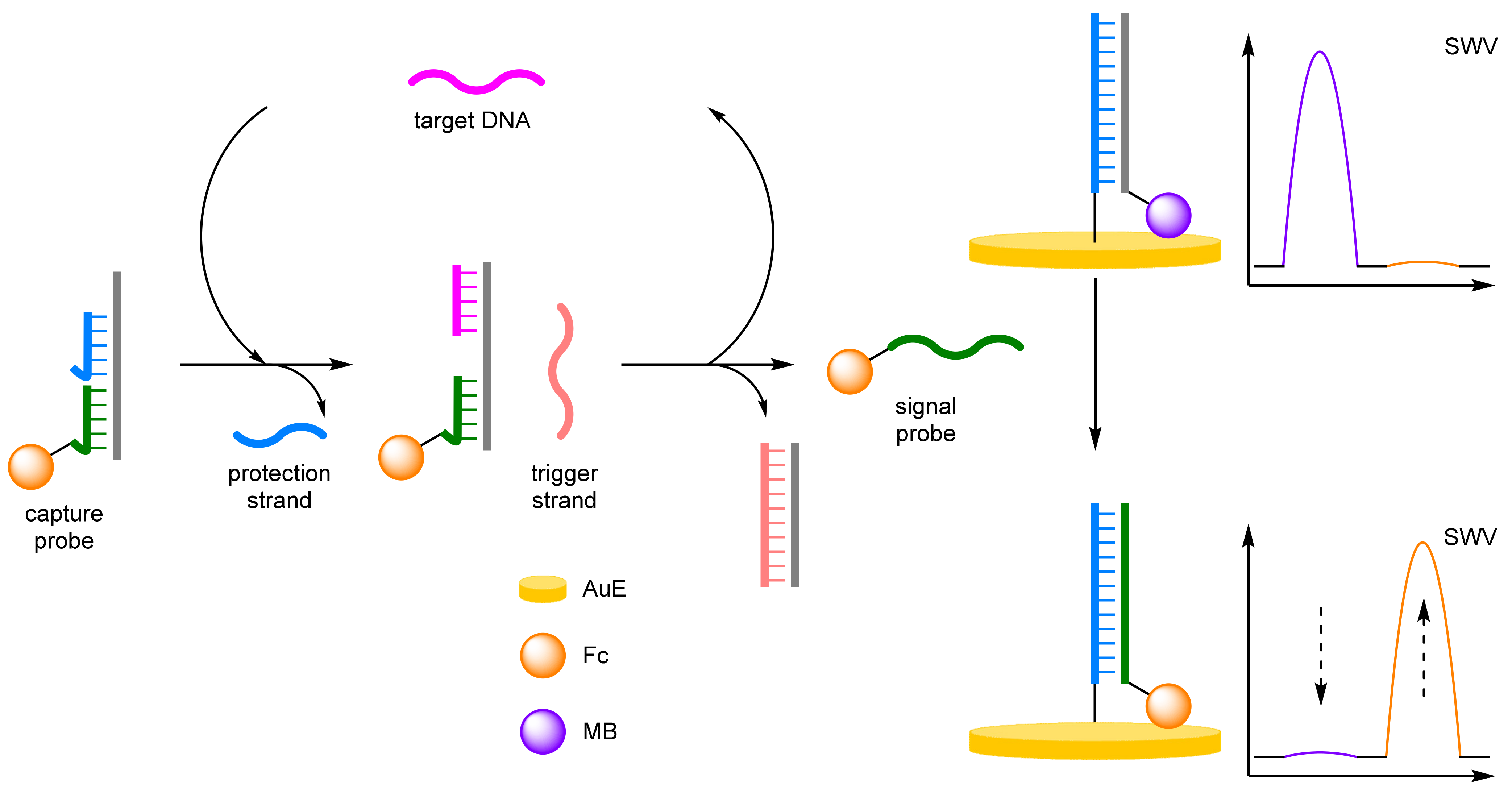
- Toehold Mediated Strand Displacement Reaction
- (c)
- Catalytic Hairpin Assembly
- (d)
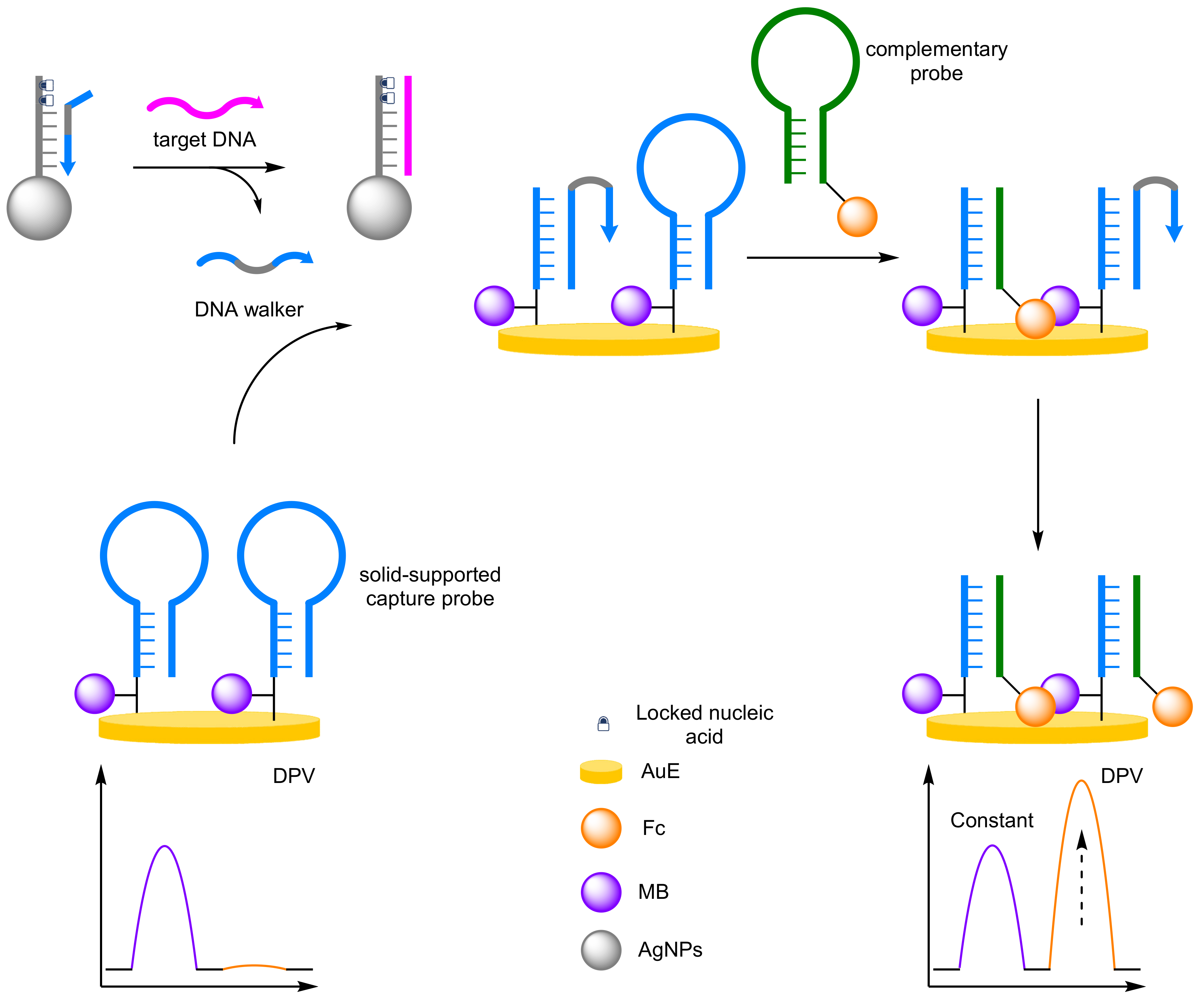
- DNA Walkers
- (e)
- Metal Nanoparticles
- (f)
- Multiple Label Intercalation
- (g)
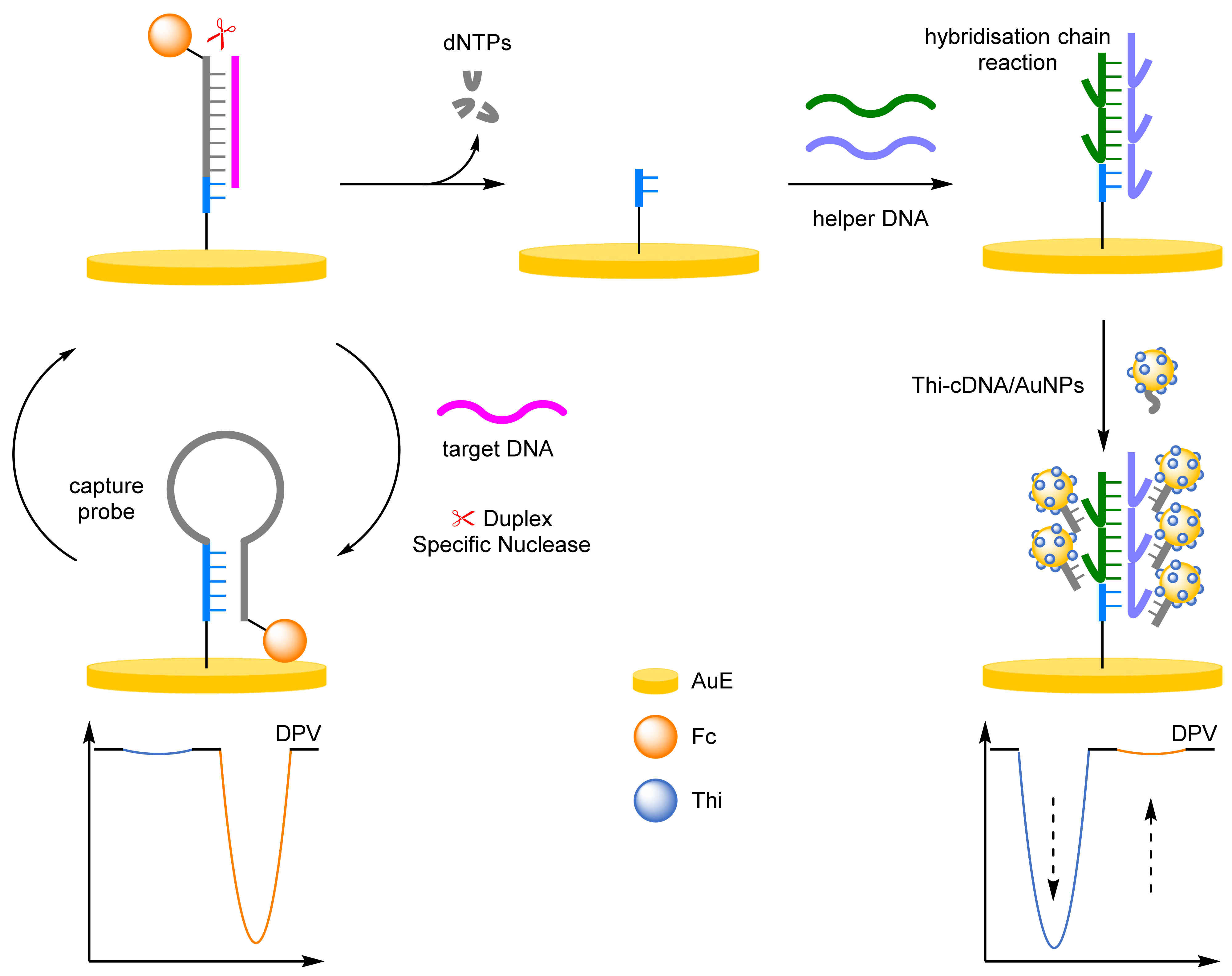
- Hybridisation Chain Reaction
2.1.4. Summary
2.2. Modified Electrodes
2.2.1. Direct Labelling of Electrodes
- (a)
- Nanoparticles
- (b)
- Metal Organic Frameworks
- (c)
- Graphene Oxide
- (d)
- Carbon Nanotubes
- (e)
- Biocompatible Electrodes
2.2.2. Selectivity Strategies
DNA
Antibodies
Aptamer
Recognition Sites
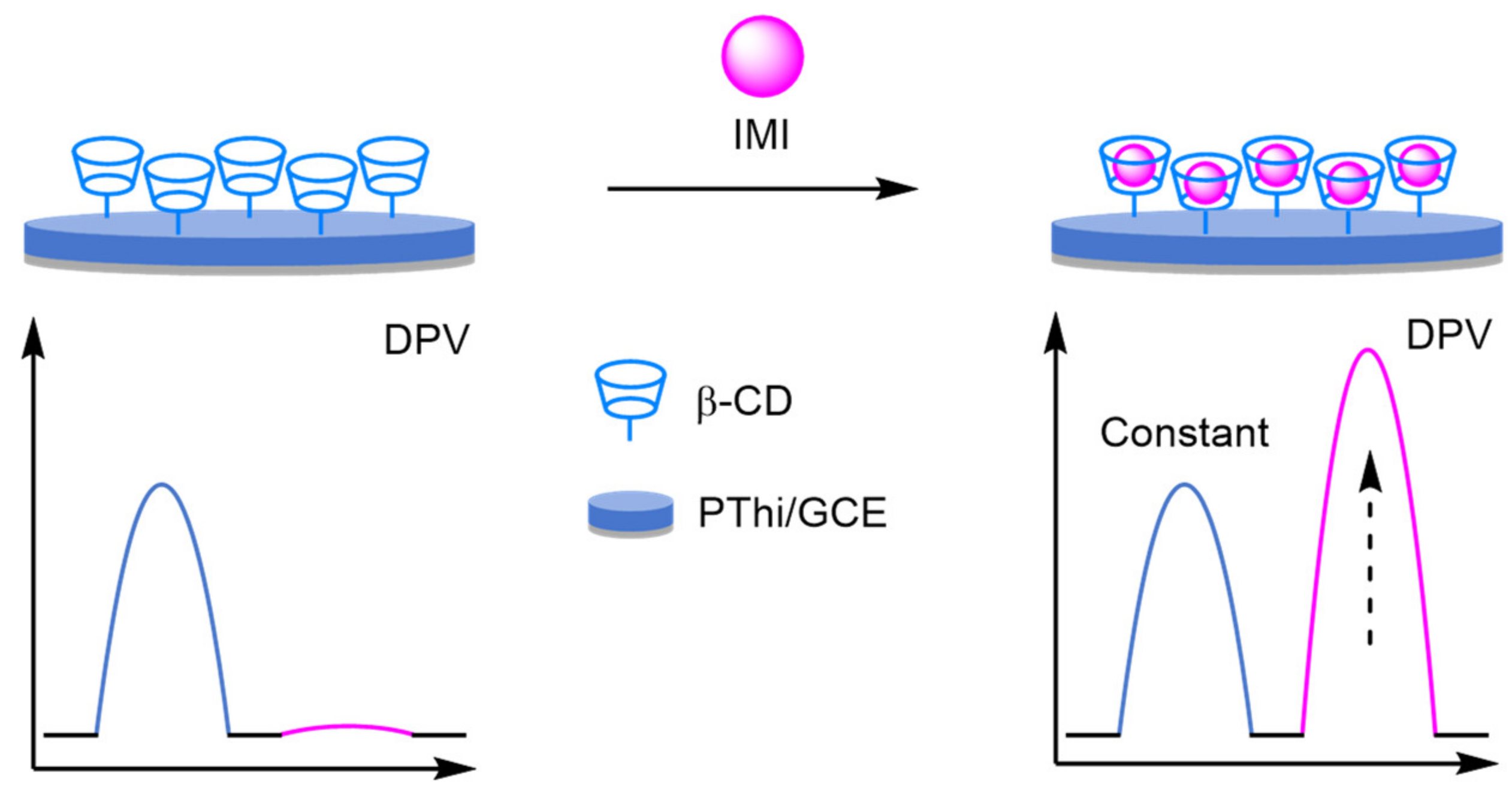
Molecular Imprinted Polymers
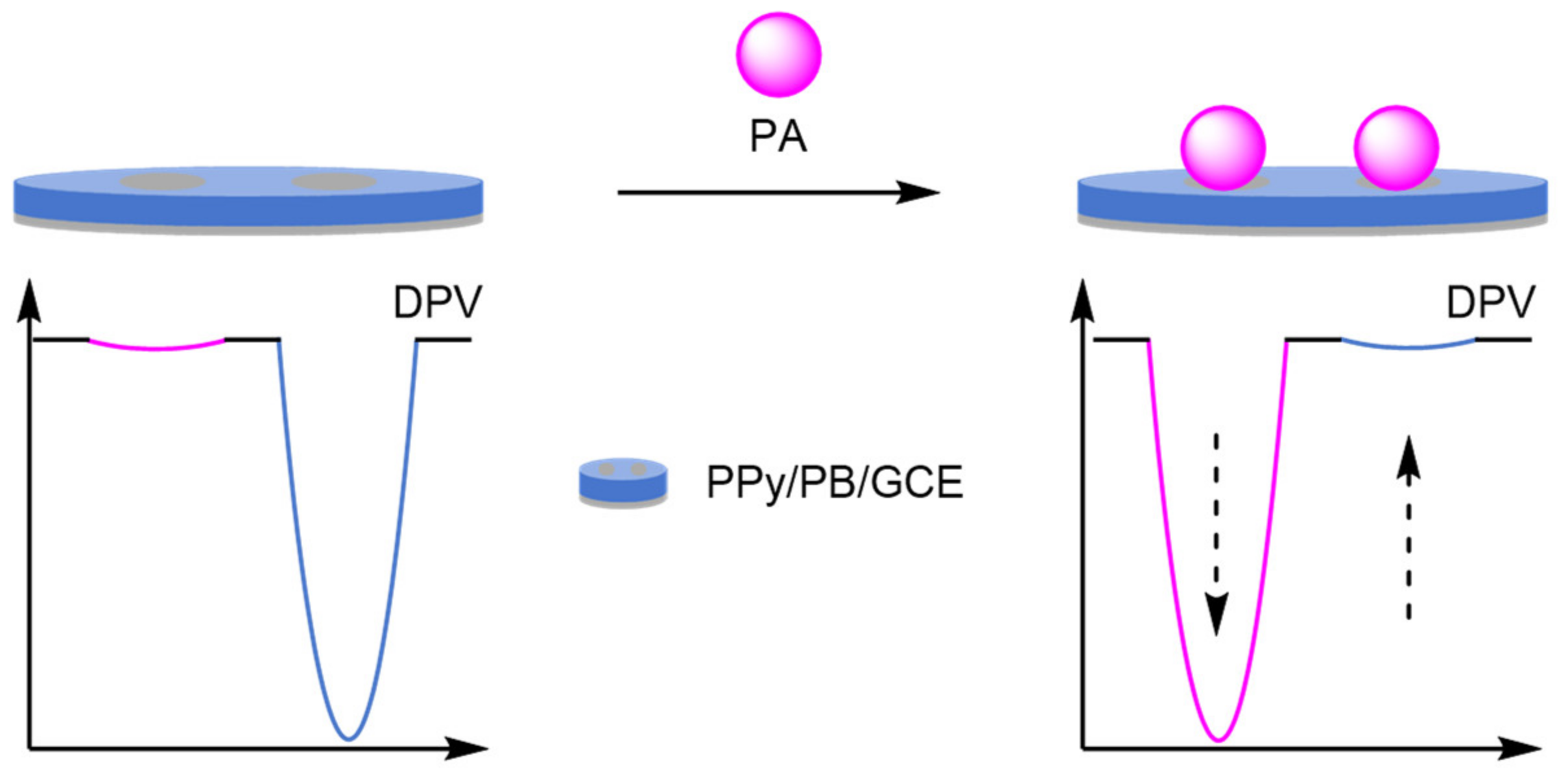
2.2.3. Host–Guest Displacement Assays
2.2.4. Oxidation Catalysts
Enzymes
Ketjen Black
2.2.5. Summary
2.3. Unmodified Electrodes
3. Chemodosimeters
3.1. Improving Sensitivity
3.2. Summary
4. Dual Channel
4.1. One Reference Electrode
4.2. Two Working Electrodes
4.3. Summary
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Marsh, B.J.; Hampton, L.; Goggins, S.; Frost, C.G. Fine-tuning of ferrocene redox potentials towards multiplex DNA detection. New J. Chem. 2014, 38, 5260–5263. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Y.; Matharu, Z.; Rahimian, A.; Revzin, A. Detecting multiple cell-secreted cytokines from the same aptamer-functionalized electrode. Biosens. Bioelectron. 2015, 64, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Simonova, A.; Magriñá, I.; Sýkorová, V.; Pohl, R.; Ortiz, M.; Havran, L.; Fojta, M.; O’Sullivan, C.K.; Hocek, M. Tuning of Oxidation Potential of Ferrocene for Ratiometric Redox Labeling and Coding of Nucleotides and DNA. Chem. A Eur. J. 2020, 26, 1286–1291. [Google Scholar] [CrossRef]
- Wang, J. Electrochemical biosensors: Towards point-of-care cancer diagnostics. Biosens. Bioelectron. 2006, 21, 1887–1892. [Google Scholar] [CrossRef]
- Wei, B.; Zhang, J.; Wang, H.; Xia, F. A new electrochemical aptasensor based on a dual-signaling strategy and supersandwich assay. Analyst 2016, 141, 4313–4318. [Google Scholar] [CrossRef]
- Cao, X.; Xia, J.; Liu, H.; Zhang, F.; Wang, Z.; Lu, L. A new dual-signalling electrochemical aptasensor with the integration of “signal on/off” and “labeling/label-free” strategies. Sens. Actuators B Chem. 2017, 239, 166–171. [Google Scholar] [CrossRef]
- Jin, H.; Gui, R.; Yu, J.; Lv, W.; Wang, Z. Fabrication strategies, sensing modes and analytical applications of ratiometric electrochemical biosensors. Biosens. Bioelectron. 2017, 91, 523–537. [Google Scholar] [CrossRef] [PubMed]
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763. [Google Scholar] [CrossRef]
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical Biosensors—Sensor Principles and Architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, X.; Liu, W.; Xiong, E.; Chen, J. Sensitive electrochemical aptasensor by coupling “signal-on” and “signal-off” strategies. Anal. Chem. 2013, 85, 8397–8402. [Google Scholar] [CrossRef]
- Lu, J.Y.; Zhang, X.X.; Huang, W.T.; Zhu, Q.Y.; Ding, X.Z.; Xia, L.Q.; Luo, H.Q.; Li, N.B. Boolean Logic Tree of Label-Free Dual-Signal Electrochemical Aptasensor System for Biosensing, Three-State Logic Computation, and Keypad Lock Security Operation. Anal. Chem. 2017, 89, 9734–9741. [Google Scholar] [CrossRef]
- Du, Y.; Lim, B.J.; Li, B.; Jiang, Y.S.; Sessler, J.L.; Ellington, A.D. Reagentless, Ratiometric Electrochemical DNA Sensors with Improved Robustness and Reproducibility. Anal. Chem. 2014, 86, 8010–8016. [Google Scholar] [CrossRef]
- Fan, C.; Plaxco, K.W.; Heeger, A.J. Electrochemical interrogation of conformational changes as a reagentless method for the sequence-specific detection of DNA. Proc. Natl. Acad. Sci. USA 2003, 100, 9134–9137. [Google Scholar] [CrossRef] [PubMed]
- North, J.R. Immunosensors: Antibody-based biosensors. Trends Biotechnol. 1985, 3, 180–186. [Google Scholar] [CrossRef]
- Kerman, K.; Saito, M.; Tamiya, E.; Yamamura, S.; Takamura, Y. Nanomaterial-based electrochemical biosensors for medical applications. TrAC Trends Anal. Chem. 2008, 27, 585–592. [Google Scholar] [CrossRef]
- Cheng, A.K.H.; Sen, D.; Yu, H.Z. Design and testing of aptamer-based electrochemical biosensors for proteins and small molecules. Bioelectrochemistry 2009, 77, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Liu, C.; Cao, F.; Ren, J.; Qu, X. DNA metallization: Principles, methods, structures, and applications. Chem. Soc. Rev. 2018, 47, 4017–4072. [Google Scholar] [CrossRef]
- Xiong, E.; Wu, L.; Zhou, J.; Yu, P.; Zhang, X.; Chen, J. A ratiometric electrochemical biosensor for sensitive detection of Hg2+ based on thymine-Hg2+-thymine structure. Anal. Chim. Acta 2015, 853, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Chen, H.G.; Feng, J.; Lei, J.L.; Luo, H.Q.; Li, N.B. A regenerative ratiometric electrochemical biosensor for selective detecting Hg2+ based on Y-shaped/hairpin DNA transformation. Anal. Chim. Acta 2016, 908, 95–101. [Google Scholar] [CrossRef]
- Ren, K.; Wu, J.; Yan, F.; Ju, H. Ratiometric electrochemical proximity assay for sensitive one-step protein detection. Sci. Rep. 2014, 4, 4360. [Google Scholar] [CrossRef]
- Ren, K.; Wu, J.; Yan, F.; Zhang, Y.; Ju, H. Immunoreaction-triggered DNA assembly for one-step sensitive ratiometric electrochemical biosensing of protein biomarker. Biosens. Bioelectron. 2015, 66, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Hermann, T.; Patel, D.J. Adaptive recognition by nucleic acid aptamers. Science 2000, 287, 820–825. [Google Scholar] [CrossRef] [PubMed]
- Bunka, D.H.J.; Stockley, P.G. Aptamers come of age—At last. Nat. Rev. Microbiol. 2006, 4, 588–596. [Google Scholar] [CrossRef]
- Song, S.; Wang, L.; Li, J.; Zhao, J.; Fan, C. Aptamer-based biosensors. Trends Anal. Chem. 2008, 27, 108–117. [Google Scholar] [CrossRef]
- Deng, C.; Pi, X.; Qian, P.; Chen, X.; Wu, W.; Xiang, J. High-Performance Ratiometric Electrochemical Method Based on the Combination of Signal Probe and Inner Reference Probe in One Hairpin-Structured DNA. Anal. Chem. 2017, 89, 966–973. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Lu, M.; Li, Y.; Tang, B.; Zhang, C. yang A reusable ratiometric electrochemical biosensor on the basis of the binding of methylene blue to DNA with alternating AT base sequence for sensitive detection of adenosine. Biosens. Bioelectron. 2018, 102, 87–93. [Google Scholar] [CrossRef]
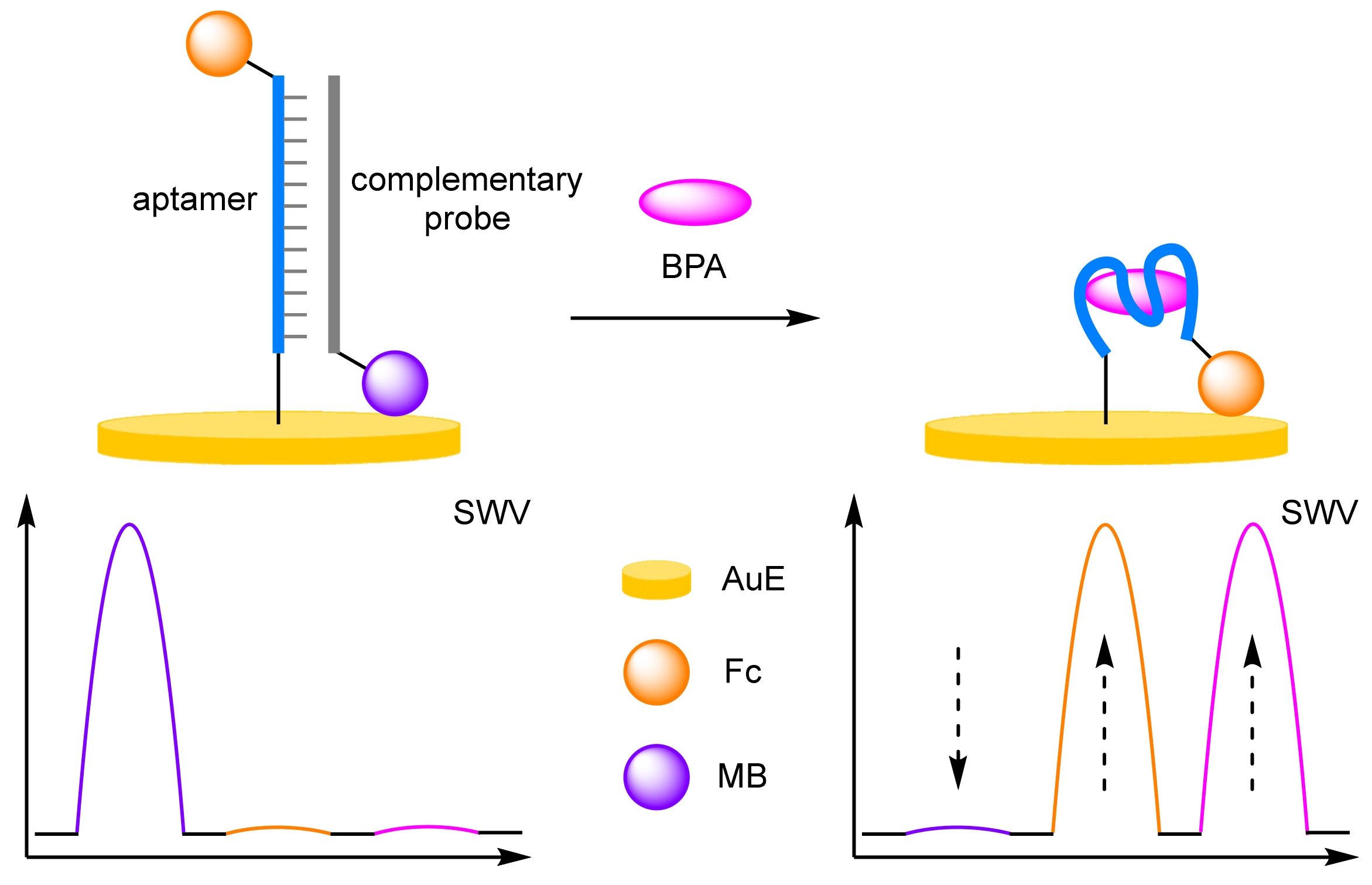
- Yu, P.; Liu, Y.; Zhang, X.; Zhou, J.; Xiong, E.; Li, X.; Chen, J. A novel electrochemical aptasensor for bisphenol A assay based on triple-signaling strategy. Biosens. Bioelectron. 2016, 79, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, X.; Yang, J.; Xiong, E.; Zhang, X.; Chen, J. Sensitive detection of bisphenol A based on a ratiometric electrochemical aptasensor. Can. J. Chem. 2016, 94, 509–514. [Google Scholar] [CrossRef]
- Li, H.; Arroyo-Currás, N.; Kang, D.; Ricci, F.; Plaxco, K.W. Dual-Reporter Drift Correction To Enhance the Performance of Electrochemical Aptamer-Based Sensors in Whole Blood. J. Am. Chem. Soc. 2016, 138, 15809–15812. [Google Scholar] [CrossRef] [PubMed]
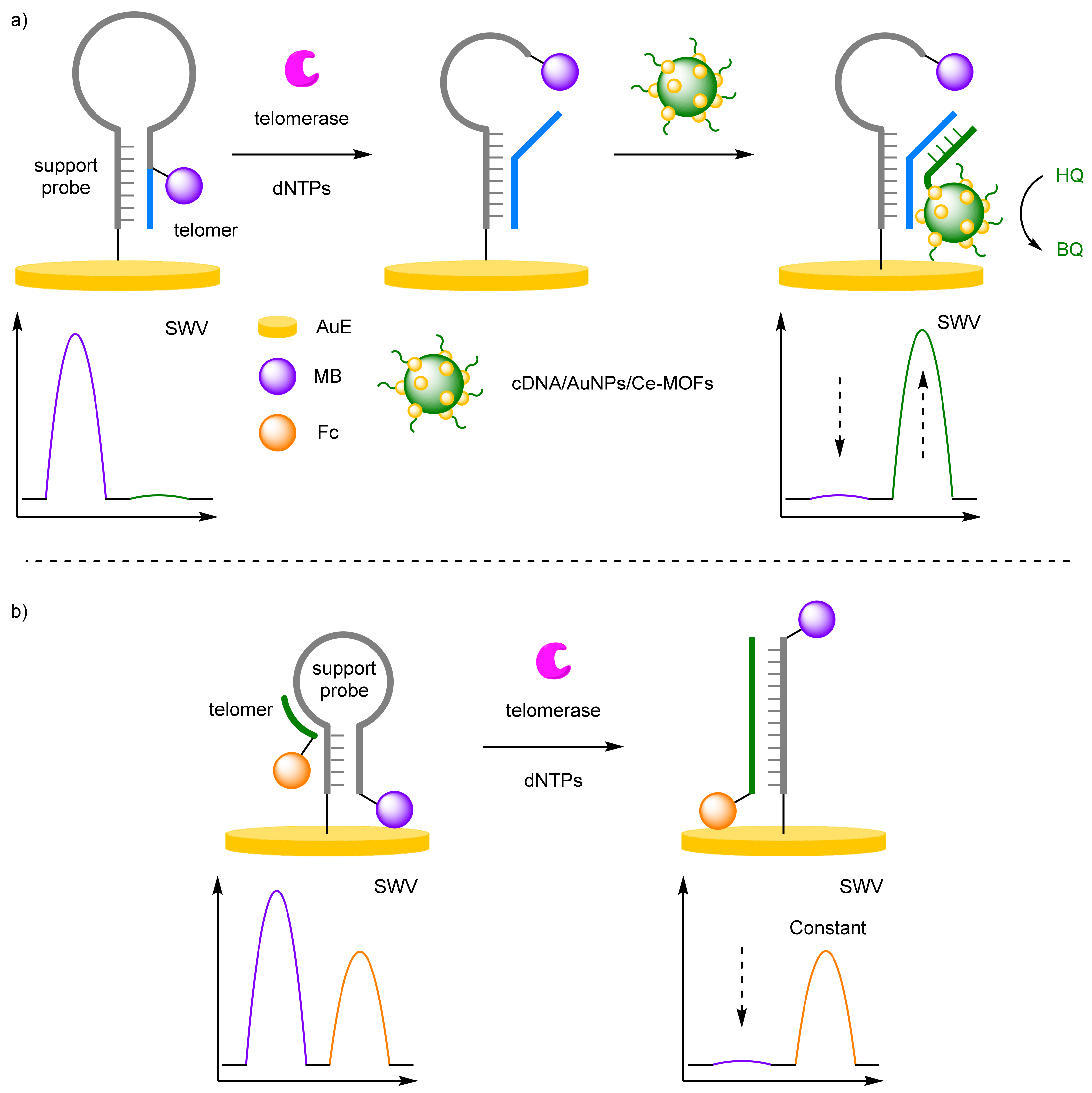
- Dong, P.; Zhu, L.; Huang, J.; Ren, J.; Lei, J. Electrocatalysis of cerium metal-organic frameworks for ratiometric electrochemical detection of telomerase activity. Biosens. Bioelectron. 2019, 138, 111313. [Google Scholar] [CrossRef]
- Meng, F.; Chen, X.; Cheng, W.; Hu, W.; Tang, Y.; Miao, P. Ratiometric Electrochemical Sensing Strategy for the Ultrasensitive Detection of Telomerase Activity. ChemElectroChem 2019, 6, 2000–2003. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Chen, S.X.; Yin, P. Optimizing the specificity of nucleic acid hybridization. Nat. Chem. 2012, 4, 208–214. [Google Scholar] [CrossRef]
- Wang, T.; Zhou, L.; Bai, S.; Zhang, Z.; Li, J.; Jing, X.; Xie, G. Ultraspecific electrochemical DNA biosensor by coupling spontaneous cascade DNA branch migration and dual-signaling sensing strategy. Biosens. Bioelectron. 2016, 78, 464–470. [Google Scholar] [CrossRef]
- Singh, S.K.; Nielsen, P.; Koshkin, A.A.; Wengel, J. LNA (locked nucleic acids): Synthesis and high-affinity nucleic acid recognition. Chem. Commun. 1998, 455–456. [Google Scholar] [CrossRef]
- Wang, L.; Yang, C.J.; Medley, C.D.; Benner, S.A.; Tan, W. Locked nucleic acid molecular beacons. J. Am. Chem. Soc. 2005, 127, 15664–15665. [Google Scholar] [CrossRef] [PubMed]
- Briones, C.; Moreno, M. Applications of peptide nucleic acids (PNAs) and locked nucleic acids (LNAs) in biosensor development. Anal. Bioanal. Chem. 2012, 402, 3071–3089. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Wang, L.; Zeng, L.; Wang, Y.; Weng, Y.; Liao, Y.; Chen, T.; Xia, Y.; Zhang, J.; Chen, J. A ratiometric electrochemical DNA biosensor for detection of exosomal MicroRNA. Talanta 2020, 207, 120298. [Google Scholar] [CrossRef]
- Bagheri, E.; Abnous, K.; Alibolandi, M.; Ramezani, M.; Taghdisi, S.M. Triple-helix molecular switch-based aptasensors and DNA sensors. Biosens. Bioelectron. 2018, 111, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Xiong, E.; Li, Z.; Zhang, X.; Zhou, J.; Yan, X.; Liu, Y.; Chen, J. Triple-Helix Molecular Switch Electrochemical Ratiometric Biosensor for Ultrasensitive Detection of Nucleic Acids. Anal. Chem. 2017, 89, 8830–8835. [Google Scholar] [CrossRef]
- Dou, B.; Li, J.; Jiang, B.; Yuan, R.; Xiang, Y. Electrochemical screening of single nucleotide polymorphisms with significantly enhanced discrimination factor by an amplified ratiometric sensor. Anal. Chim. Acta 2018, 1038, 166–172. [Google Scholar] [CrossRef]
- Xiao, Q.; Feng, J.; Feng, M.; Li, J.; Liu, Y.; Wang, D.; Huang, S. A ratiometric electrochemical aptasensor for ultrasensitive determination of adenosine triphosphate via a triple-helix molecular switch. Microchim. Acta 2019, 186, 478. [Google Scholar] [CrossRef]
- Eis, P.S.; Millar, D.P. Conformational Distributions of a Four-Way DNA Junction Revealed by Time-Resolved Fluorescence Resonance Energy Transfer. Biochemistry 1993, 32, 13852–13860. [Google Scholar] [CrossRef] [PubMed]
- McKinney, S.A.; Tan, E.; Wilson, T.J.; Nahas, M.K.; Déclais, A.C.; Clegg, R.M.; Lilley, D.M.J.; Ha, T. Single-molecule studies of DNA and RNA four-way junctions. In Biochemical Society Transactions; Portland Press: London, UK, 2004; Volume 32, pp. 41–45. [Google Scholar]
- Nowakowski, J.; Shim, P.J.; Stout, C.D.; Joyce, G.F. Alternative conformations of a nucleic acid four-way junction. J. Mol. Biol. 2000, 300, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Lu, M.; Yang, X.Y.; Tang, B.; Zhang, C.Y. A sensitive ratiometric electrochemical biosensor based on DNA four-way junction formation and enzyme-assisted recycling amplification. Analyst 2017, 142, 1562–1568. [Google Scholar] [CrossRef] [PubMed]
- Scrimin, P.; Prins, L.J. Sensing through signal amplification. Chem. Soc. Rev. 2011, 40, 4488–4505. [Google Scholar] [CrossRef]
- Goggins, S.; Frost, C.G. Approaches towards molecular amplification for sensing. Analyst 2016, 141, 3157–3218. [Google Scholar] [CrossRef]
- Wang, J. Nanomaterial-based electrochemical biosensors. Analyst 2005, 130, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Pingarrón, J.M.; Yáñez-Sedeño, P.; González-Cortés, A. Gold nanoparticle-based electrochemical biosensors. Electrochim. Acta 2008, 53, 5848–5866. [Google Scholar] [CrossRef]
- Cao, X.; Ye, Y.; Liu, S. Gold nanoparticle-based signal amplification for biosensing. Anal. Biochem. 2011, 417, 1–16. [Google Scholar] [CrossRef]
- Lei, J.; Ju, H. Signal amplification using functional nanomaterials for biosensing. Chem. Soc. Rev. 2012, 41, 2122–2134. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Zhou, J.; Wu, L.; Xiong, E.; Zhang, X.; Chen, J. A ratiometric electrochemical aptasensor for sensitive detection of protein based on aptamer-target-aptamer sandwich structure. J. Electroanal. Chem. 2014, 732, 61–65. [Google Scholar] [CrossRef]
- Wang, L.; Ma, R.; Jiang, L.; Jia, L.; Jia, W.; Wang, H. A novel “signal-on/off” sensing platform for selective detection of thrombin based on target-induced ratiometric electrochemical biosensing and bio-bar-coded nanoprobe amplification strategy. Biosens. Bioelectron. 2017, 92, 390–395. [Google Scholar] [CrossRef]
- Gao, F.; Qian, Y.; Zhang, L.; Dai, S.; Lan, Y.; Zhang, Y.; Du, L.; Tang, D. Target catalyzed hairpin assembly for constructing a ratiometric electrochemical aptasensor. Biosens. Bioelectron. 2015, 71, 158–163. [Google Scholar] [CrossRef]
- Xiong, E.; Zhang, X.; Liu, Y.; Zhou, J.; Yu, P.; Li, X.; Chen, J. Ultrasensitive Electrochemical Detection of Nucleic Acids Based on the Dual-Signaling Electrochemical Ratiometric Method and Exonuclease III-Assisted Target Recycling Amplification Strategy. Anal. Chem. 2015, 87, 7291–7296. [Google Scholar] [CrossRef]
- Ma, R.N.; Wang, L.L.; Wang, H.S.F.; Jia, L.P.; Zhang, W.; Shang, L.; Xue, Q.W.; Jia, W.L.; Liu, Q.Y.; Wang, H.S.F. Highly sensitive ratiometric electrochemical DNA biosensor based on homogeneous exonuclease III-assisted target recycling amplification and one-step triggered dual-signal output. Sens. Actuators B Chem. 2018, 269, 173–179. [Google Scholar] [CrossRef]
- Gao, F.; Du, L.; Zhang, Y.; Tang, D.; Du, Y. Molecular beacon mediated circular strand displacement strategy for constructing a ratiometric electrochemical deoxyribonucleic acid sensor. Anal. Chim. Acta 2015, 883, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Xie, P.; Yang, Z.H.; Yuan, R.; Zhang, K. Highly sensitive electrochemical nuclear factor kappa B aptasensor based on target-induced dual-signal ratiometric and polymerase-assisted protein recycling amplification strategy. Biosens. Bioelectron. 2018, 102, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Lizardi, P.M.; Huang, X.; Zhu, Z.; Bray-Ward, P.; Thomas, D.C.; Ward, D.C. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 1998, 19, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Feng, M.; Li, J.; Liu, Y.; Xiao, Q. Voltammetric determination of attomolar levels of a sequence derived from the genom of hepatitis B virus by using molecular beacon mediated circular strand displacement and rolling circle amplification. Microchim. Acta 2018, 185, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Feng, J.; Li, J.; Liu, Y.; Wang, D.; Huang, S. A ratiometric electrochemical biosensor for ultrasensitive and highly selective detection of the K-ras gene via exonuclease III-assisted target recycling and rolling circle amplification strategies. Anal. Methods 2019, 11, 4146–4156. [Google Scholar] [CrossRef]
- Shen, W.J.; Zhuo, Y.; Chai, Y.Q.; Yuan, R. Cu-Based Metal-Organic Frameworks as a Catalyst to Construct a Ratiometric Electrochemical Aptasensor for Sensitive Lipopolysaccharide Detection. Anal. Chem. 2015, 87, 11345–11352. [Google Scholar] [CrossRef]
- Yurke, B.; Turberfield, A.J.; Mills, A.P.; Simmel, F.C.; Neumann, J.L. A DNA-fuelled molecular machine made of DNA. Nature 2000, 406, 605–608. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Tao, Y.; Tang, L.; Li, W.; Zhang, Z.; Li, J.; Xie, G. Cascade toehold-mediated strand displacement along with non-enzymatic target recycling amplification for the electrochemical determination of the HIV-1 related gene. Microchim. Acta 2017, 184, 3721–3728. [Google Scholar] [CrossRef]
- Jiang, Y.S.; Bhadra, S.; Li, B.; Ellington, A.D. Mismatches Improve the Performance of Strand-Displacement Nucleic Acid Circuits. Angew. Chem. 2014, 53, 1845–1848. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Li, B.; Milligan, J.N.; Bhadra, S.; Ellington, A.D. Real-time detection of isothermal amplification reactions with thermostable catalytic hairpin assembly. J. Am. Chem. Soc. 2013, 135, 7430–7433. [Google Scholar] [CrossRef]
- Li, X.; Dou, B.; Yuan, R.; Xiang, Y. Mismatched catalytic hairpin assembly and ratiometric strategy for highly sensitive electrochemical detection of microRNA from tumor cells. Sens. Actuators B Chem. 2019, 286, 191–197. [Google Scholar] [CrossRef]
- Cha, T.G.; Pan, J.; Chen, H.; Robinson, H.N.; Li, X.; Mao, C.; Choi, J.H. Design Principles of DNA Enzyme-Based Walkers: Translocation Kinetics and Photoregulation. J. Am. Chem. Soc. 2015, 137, 9429–9437. [Google Scholar] [CrossRef] [PubMed]
- You, M.; Chen, Y.; Zhang, X.; Liu, H.; Wang, R.; Wang, K.; Williams, K.R.; Tan, W. An Autonomous and Controllable Light-Driven DNA Walking Device. Angew. Chem. 2012, 51, 2457–2460. [Google Scholar] [CrossRef]
- Jung, C.; Allen, P.B.; Ellington, A.D. A stochastic DNA walker that traverses a microparticle surface. Nat. Nanotechnol. 2016, 11, 157–163. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, L.L.; Hou, M.F.; Xia, Y.K.; He, W.H.; Yan, A.; Weng, Y.P.; Zeng, L.P.; Chen, J.H. A ratiometric electrochemical biosensor for the exosomal microRNAs detection based on bipedal DNA walkers propelled by locked nucleic acid modified toehold mediate strand displacement reaction. Biosens. Bioelectron. 2018, 102, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Pu, Q.; Li, J.; Qiu, J.; Yang, X.; Li, Y.; Yin, D.; Zhang, X.; Tao, Y.; Sheng, S.; Xie, G. Universal ratiometric electrochemical biosensing platform based on mesoporous platinum nanocomposite and nicking endonuclease assisted DNA walking strategy. Biosens. Bioelectron. 2017, 94, 719–727. [Google Scholar] [CrossRef]
- Zhu, C.; Liu, M.; Li, X.; Zhang, X.; Chen, J. A new electrochemical aptasensor for sensitive assay of a protein based on the dual-signaling electrochemical ratiometric method and DNA walker strategy. Chem. Commun. 2018, 54, 10359–10362. [Google Scholar] [CrossRef]
- Zhao, L.; Sun, R.; He, P.; Zhang, X. Ultrasensitive Detection of Exosomes by Target-Triggered Three-Dimensional DNA Walking Machine and Exonuclease III-Assisted Electrochemical Ratiometric Biosensing. Anal. Chem. 2019, 91, 14773–14779. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, F.; Liang, Q.; Wang, Z. A three-dimensional graphene-based ratiometric signal amplification aptasensor for MUC1 detection. Biosens. Bioelectron. 2018, 120, 85–92. [Google Scholar] [CrossRef]
- Zhang, F.T.; Nie, J.; Zhang, D.W.; Chen, J.T.; Zhou, Y.L.; Zhang, X.X. Methylene blue as a G-quadruplex binding probe for label-free homogeneous electrochemical biosensing. Anal. Chem. 2014, 86, 9489–9495. [Google Scholar] [CrossRef]
- Zhu, C.; Liu, D.; Li, Y.; Shen, X.; Ma, S.; Liu, Y.; You, T. Ratiometric electrochemical aptasensor for ultrasensitive detection of Ochratoxin A based on a dual signal amplification strategy: Engineering the binding of methylene blue to DNA. Biosens. Bioelectron. 2020, 150, 111814. [Google Scholar] [CrossRef]
- Dirks, R.M.; Pierce, N.A. Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. USA 2004, 101, 15275–15278. [Google Scholar] [CrossRef]
- Evanko, D. Hybridization chain reaction. Nat. Methods 2004, 1, 186–187. [Google Scholar] [CrossRef]
- Huang, J.; Wu, Y.; Chen, Y.; Zhu, Z.; Yang, X.; Yang, C.J.; Wang, K.; Tan, W. Pyrene-Excimer Probes Based on the Hybridization Chain Reaction for the Detection of Nucleic Acids in Complex Biological Fluids. Angew. Chem. 2011, 50, 401–404. [Google Scholar] [CrossRef]
- Yuan, Y.H.; Chi, B.Z.; Wen, S.H.; Liang, R.P.; Li, Z.M.; Qiu, J.D. Ratiometric electrochemical assay for sensitive detecting microRNA based on dual-amplification mechanism of duplex-specific nuclease and hybridization chain reaction. Biosens. Bioelectron. 2018, 102, 211–216. [Google Scholar] [CrossRef]
- Yang, D.; Mei, Q.; Tang, Y.; Miao, P. A ratiometric electrochemical assay for human 8-oxoguanine DNA glycosylase amplified by hybridization chain reaction. Electrochem. Commun. 2019, 103, 37–41. [Google Scholar] [CrossRef]
- Berti, L.; Alessandrini, A.; Facci, P. DNA-templated photoinduced silver deposition. J. Am. Chem. Soc. 2005, 127, 11216–11217. [Google Scholar] [CrossRef]
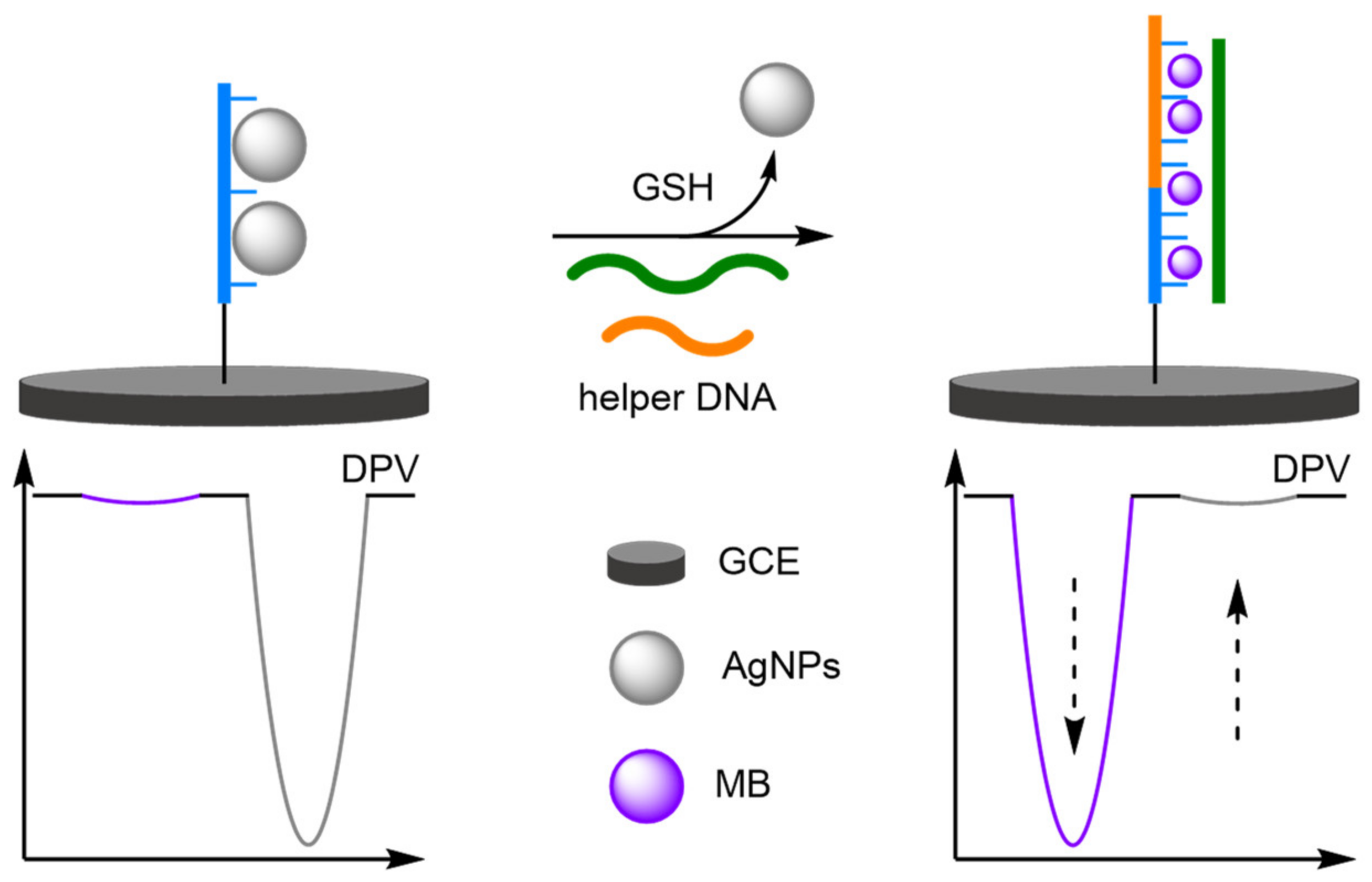
- Liu, X.; Yan, Z.; Sun, Y.; Ren, J.; Qu, X. A label-free ratiometric electrochemical DNA sensor for monitoring intracellular redox homeostasis. Chem. Commun. 2017, 53, 6215–6218. [Google Scholar] [CrossRef]
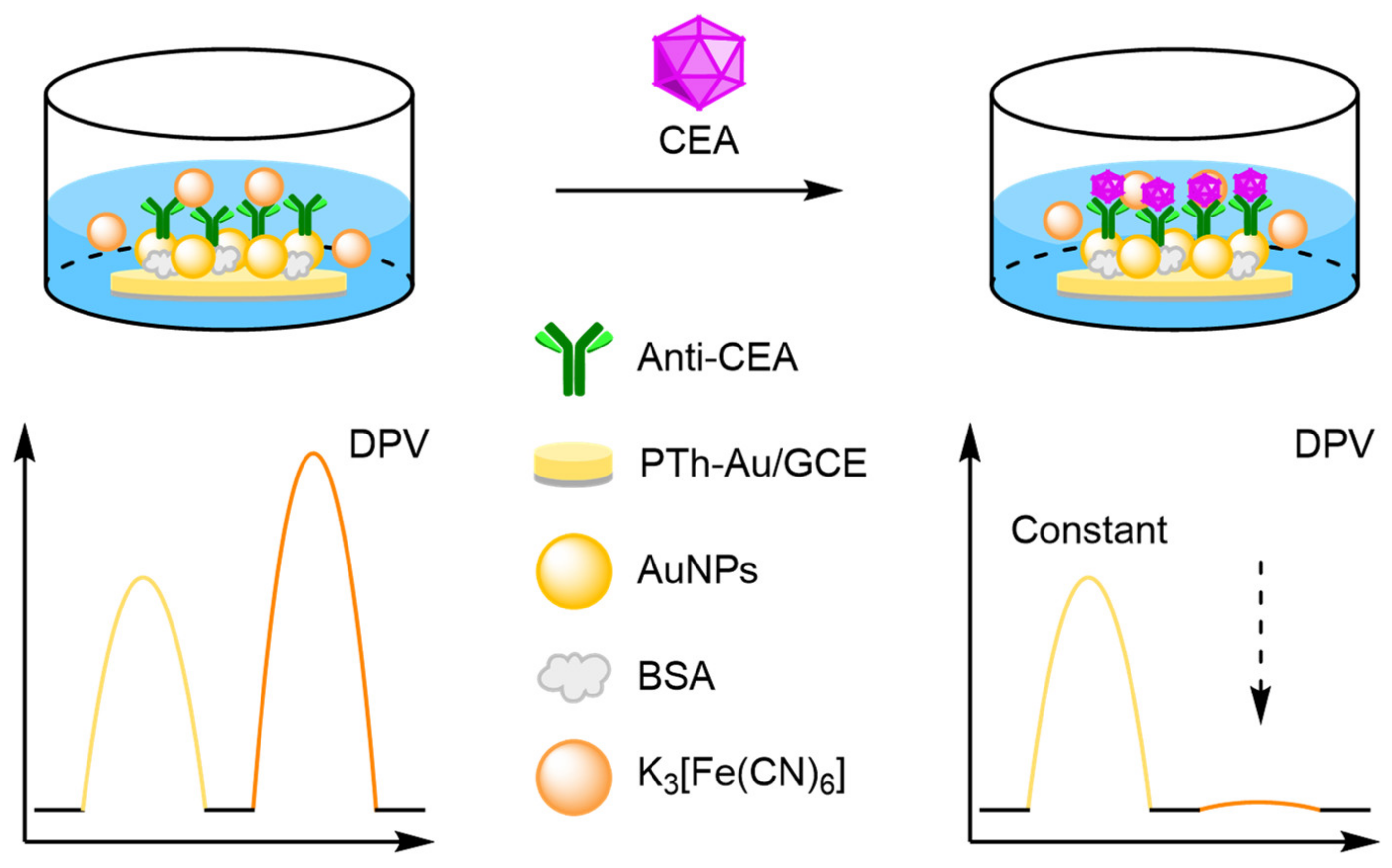
- Cai, X.; Weng, S.; Guo, R.; Lin, L.; Chen, W.; Zheng, Z.; Huang, Z.; Lin, X. Ratiometric electrochemical immunoassay based on internal reference value for reproducible and sensitive detection of tumor marker. Biosens. Bioelectron. 2016, 81, 173–180. [Google Scholar] [CrossRef]
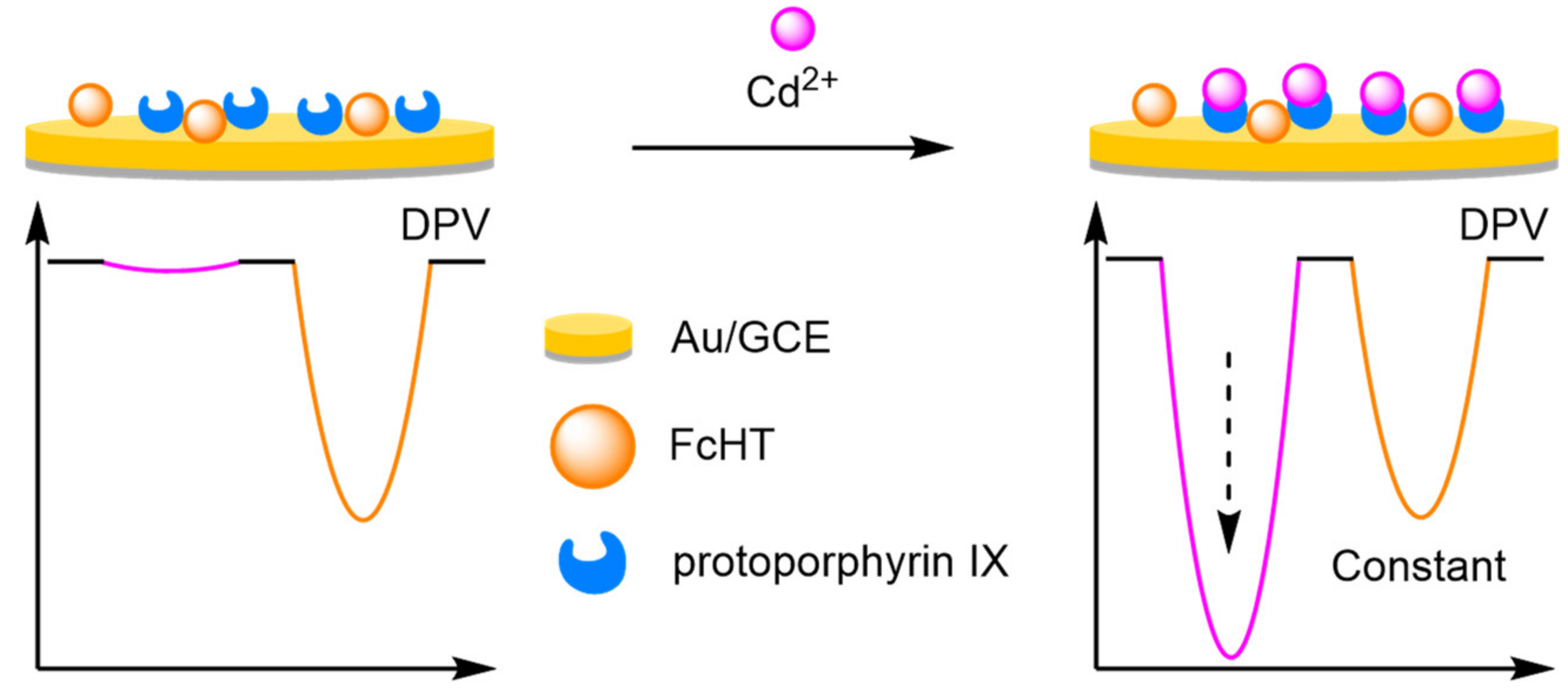
- Chai, X.; Zhang, L.; Tian, Y. Ratiometric electrochemical sensor for selective monitoring of cadmium ions using biomolecular recognition. Anal. Chem. 2014, 86, 10668–10673. [Google Scholar] [CrossRef]
- Zhang, L.; Han, Y.; Zhao, F.; Shi, G.; Tian, Y. A selective and accurate ratiometric electrochemical biosensor for monitoring of Cu2+ ions in a rat brain. Anal. Chem. 2015, 87, 2931–2936. [Google Scholar] [CrossRef]
- Luo, X.; Morrin, A.; Killard, A.J.; Smyth, M.R. Application of Nanoparticles in Electrochemical Sensors and Biosensors. Electroanalysis 2006, 18, 319–326. [Google Scholar] [CrossRef]
- Wang, F.; Hu, S. Electrochemical sensors based on metal and semiconductor nanoparticles. Microchim. Acta 2009, 165, 1–22. [Google Scholar] [CrossRef]
- Rassaei, L.; Marken, F.; Sillanpää, M.; Amiri, M.; Cirtiu, C.M.; Sillanpää, M. Nanoparticles in electrochemical sensors for environmental monitoring. TrAC—Trends Anal. Chem. 2011, 30, 1704–1715. [Google Scholar] [CrossRef]
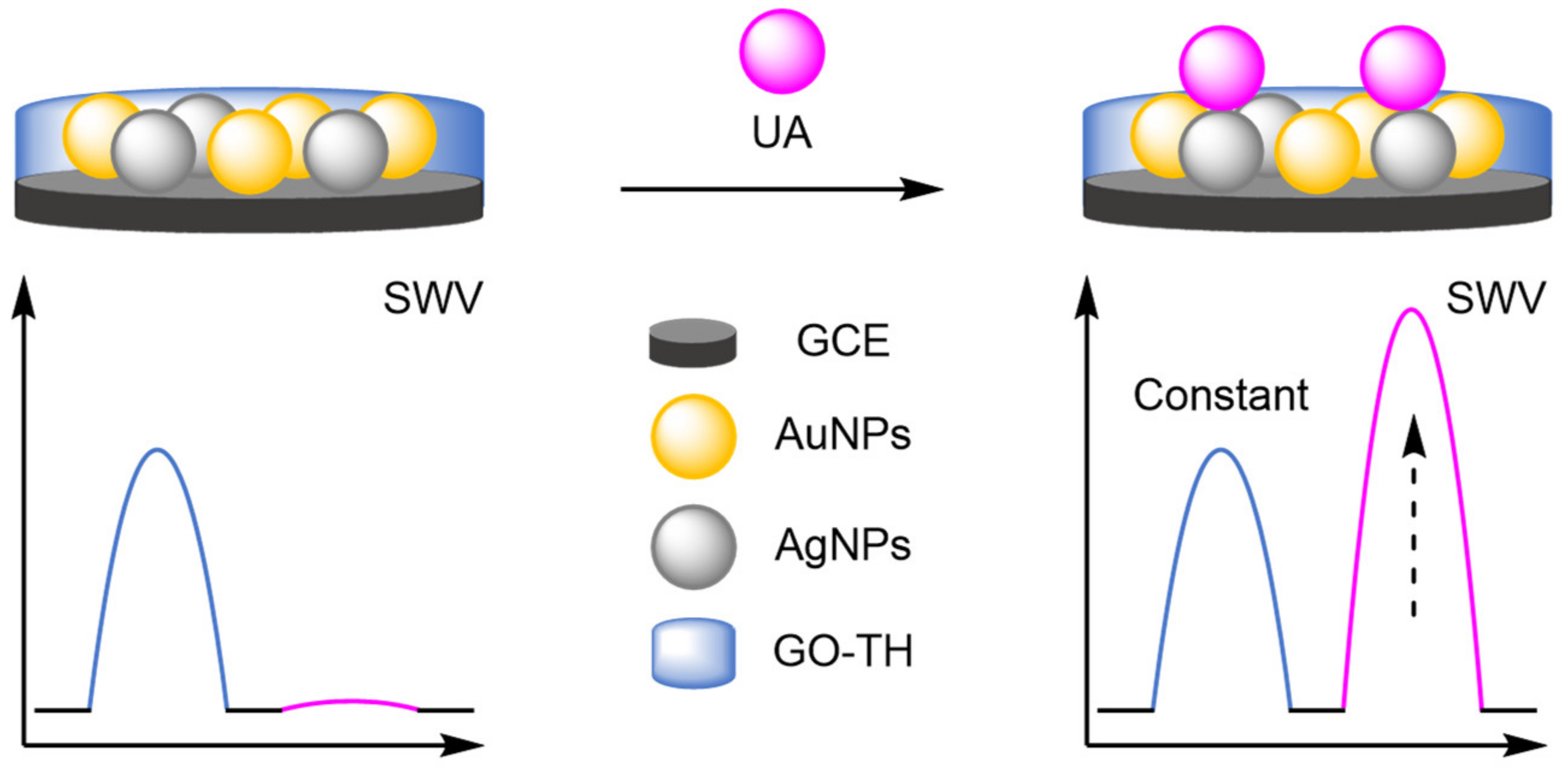
- Gao, X.; Gui, R.; Xu, K.Q.; Guo, H.; Jin, H.; Wang, Z. A bimetallic nanoparticle/graphene oxide/thionine composite-modified glassy carbon electrode used as a facile ratiometric electrochemical sensor for sensitive uric acid determination. New J. Chem. 2018, 42, 14796–14804. [Google Scholar] [CrossRef]
- Gao, X.; Gui, R.; Guo, H.; Wang, Z.; Liu, Q. Creatinine-induced specific signal responses and enzymeless ratiometric electrochemical detection based on copper nanoparticles electrodeposited on reduced graphene oxide-based hybrids. Sens. Actuators B Chem. 2019, 285, 201–208. [Google Scholar] [CrossRef]
- Ren, H.; Wang, J.; Feng, H.; Li, Y.; Ye, B.C. A versatile ratiometric electrochemical sensing platform based on N-Mo2C for detection of m-nitrophenol. Biosens. Bioelectron. 2019, 144, 111663. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Wang, Y.; Zhuang, Q.; Wang, L.; Ni, Y. Developing an electrochemical sensor for the detection of tert-butylhydroquinone. Sens. Actuators B Chem. 2019, 293, 321–328. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Zhuang, Q. A ratiometric electrochemical sensor for dopamine detection based on hierarchical manganese dioxide nanoflower/multiwalled carbon nanotube nanocomposite modified glassy carbon electrode. J. Alloy. Compd. 2019, 802, 326–334. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, J.; Li, Y. MXene with Great Adsorption Ability toward Organic Dye: An Excellent Material for Constructing a Ratiometric Electrochemical Sensing Platform. ACS Sens. 2019, 4, 2058–2064. [Google Scholar] [CrossRef]
- Wang, X.; Liu, G.; Qi, Y.; Yuan, Y.; Gao, J.; Luo, X.; Yang, T. Embedded Au Nanoparticles-Based Ratiometric Electrochemical Sensing Strategy for Sensitive and Reliable Detection of Copper Ions. Anal. Chem. 2019, 91, 12006–12013. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, D.; Li, Y. Ratiometric Electrochemical Sensors Associated with Self-Cleaning Electrodes for Simultaneous Detection of Adrenaline, Serotonin, and Tryptophan. ACS Appl. Mater. Interfaces 2019, 11, 13557–13563. [Google Scholar] [CrossRef] [PubMed]
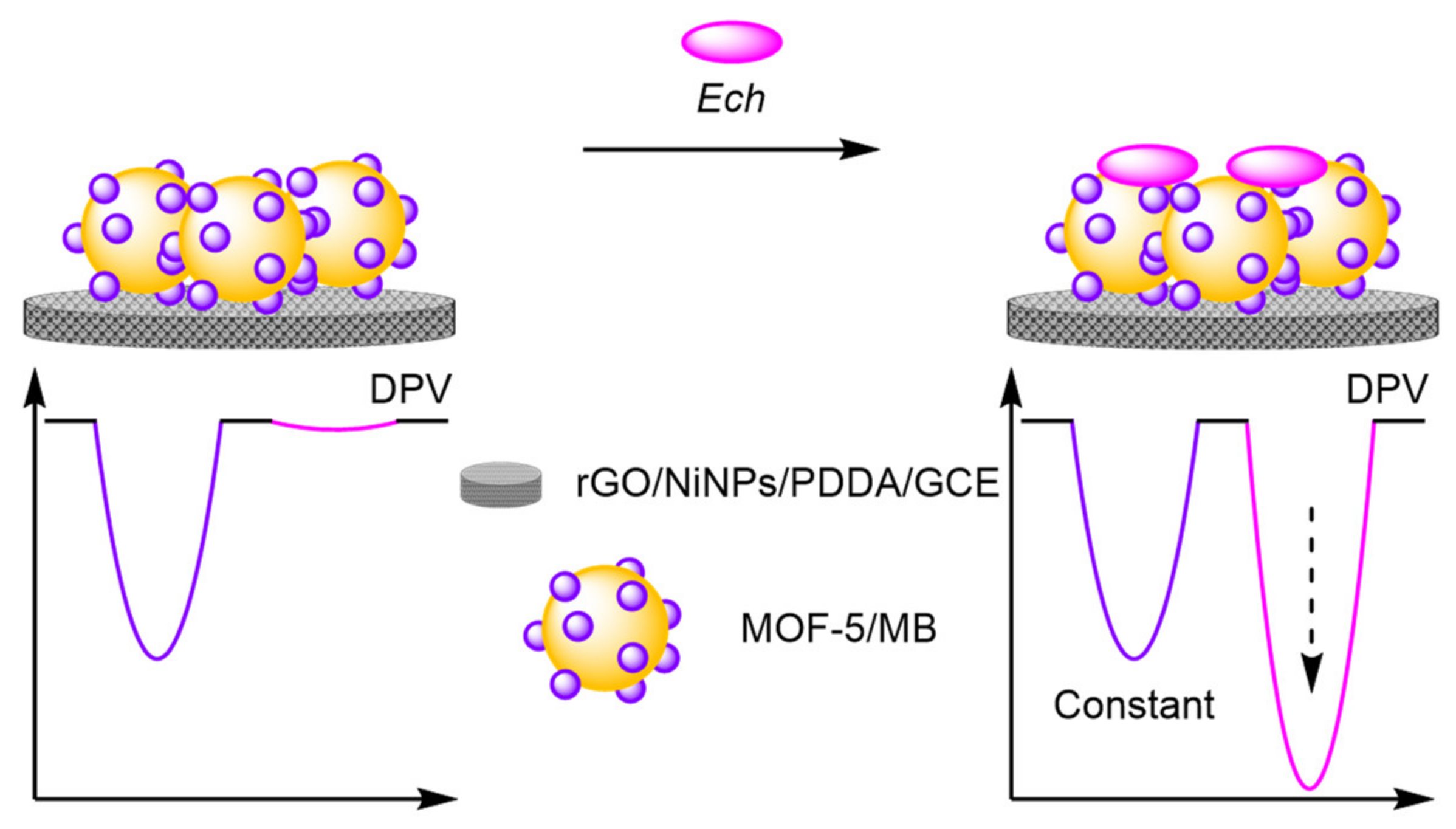
- Li, S.; Duan, Y.; Lei, S.; Qiao, J.; Li, G.; Ye, B. A new electrochemical sensing strategy for echinacoside based on an original nanocomposite. Sens. Actuators B Chem. 2018, 274, 218–227. [Google Scholar] [CrossRef]
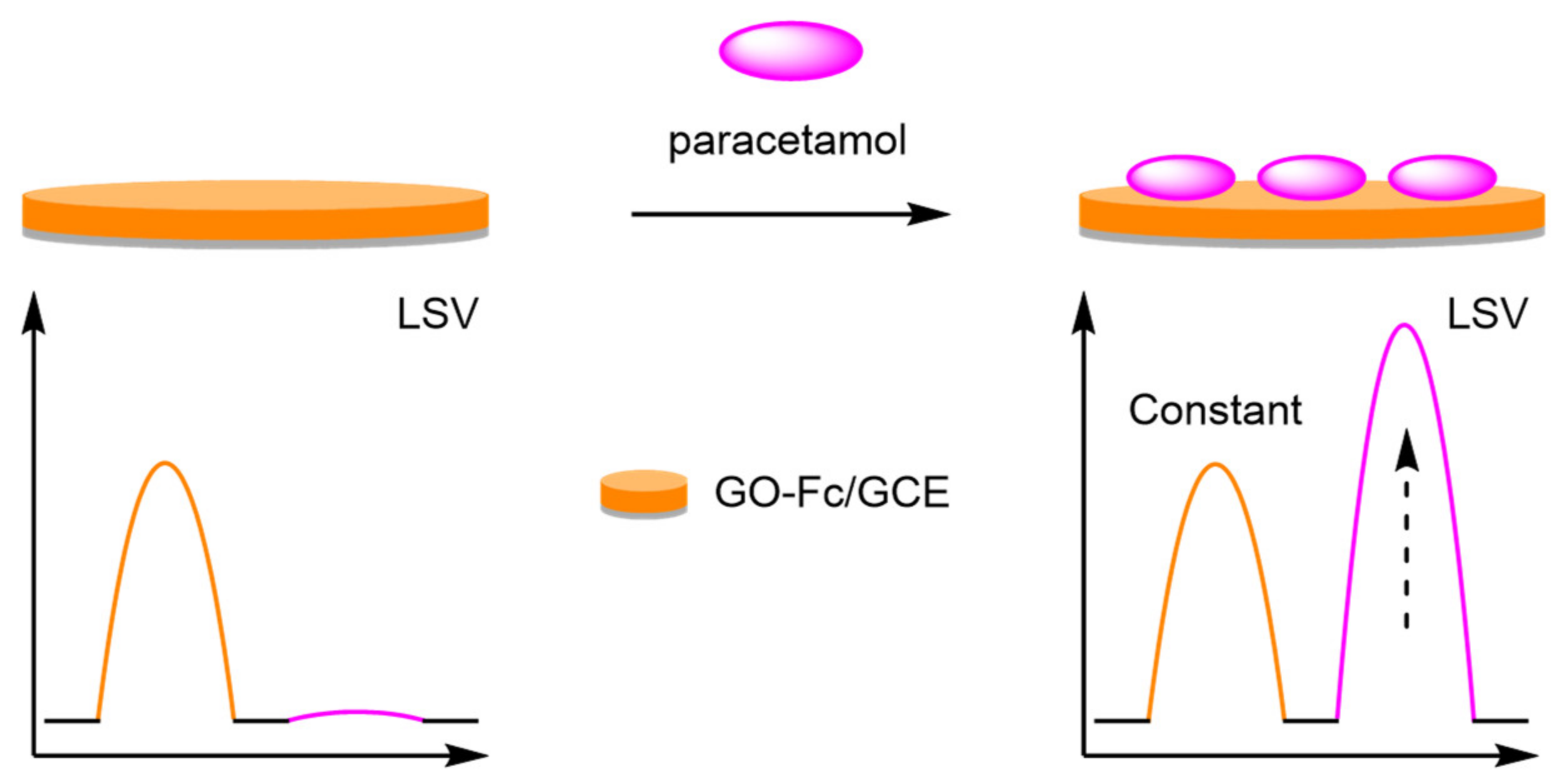
- Guo, H.; Wang, Z.; Yang, W.; Li, J.; Jiang, D. A facile ratiometric electrochemical sensor for sensitive 4-acetamidophenol determination based on ferrocene–graphene oxide–Nafion modified electrode. Anal. Methods 2020, 12, 1353–1359. [Google Scholar] [CrossRef]
- Jiang, Y.; Xiao, X.; Li, C.; Luo, Y.; Chen, S.; Shi, G.; Han, K.; Gu, H. A Facile Ratiometric Electrochemical Sensor for In Vivo/On-line Repetitive Measurements of Cerebral Ascorbic Acid in Brain Microdiaysate. Anal. Chem. 2020, 92, 3981–3989. [Google Scholar] [CrossRef]
- Jacobs, C.B.; Peairs, M.J.; Venton, B.J. Review: Carbon nanotube based electrochemical sensors for biomolecules. Anal. Chim. Acta 2010, 662, 105–127. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Denno, M.E.; Pyakurel, P.; Venton, B.J. Recent trends in carbon nanomaterial-based electrochemical sensors for biomolecules: A review. Anal. Chim. Acta 2015, 887, 17–37. [Google Scholar] [CrossRef]
- Yu, Y.; Yu, C.; Yin, T.; Ou, S.; Sun, X.; Wen, X.; Zhang, L.; Tang, D.; Yin, X. Functionalized poly (ionic liquid) as the support to construct a ratiometric electrochemical biosensor for the selective determination of copper ions in AD rats. Biosens. Bioelectron. 2017, 87, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, P.; Zhu, X.; Peng, Q.; Zhou, Y.; Yin, T.; Liang, Y.; Yin, X. Combined determination of copper ions and β-amyloid peptide by a single ratiometric electrochemical biosensor. Analyst 2018, 143, 323–331. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Zhuang, Q. Simple self-referenced ratiometric electrochemical sensor for dopamine detection using electrochemically pretreated glassy carbon electrode modified by acid-treated multiwalled carbon nanotube. J. Electroanal. Chem. 2019, 851, 113446. [Google Scholar] [CrossRef]
- Pang, S.; Kan, X. Reliable detection of O-nitrophenol and p-nitrophenol based on carbon nanotubes covalently functionalized with ferrocene as an inner reference. New J. Chem. 2019, 43, 10517–10522. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, X.; Wang, C.; Hou, P.; Dong, H.; Luo, B.; Li, A. A multifunctional ratiometric electrochemical sensor for combined determination of indole-3-acetic acid and salicylic acid. RSC Adv. 2020, 10, 3115–3121. [Google Scholar] [CrossRef]
- Yu, X.-H.; Wang, Z.-X.; Gao, Y.-F.; Kong, F.-Y.; Lv, W.-X.; Ma, H.-F.; Wang, W. Novel Ratiometric Electrochemical Sensor for Sensitive Detection of Ag + Ion Using High Nitrogen Doped Carbon Nanosheets. Int. J. Electrochem. Sci 2018, 13, 2875–2886. [Google Scholar] [CrossRef]
- Yu, L.; Cui, X.; Li, H.; Lu, J.; Kang, Q.; Shen, D. A ratiometric electrochemical sensor for multiplex detection of cancer biomarkers using bismuth as an internal reference and metal sulfide nanoparticles as signal tags. Analyst 2019, 144, 4073–4080. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, L.; Liu, W.; Yu, Y.; Tian, Y. A Single Biosensor for Evaluating the Levels of Copper Ion and L -Cysteine in a Live Rat Brain with Alzheimer’s Disease. Angew. Chem. 2015, 54, 14053–14056. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, F.; Liu, W.; Zhu, T.; Zhang, J.Z.H.; Chen, C.; Dai, Z.; Peng, H.; Huang, J.L.; Hu, Q.; et al. An Electrochemical Biosensor with Dual Signal Outputs: Toward Simultaneous Quantification of pH and O2 in the Brain upon Ischemia and in a Tumor during Cancer Starvation Therapy. Angew. Chem. 2017, 56, 10471–10475. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Zhao, W.; Zhang, L.; Tian, Y. A ratiometric electrochemical strategy for sensitive determination of Furin activity based on dual signal amplification and antifouling nanosurfaces. Analyst 2017, 142, 4215–4220. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Ji, H.; Sun, H.; Ding, C.; Ren, J.; Qu, X. Label-free ratiometric electrochemical detection of the mutated apolipoprotein e gene associated with Alzheimer’s disease. Chem. Commun. 2016, 52, 12080–12083. [Google Scholar] [CrossRef]
- Gai, P.; Gu, C.; Li, H.; Sun, X.; Li, F. Ultrasensitive Ratiometric Homogeneous Electrochemical MicroRNA Biosensing via Target-Triggered Ru(III) Release and Redox Recycling. Anal. Chem. 2017, 89, 12293–12298. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Wang, W.; Li, F. Electro-Grafted Electrode with Graphene-Oxide-Like DNA Affinity for Ratiometric Homogeneous Electrochemical Biosensing of MicroRNA. Anal. Chem. 2017, 89, 11560–11567. [Google Scholar] [CrossRef]
- Tang, Z.; Ma, Z. Ratiometric ultrasensitive electrochemical immunosensor based on redox substrate and immunoprobe. Sci. Rep. 2016, 6, 35440. [Google Scholar] [CrossRef]
- Wei, Y.; Ma, H.; Ren, X.; Ding, C.; Wang, H.; Sun, X.; Du, B.; Zhang, Y.; Wei, Q. A dual-signaling electrochemical ratiometric method for sensitive detection of carcinoembryonic antigen based on Au-Cu2S-CuS/graphene and Au-CeO2 supported toluidine blue complex. Sens. Actuators B Chem. 2018, 256, 504–511. [Google Scholar] [CrossRef]
- Li, M.; Jiao, L.; Liu, S.; Zhang, L.; Li, H. A new ratiometric electrochemical immunoassay for reliable detection of nuclear matrix protein 22. Anal. Chim. Acta 2019, 1086, 103–109. [Google Scholar] [CrossRef]
- Jin, H.; Zhao, C.; Gui, R.; Gao, X.; Wang, Z. Reduced graphene oxide/nile blue/gold nanoparticles complex-modified glassy carbon electrode used as a sensitive and label-free aptasensor for ratiometric electrochemical sensing of dopamine. Anal. Chim. Acta 2018, 1025, 154–162. [Google Scholar] [CrossRef]
- Liu, X.; Deng, K.; Wang, H.; Li, C.; Zhang, S.; Huang, H. Aptamer based ratiometric electrochemical sensing of 17β-estradiol using an electrode modified with gold nanoparticles, thionine, and multiwalled carbon nanotubes. Microchim. Acta 2019, 186, 347. [Google Scholar] [CrossRef]
- Li, X.; Kan, X. A ratiometric strategy -based electrochemical sensing interface for the sensitive and reliable detection of imidacloprid. Analyst 2018, 143, 2150–2156. [Google Scholar] [CrossRef]
- Dai, Y.; Li, X.; Lu, X.; Kan, X. Voltammetric determination of paracetamol using a glassy carbon electrode modified with Prussian Blue and a molecularly imprinted polymer, and ratiometric read-out of two signals. Microchim. Acta 2016, 183, 2771–2778. [Google Scholar] [CrossRef]
- Dai, Y.; Li, X.; Fan, L.; Lu, X.; Kan, X. “Sign-on/off” sensing interface design and fabrication for propyl gallate recognition and sensitive detection. Biosens. Bioelectron. 2016, 86, 741–747. [Google Scholar] [CrossRef]
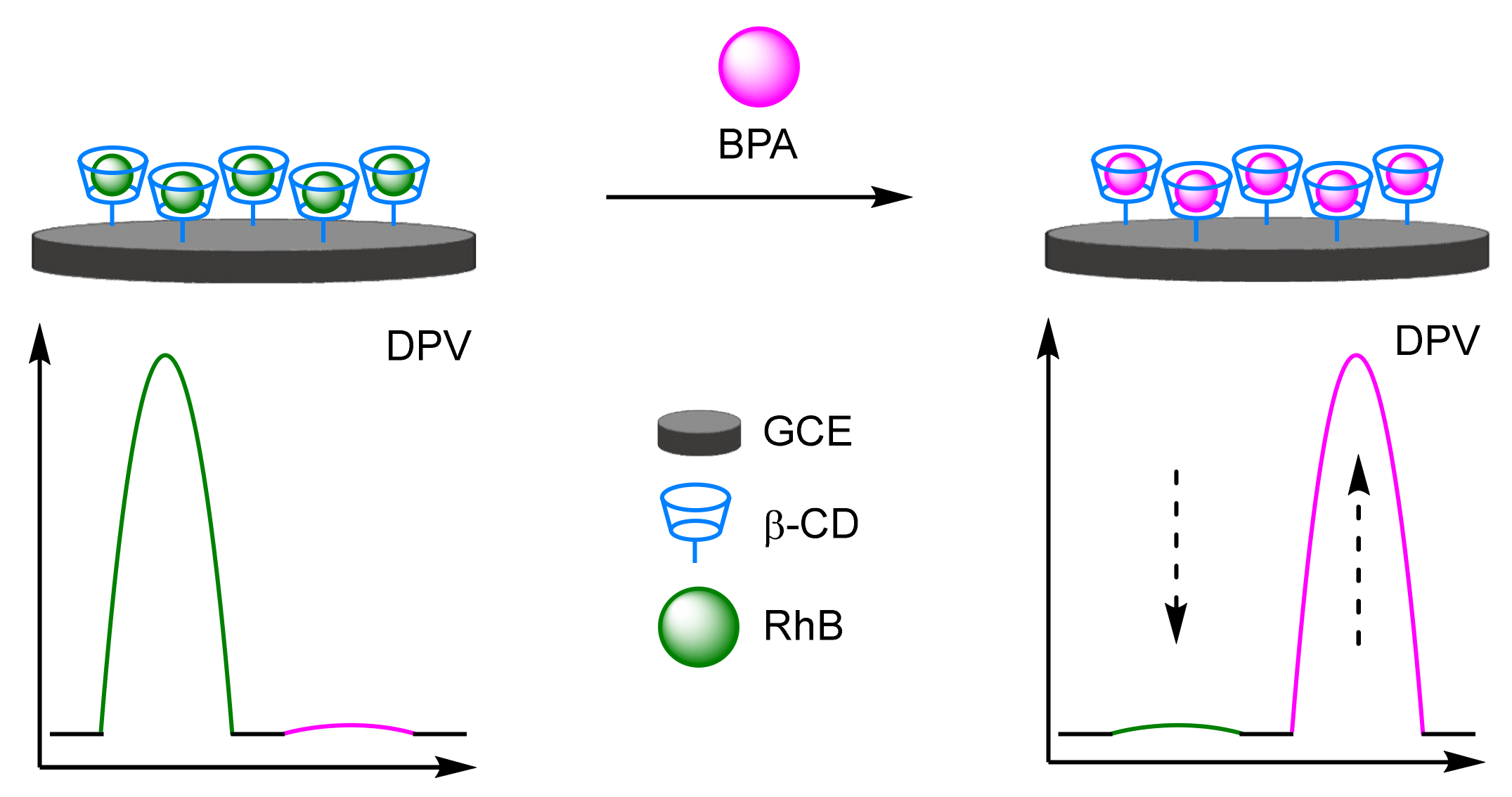
- Zhang, X.; Wu, L.; Zhou, J.; Zhang, X.; Chen, J. A new ratiometric electrochemical sensor for sensitive detection of bisphenol A based on poly-β-cyclodextrin/electroreduced graphene modified glassy carbon electrode. J. Electroanal. Chem. 2015, 742, 97–103. [Google Scholar] [CrossRef]
- Yu, P.; Zhang, X.; Zhou, J.; Xiong, E.; Li, X.; Chen, J. Smart protein biogate as a mediator to regulate competitive host-guest interaction for sensitive ratiometric electrochemical assay of prion. Sci. Rep. 2015, 5, 16015. [Google Scholar] [CrossRef]
- Wang, Y.; Ning, G.; Bi, H.; Wu, Y.; Liu, G.; Zhao, Y. A novel ratiometric electrochemical assay for ochratoxin A coupling Au nanoparticles decorated MoS2 nanosheets with aptamer. Electrochim. Acta 2018, 285, 120–127. [Google Scholar] [CrossRef]
- Ma, X.; Chen, D.; Tu, X.; Gao, F.; Xie, Y.; Dai, R.; Lu, L.; Wang, X.; Qu, F.; Yu, Y.; et al. Ratiometric electrochemical sensor for sensitive detection of sunset yellow based on three-dimensional polyethyleneimine functionalized reduced graphene oxide aerogels@Au nanoparticles/SH-β-cyclodextrin. Nanotechnology 2019, 30, 475503. [Google Scholar] [CrossRef]
- Hao, Q.; Lu, L.; Kan, X. Probe and analogue: Double roles of thionine for aloe-emodin selective and sensitive ratiometric detection. Sens. Actuators B Chem. 2019, 292, 247–253. [Google Scholar] [CrossRef]
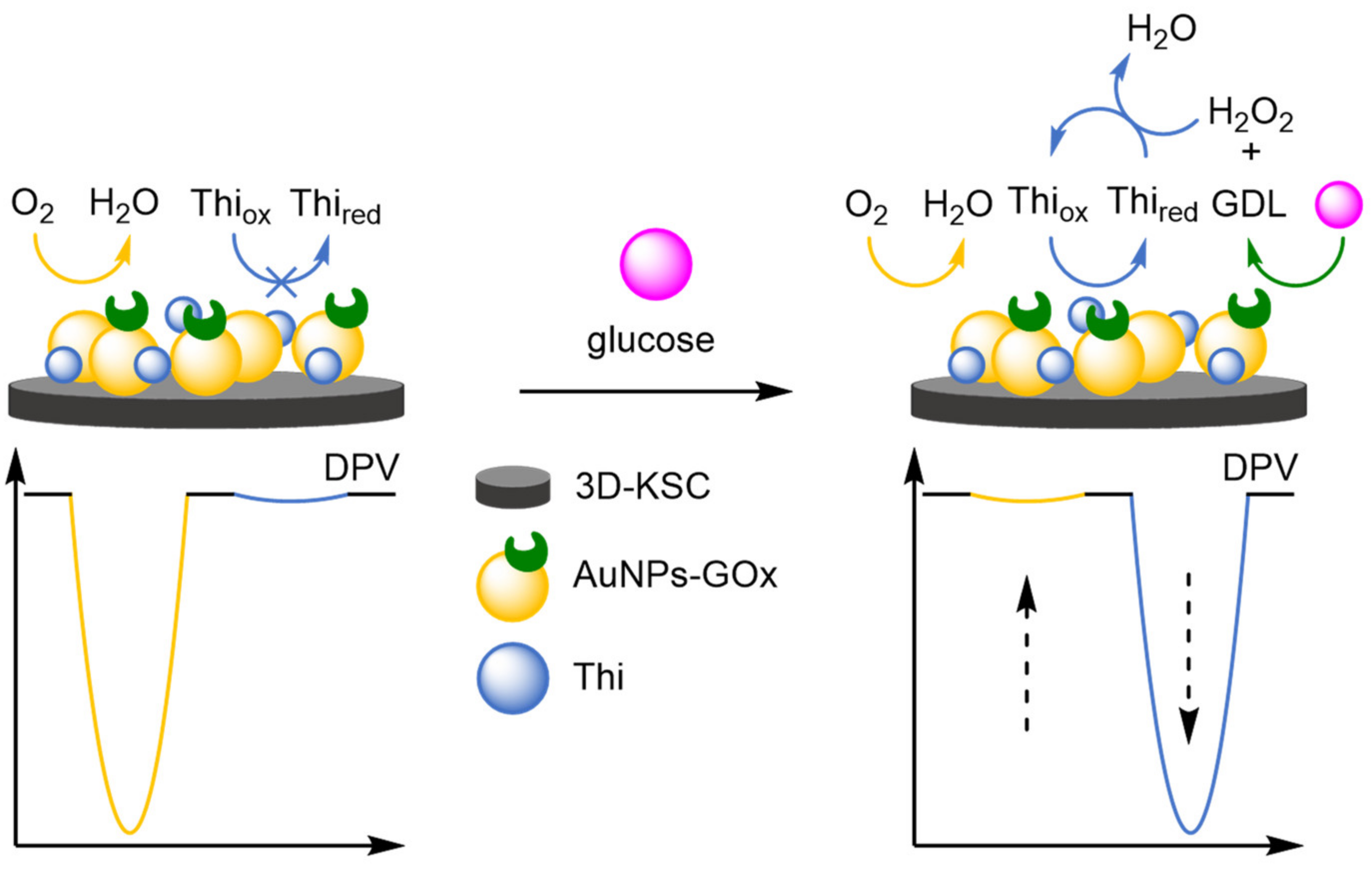
- Gong, C.; Shen, Y.; Song, Y.; Wang, L. On-Off Ratiometric Electrochemical Biosensor for Accurate Detection of Glucose. Electrochim. Acta 2017, 235, 488–494. [Google Scholar] [CrossRef]
- Song, Y.; Xu, M.; Gong, C.; Shen, Y.; Wang, L.; Xie, Y.; Wang, L. Ratiometric electrochemical glucose biosensor based on GOD/AuNPs/Cu-BTC MOFs/macroporous carbon integrated electrode. Sens. Actuators B Chem. 2018, 257, 792–799. [Google Scholar] [CrossRef]
- Wang, L.; Xu, M.; Xie, Y.; Qian, C.; Ma, W.; Wang, L.; Song, Y. Ratiometric electrochemical glucose sensor based on electroactive Schiff base polymers. Sens. Actuators B Chem. 2019, 285, 264–270. [Google Scholar] [CrossRef]
- Wang, L.; Liang, H.; Xu, M.; Wang, L.; Xie, Y.; Song, Y. Ratiometric electrochemical biosensing based on double-enzymes loaded on two-dimensional dual-pore COFETTA-TPAL. Sens. Actuators B Chem. 2019, 298, 126859. [Google Scholar] [CrossRef]
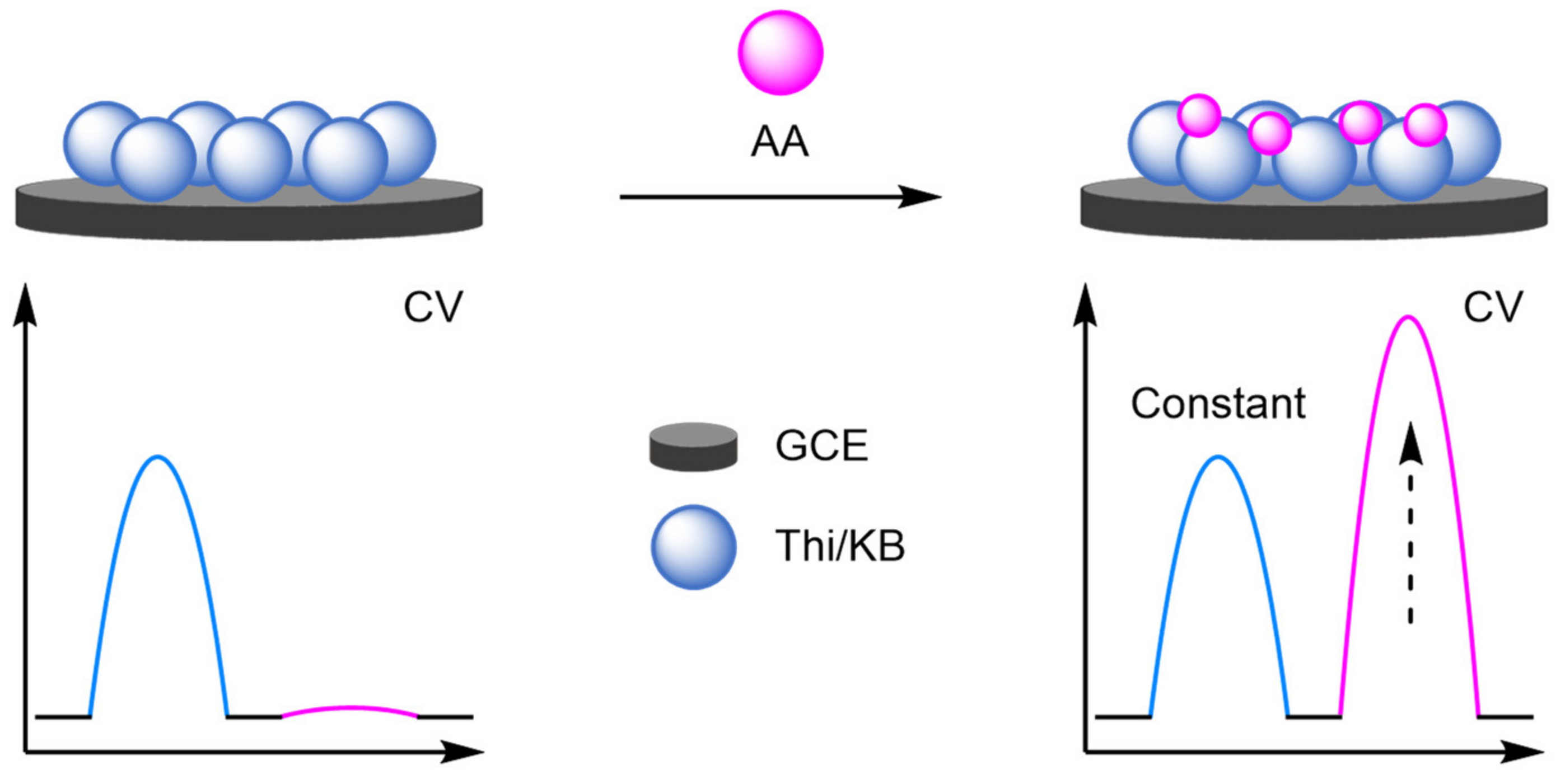
- Cheng, H.; Wang, X.; Wei, H. Ratiometric Electrochemical Sensor for Effective and Reliable Detection of Ascorbic Acid in Living Brains. Anal. Chem. 2015, 87, 8889–8895. [Google Scholar] [CrossRef] [PubMed]
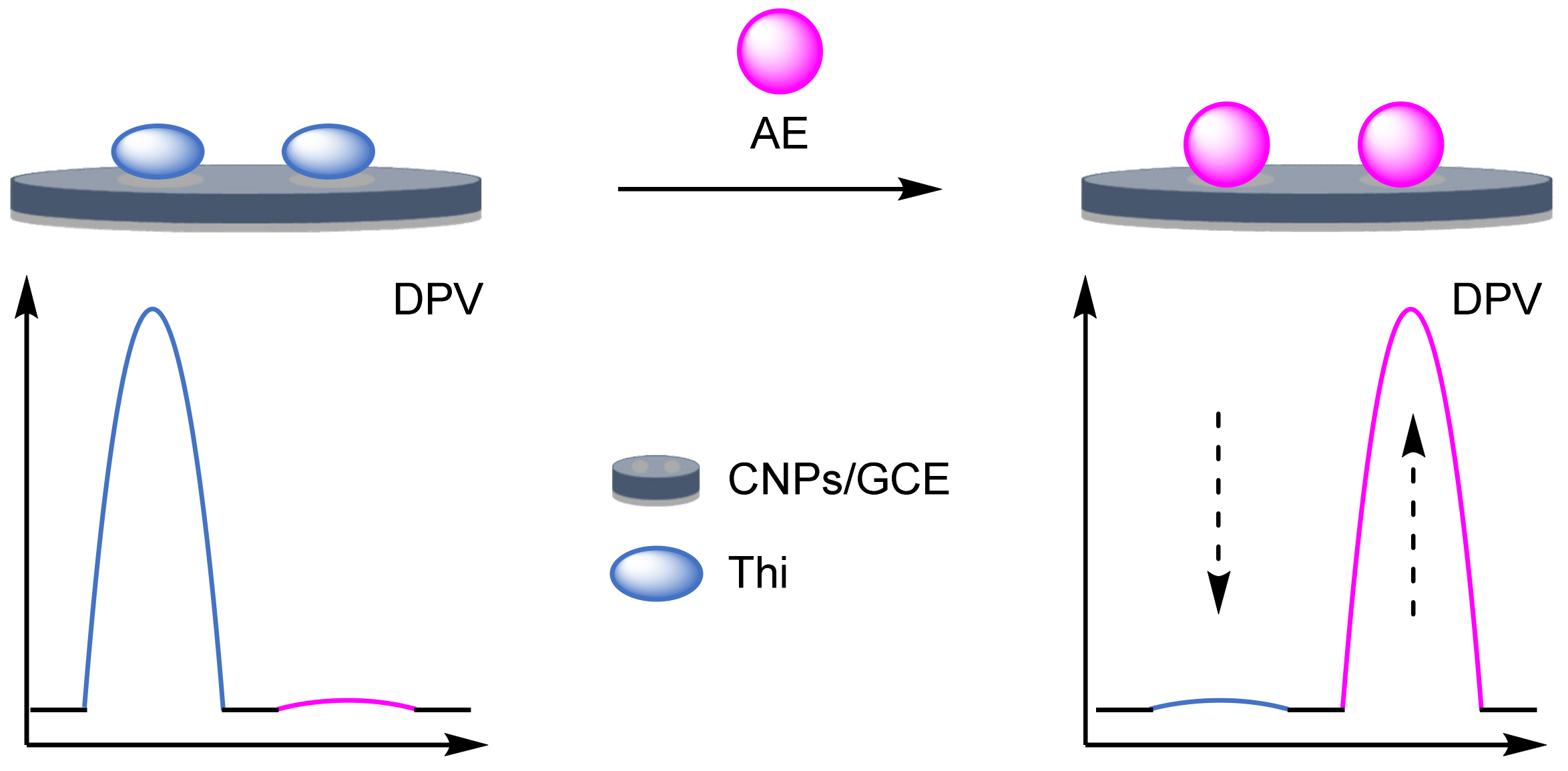
- Sun, Y.; Jiang, X.; Jin, H.; Gui, R. Ketjen black/ferrocene dual-doped MOFs and aptamer-coupling gold nanoparticles used as a novel ratiometric electrochemical aptasensor for vanillin detection. Anal. Chim. Acta 2019, 1083, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Jin, H.; Gui, R.; Wang, Z.; Ge, F. A general strategy to facilely design ratiometric electrochemical sensors in electrolyte solution by directly using a bare electrode for dual-signal sensing of analytes. Talanta 2017, 162, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Jin, H.; Gui, R.; Wang, Z. Facile fabrication of dual-ratiometric electrochemical sensors based on a bare electrode for dual-signal sensing of analytes in electrolyte solution. Sens. Actuators B Chem. 2017, 242, 71–78. [Google Scholar] [CrossRef]
- Yu, J.; Jin, H.; Gui, R.; Lv, W.; Wang, Z. A facile strategy for ratiometric electrochemical sensing of quercetin in electrolyte solution directly using bare glassy carbon electrode. J. Electroanal. Chem. 2017, 795, 97–102. [Google Scholar] [CrossRef]
- Chang, J.; Li, H.; Li, F. Diffusivity and intercalation of electroactive dyes-mediated truly ratiometric homogeneous electrochemical strategy for highly sensitive biosensing. Chem. Commun. 2019, 55, 10603–10606. [Google Scholar] [CrossRef]
- Germain, M.E.; Knapp, M.J. Optical explosives detection: From color changes to fluorescence turn-on. Chem. Soc. Rev. 2009, 38, 2543–2555. [Google Scholar] [CrossRef]
- Kim, H.N.; Guo, Z.; Zhu, W.; Yoon, J.; Tian, H. Recent progress on polymer-based fluorescent and colorimetric chemosensors. Chem. Soc. Rev. 2011, 40, 79–93. [Google Scholar] [CrossRef]
- Li, Z.; Askim, J.R.; Suslick, K.S. The Optoelectronic Nose: Colorimetric and Fluorometric Sensor Arrays. Chem. Rev. 2019, 119, 231–292. [Google Scholar] [CrossRef]
- Sagi, A.; Rishpon, J.; Shabat, D. Amperometric Assay for Aldolase Activity: Antibody-Catalyzed Ferrocenylamine Formation. Anal. Chem. 2006, 78, 1459–1461. [Google Scholar] [CrossRef] [PubMed]
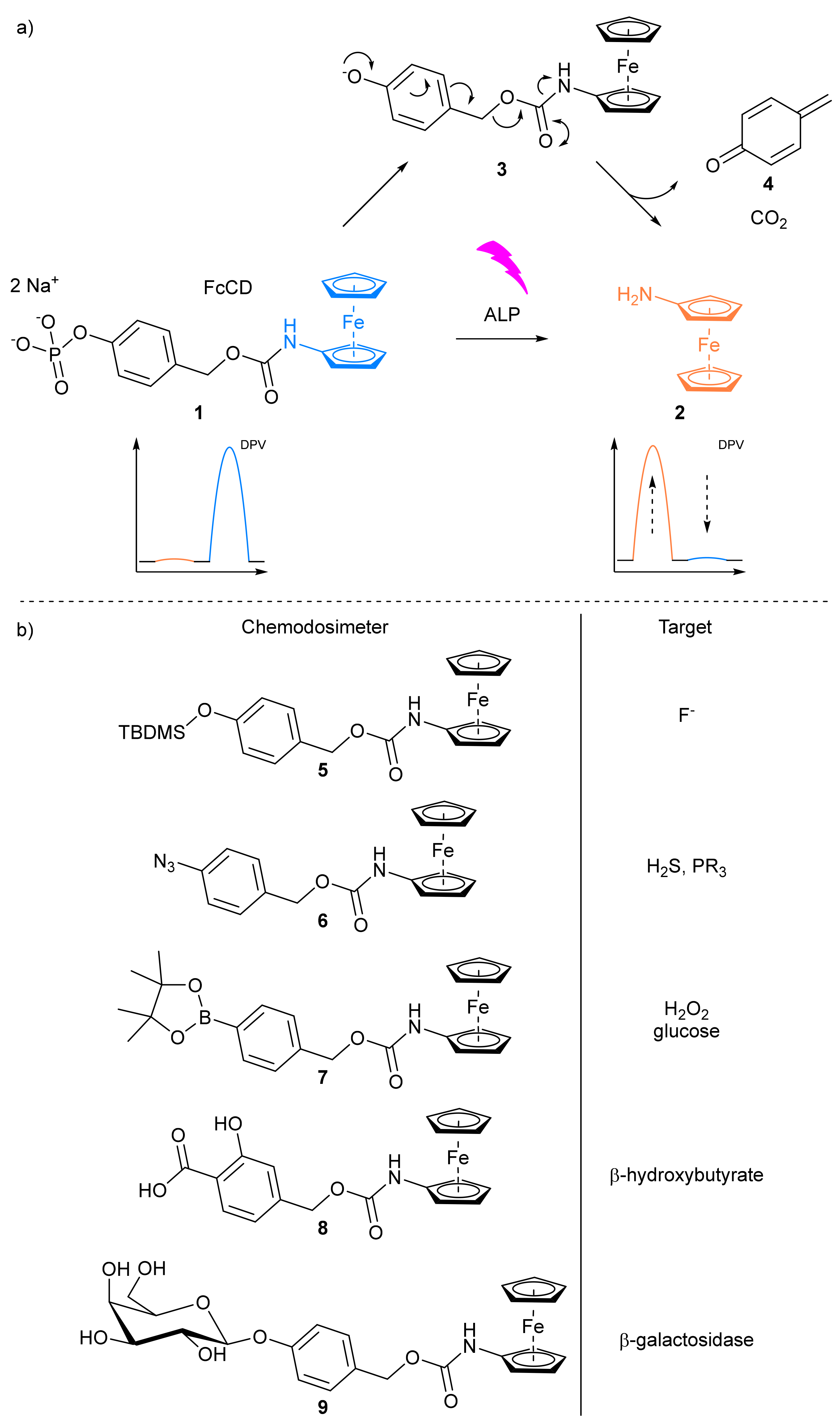
- Goggins, S.; Naz, C.; Marsh, B.J.; Frost, C.G. Ratiometric electrochemical detection of alkaline phosphatase. Chem. Commun. 2015, 51, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Goggins, S.; Apsey, E.A.; Mahon, M.F.; Frost, C.G. Ratiometric electrochemical detection of hydrogen peroxide and glucose. Org. Biomol. Chem. 2017, 15, 2459–2466. [Google Scholar] [CrossRef] [PubMed]
- Spring, S.A.; Goggins, S.; Frost, C.G. Ratiometric electrochemical detection of β-galactosidase. Org. Biomol. Chem. 2017, 15, 7122–7126. [Google Scholar] [CrossRef]
- Manibalan, K.; Mani, V.; Huang, S.-T. A switchable electrochemical redox ratiometric substrate based on ferrocene for highly selective and sensitive fluoride detection. RSC Adv. 2016, 6, 71727–71732. [Google Scholar] [CrossRef]
- Manibalan, K.; Mani, V.; Chang, P.C.; Huang, C.H.; Huang, S.T.; Marchlewicz, K.; Neethirajan, S. Electrochemical latent redox ratiometric probes for real-time tracking and quantification of endogenous hydrogen sulfide production in living cells. Biosens. Bioelectron. 2017, 96, 233–238. [Google Scholar] [CrossRef]
- Chen, T.Y.; Mani, V.; Huang, S.T.; Chang, P.C.; Huang, C.H.; Huang, N.M. Synthesis of robust electrochemical substrate and fabrication of immobilization free biosensors for rapid sensing of salicylate and β-hydroxybutyrate in whole blood. Anal. Chim. Acta 2017, 990, 78–83. [Google Scholar] [CrossRef]
- Spring, S.A.; Goggins, S.; Frost, C.G. An Organophosphorus(III)-Selective Chemodosimeter for the Ratiometric Electrochemical Detection of Phosphines. Chemosensors 2019, 7, 19. [Google Scholar] [CrossRef]
- Goggins, S.; Stark, O.P.; Naz, C.; Marsh, B.J.; Frost, C.G. Ratiometric electrochemical detection of Pd•••π interactions: Application towards electrochemical molecular logic gates. Supramol. Chem. 2017, 29, 749–757. [Google Scholar] [CrossRef][Green Version]
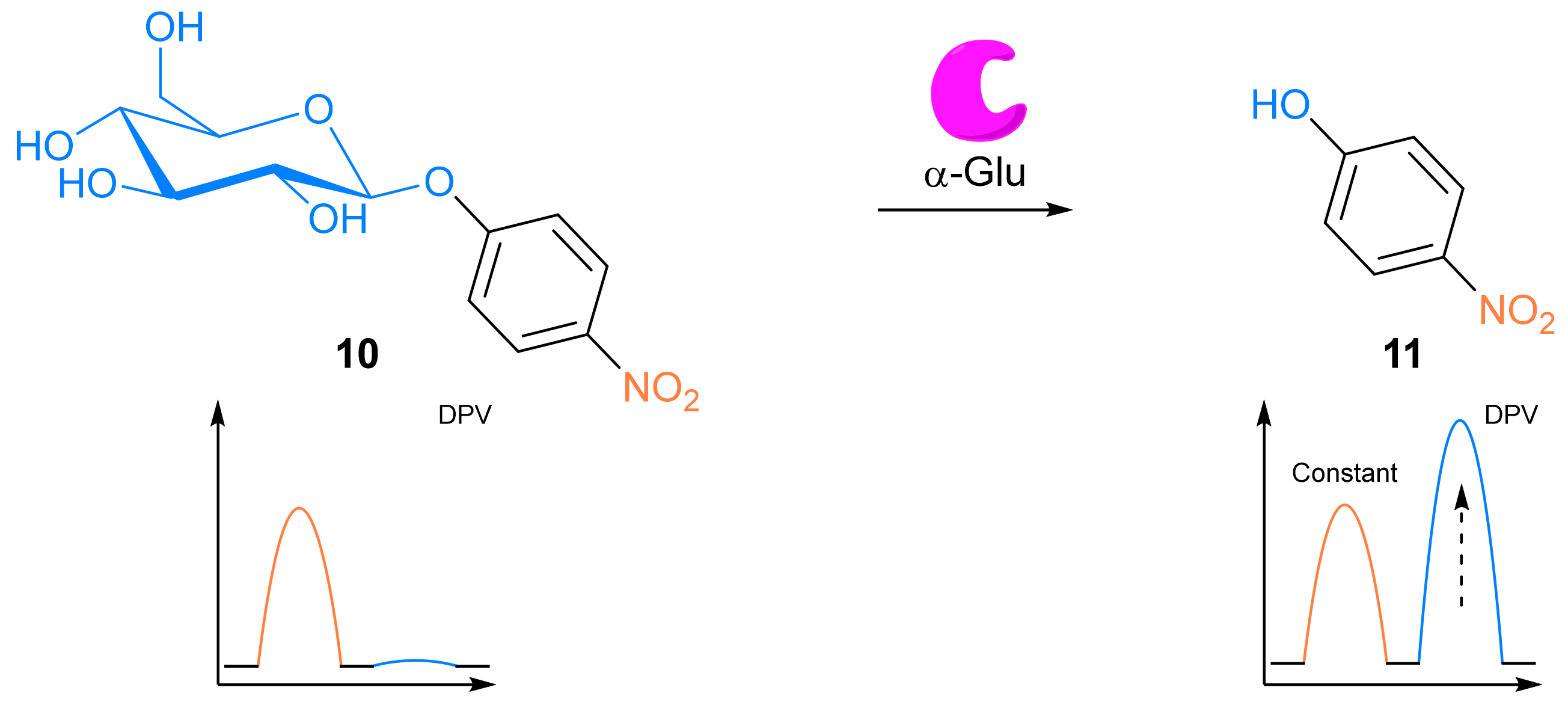
- Liu, Y.; Li, H.; Lu, L.; Sun, B.; Huang, L.; Chen, H.; Qiu, W.; Tao, J.; Zhao, P. A Ratiometric Electrochemical Sensor with Integrated Probe for the Assay of α-glucosidase Activity and Screening of Its Inhibitors. J. Electrochem. Soc. 2019, 166, B133–B140. [Google Scholar] [CrossRef]
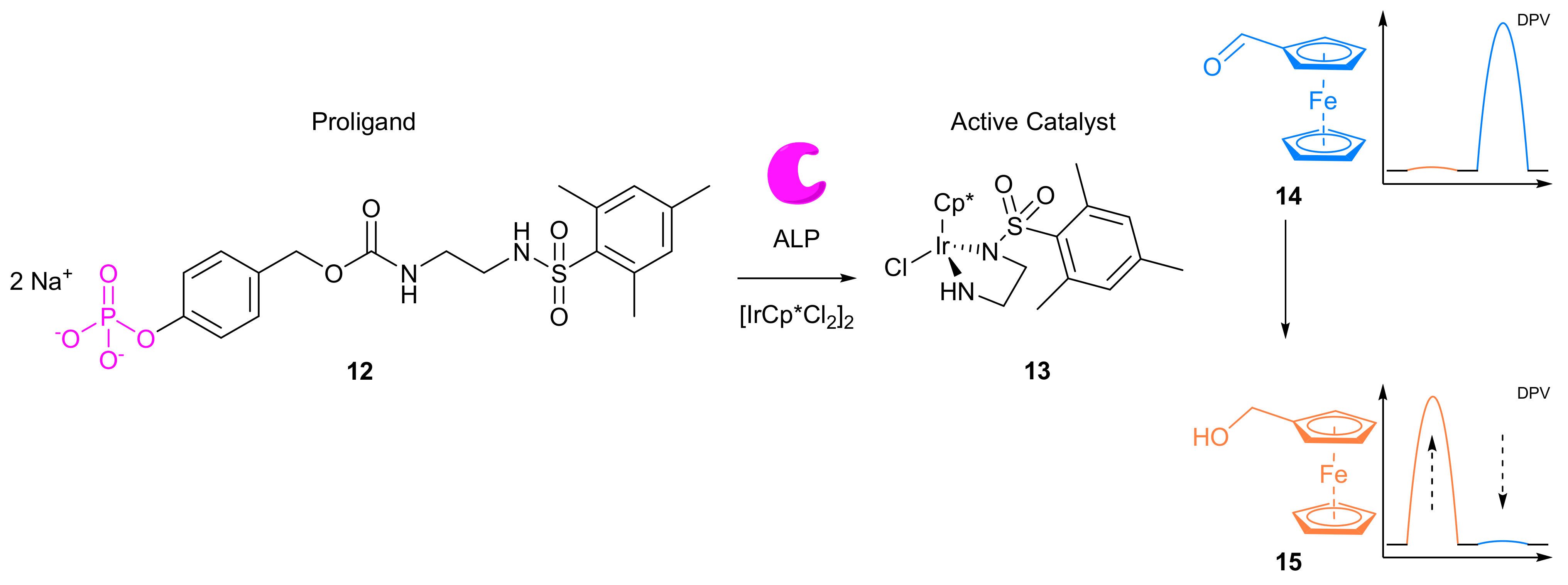
- Goggins, S.; Marsh, B.J.; Lubben, A.T.; Frost, C.G. Signal transduction and amplification through enzyme-triggered ligand release and accelerated catalysis. Chem. Sci. 2015, 6, 4978–4985. [Google Scholar] [CrossRef] [PubMed]
- Chai, X.; Zhou, X.; Zhu, A.; Zhang, L.; Qin, Y.; Shi, G.; Tian, Y. A two-channel ratiometric electrochemical biosensor for in vivo monitoring of copper ions in a rat brain using gold truncated octahedral microcages. Angew. Chem. 2013, 52, 8129–8133. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Zhang, L.; Zhu, A.; Shi, G.; Tian, Y. In vivo monitoring of local pH values in a live rat brain based on the design of a specific electroactive molecule for H+. Chem. Commun. 2016, 52, 3717–3720. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Hou, Q.; Liu, Y.; Cai, Y.; Guo, Y.; Xiang, H.; Chen, S. On-line regeneration of electrochemical biosensor for in vivo repetitive measurements of striatum Cu2+ under global cerebral ischemia/reperfusion events. Biosens. Bioelectron. 2019, 135, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Liu, Z.; Fu, J.; Zhao, W.; Guo, Y.; Sun, X.; Zhang, H. Ratiometric electrochemical aptasensor based on ferrocene and carbon nanofibers for highly specific detection of tetracycline residues. Sci. Rep. 2017, 7, 14729. [Google Scholar] [CrossRef]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spring, S.A.; Goggins, S.; Frost, C.G. Ratiometric Electrochemistry: Improving the Robustness, Reproducibility and Reliability of Biosensors. Molecules 2021, 26, 2130. https://doi.org/10.3390/molecules26082130
Spring SA, Goggins S, Frost CG. Ratiometric Electrochemistry: Improving the Robustness, Reproducibility and Reliability of Biosensors. Molecules. 2021; 26(8):2130. https://doi.org/10.3390/molecules26082130
Chicago/Turabian StyleSpring, Sam A., Sean Goggins, and Christopher G. Frost. 2021. "Ratiometric Electrochemistry: Improving the Robustness, Reproducibility and Reliability of Biosensors" Molecules 26, no. 8: 2130. https://doi.org/10.3390/molecules26082130
APA StyleSpring, S. A., Goggins, S., & Frost, C. G. (2021). Ratiometric Electrochemistry: Improving the Robustness, Reproducibility and Reliability of Biosensors. Molecules, 26(8), 2130. https://doi.org/10.3390/molecules26082130




