Solution and Solid State Studies of Urea Derivatives of DITIPIRAM Acting as Powerful Anion Receptors
Abstract
1. Introduction
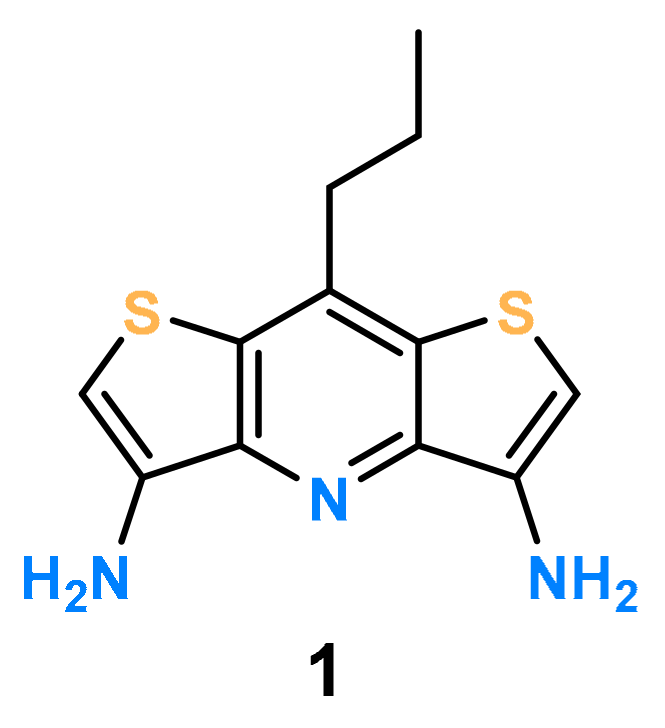
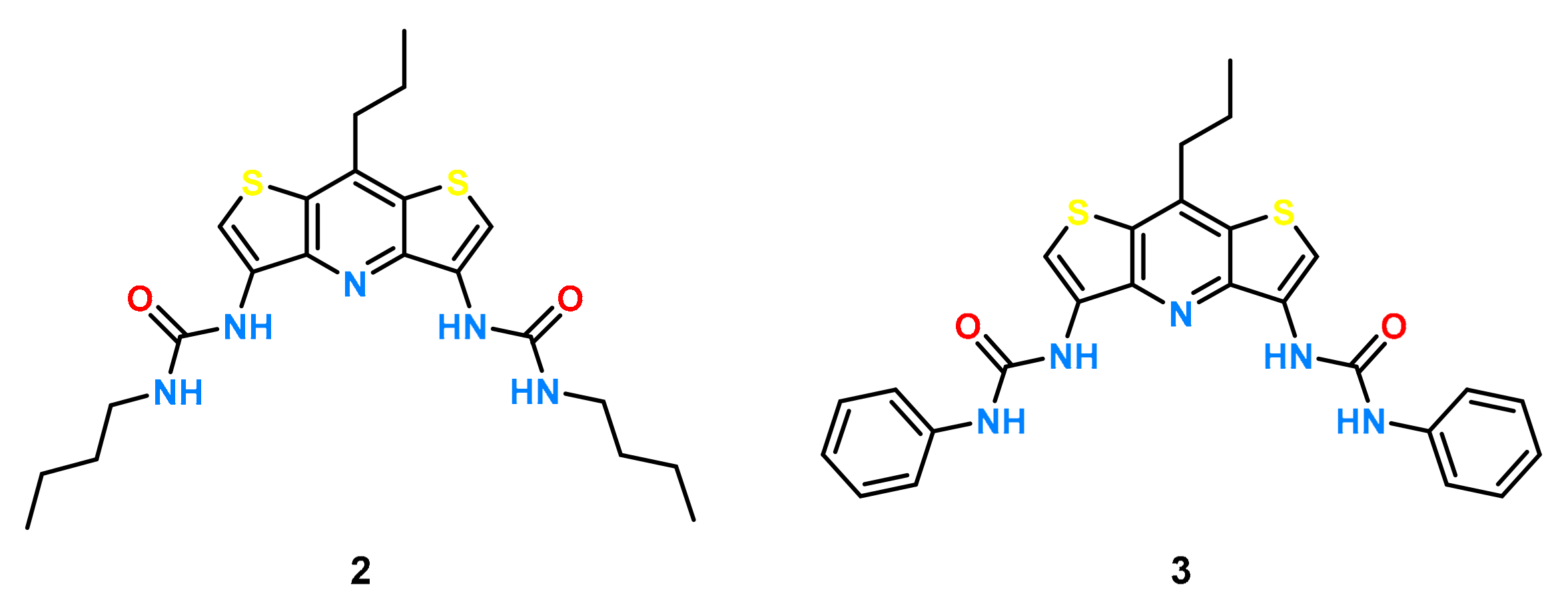
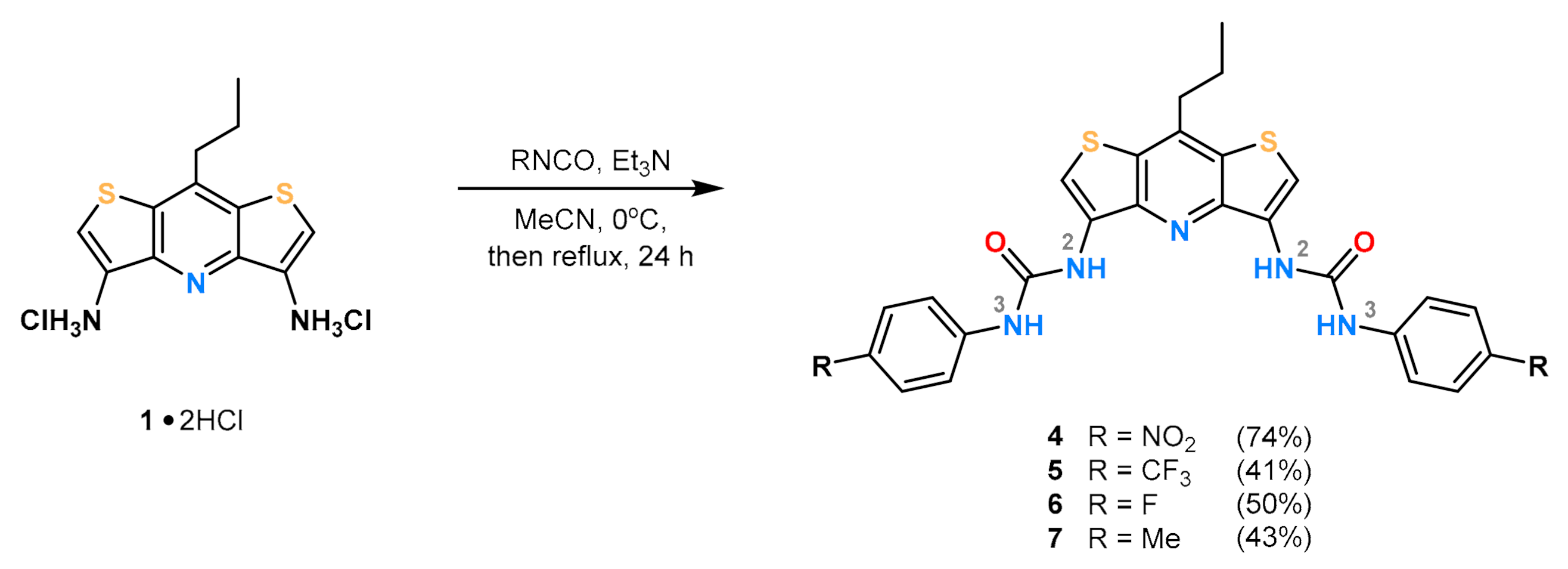
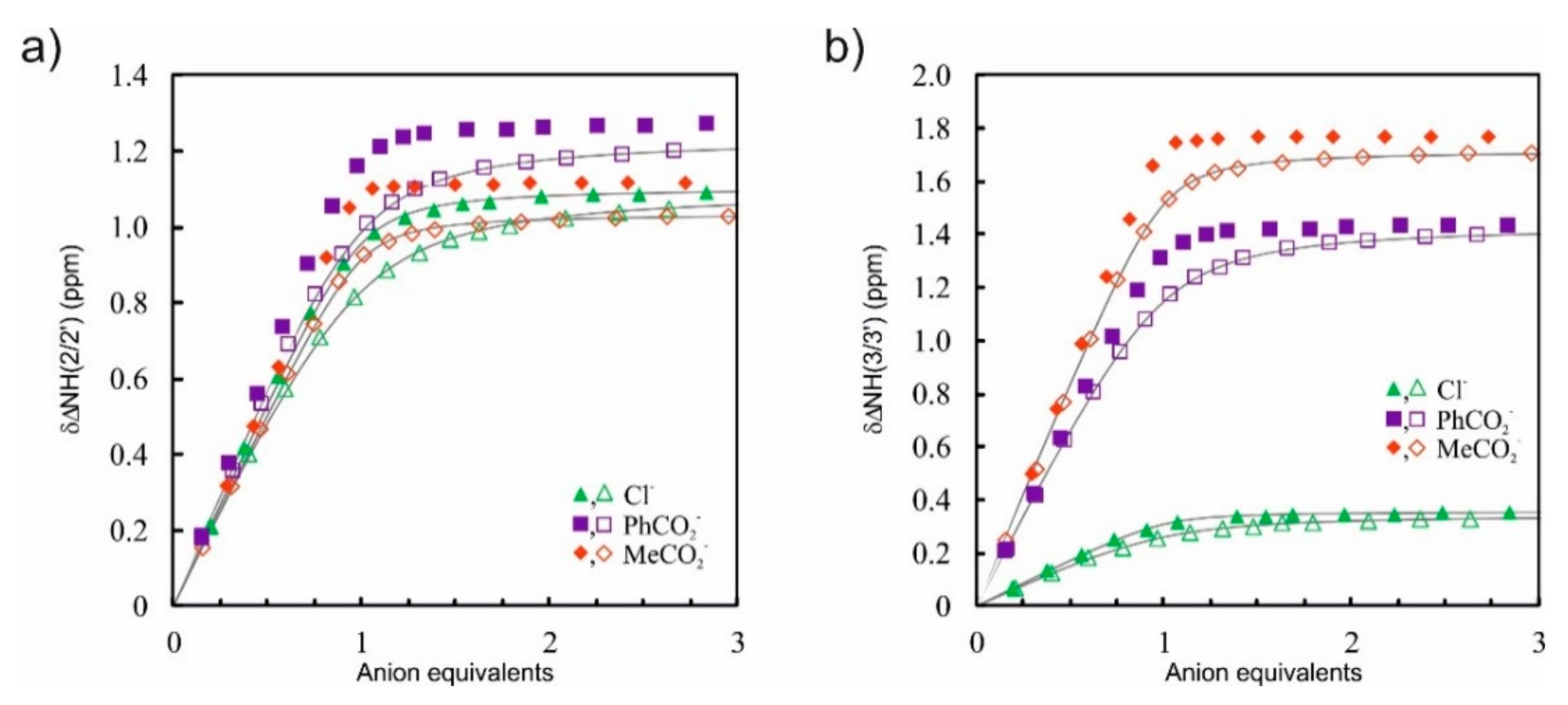
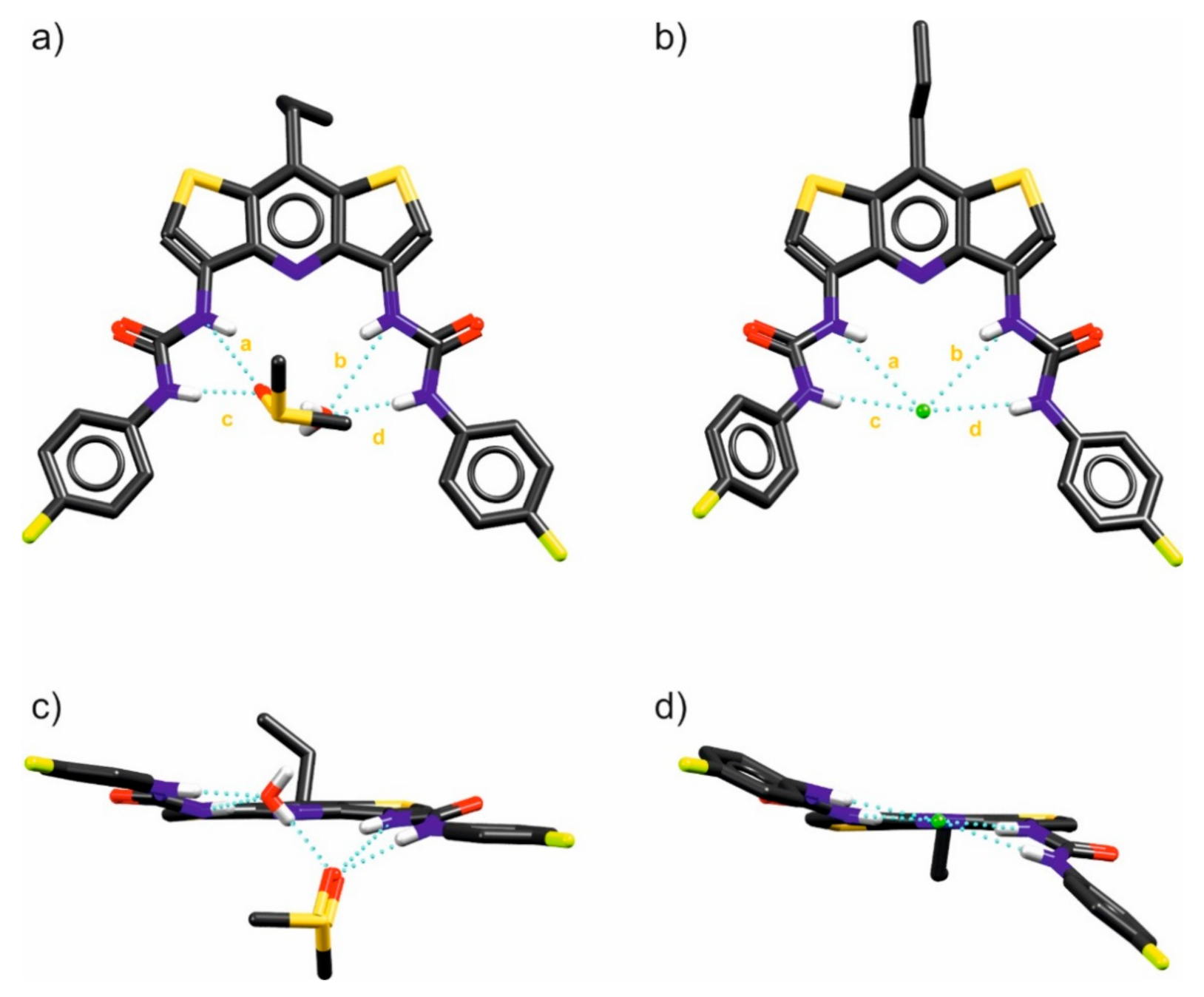
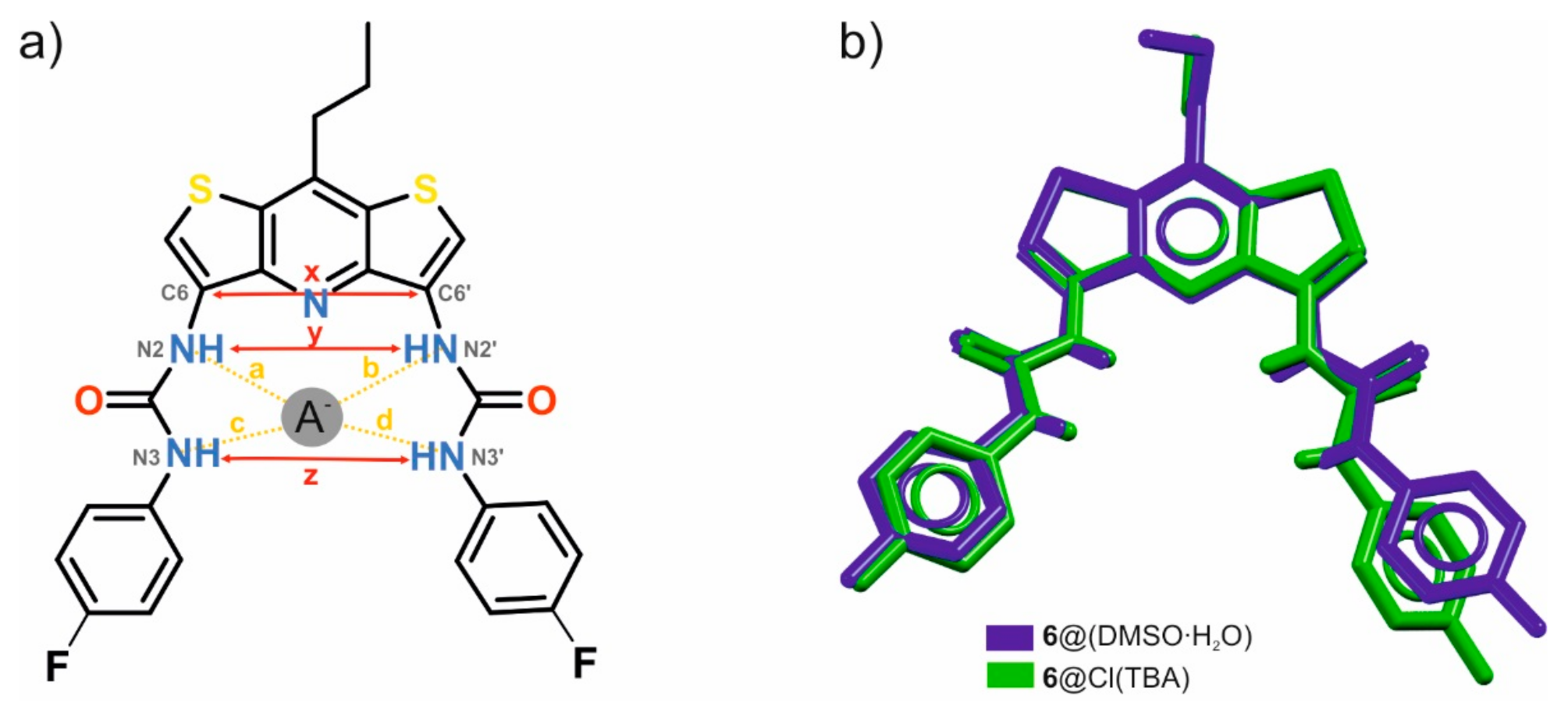
2. Results and Discussion
3. Materials and Methods
3.1. Reagents and General Methods
3.2. Synthetic Procedures
3.2.1. Synthesis of 1-(4-nitrophenyl)-3-(12-{[(4-nitrophenyl)carbamoyl]amino}-8-propyl-6,10-dithia-2-azatricyclo[7.3.0.03,⁷]dodeca-1,3(7),4,8,11-pentaen-4-yl)urea (4)
3.2.2. Synthesis of 3-[8-propyl-12-({[4-(trifluoromethyl)phenyl]carbamoyl}amino)-6,10-dithia-2-azatricyclo[7.3.0.03,7]dodeca-1,3(7),4,8,11-pentaen-4-yl]-1-[4-(trifluoromethyl)phenyl]urea (5)
3.2.3. Synthesis of 1-(4-fluorophenyl)-3-(12-{[(4-fluorophenyl)carbamoyl]amino}-8-propyl-6,10-dithia-2-azatricyclo[7.3.0.03,7]dodeca-1,3(7),4,8,11-pentaen-4-yl)urea (6)
3.2.4. Synthesis of 1-(4-methylphenyl)-3-(12-{[(4-methylphenyl)carbamoyl]amino}-8-propyl-6,10-dithia-2-azatricyclo[7.3.0.03,7]dodeca-1,3(7),4,8,11-pentaen-4-yl)urea (7)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Pina, F.; Bernardo, M.A.; García-España, E. Fluorescent Chemosensors Containing Polyamine Receptors. Eur. J. Inorg. Chem. 2000, 2000, 2143–2157. [Google Scholar] [CrossRef]
- Amendola, V.; Bonizzoni, M.; Esteban-Gómez, D.; Fabbrizzi, L.; Licchelli, M.; Sancenón, F.; Taglietti, A. Some guidelines for the design of anion receptors. Coord. Chem. Rev. 2006, 250, 1451–1470. [Google Scholar] [CrossRef]
- Li, X.; Wu, Y.-D.; Yang, D. α-Aminoxy Acids: New Possibilities from Foldamers to Anion Receptors and Channels. Acc. Chem. Res. 2008, 41, 1428–1438. [Google Scholar] [CrossRef] [PubMed]
- Dydio, P.; Lichosyt, D.; Jurczak, J. Amide- and urea-functionalized pyrroles and benzopyrroles as synthetic, neutral anion receptors. Chem. Soc. Rev. 2011, 40, 2971–2985. [Google Scholar] [CrossRef] [PubMed]
- Haridas, V.; Sahu, S.; Kumar, P.P.P.; Sapala, A.R. Triazole: A new motif for anion recognition. RSC Adv. 2012, 2, 12594–12605. [Google Scholar] [CrossRef]
- Dydio, P.; Detz, R.J.; Reek, J.N.H. Precise Supramolecular Control of Selectivity in the Rh-Catalyzed Hydroformylation of Terminal and Internal Alkenes. J. Am. Chem. Soc. 2013, 135, 10817–10828. [Google Scholar] [CrossRef]
- Evans, N.H.; Beer, P.D. Advances in Anion Supramolecular Chemistry: From Recognition to Chemical Applications. Angew. Chem. Int. Ed. 2014, 53, 11716–11754. [Google Scholar] [CrossRef]
- Langton, M.J.; Serpell, C.J.; Beer, P.D. Anion Recognition in Water: Recent Advances from a Supramolecular and Macromolecular Perspective. Angew. Chem. Int. Ed. 2015, 55, 1974–1987. [Google Scholar] [CrossRef]
- Busschaert, N.; Caltagirone, C.; Van Rossom, W.; Gale, P.A. Applications of Supramolecular Anion Recognition. Chem. Rev. 2015, 115, 8038–8155. [Google Scholar] [CrossRef]
- Gale, P.A.; Howe, E.N.W.; Wu, X. Anion Receptor Chemistry. Chem 2016, 1, 351–422. [Google Scholar] [CrossRef]
- Molina, P.; Zapata, F.; Caballero, A. Anion Recognition Strategies Based on Combined Noncovalent Interactions. Chem. Rev. 2017, 117, 9907–9972. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Vargas-Zúñiga, G.I.; Kim, S.H.; Kim, S.K.; Sessler, J.L. Macrocycles as Ion Pair Receptors. Chem. Rev. 2019, 119, 9753–9835. [Google Scholar] [CrossRef]
- Davis, J.T.; Gale, P.A.; Quesada, R. Advances in anion transport and supramolecular medicinal chemistry. Chem. Soc. Rev. 2020, 49, 6056–6086. [Google Scholar] [CrossRef]
- Zhao, W.; Flood, A.H.; White, N.G. Recognition and applications of anion–anion dimers based on anti-electrostatic hydrogen bonds (AEHBs). Chem. Soc. Rev. 2020, 49, 7893–7906. [Google Scholar] [CrossRef]
- Kumar, P.; Kumar, V.; Gupta, R. Dipicolinamide and isophthalamide based fluorescent chemosensors: Recognition and detection of assorted analytes. Dalton Trans. 2020, 49, 9544–9555. [Google Scholar] [CrossRef]
- Taylor, M.S. Anion recognition based on halogen, chalcogen, pnictogen and tetrel bonding. Coord. Chem. Rev. 2020, 413, 213270. [Google Scholar] [CrossRef]
- Malinowska, E.; Jurczak, J.; Stankiewicz, T. Macrocyclic oxamides as ionophores for lead-selective membrane electrodes. Electroanalysis 1993, 5, 489–492. [Google Scholar] [CrossRef]
- Duke, R.M.; McCabe, T.; Schmitt, W.; Gunnlaugsson, T. Recognition and Sensing of Biologically Relevant Anions in Alcohol and Mixed Alcohol–Aqueous Solutions Using Charge Neutral Cleft-Like Glycol-Derived Pyridyl–Amidothiourea Receptors. J. Org. Chem. 2012, 77, 3115–3126. [Google Scholar] [CrossRef]
- Lim, J.Y.C.; Marques, I.; Félix, V.; Beer, P.D. Enantioselective Anion Recognition by Chiral Halogen-Bonding [2]Rotaxanes. J. Am. Chem. Soc. 2017, 139, 12228–12239. [Google Scholar] [CrossRef]
- Zawada, Z.; Tatar, A.; Mocilac, P.; Buděšínský, M.; Kraus, T. Transport of Nucleoside Triphosphates into Cells by Artificial Molecular Transporters. Angew. Chem. Int. Ed. 2018, 57, 9891–9895. [Google Scholar] [CrossRef]
- Liu, W.; Oliver, A.G.; Smith, B.D. Macrocyclic Receptor for Precious Gold, Platinum, or Palladium Coordination Complexes. J. Am. Chem. Soc. 2018, 140, 6810–6813. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Chi, X.; Ahmed, M.; Long, L.; Sessler, J.L. Soft Materials Constructed Using Calix[4]pyrrole- and “Texas-Sized” Box-Based Anion Receptors. Acc. Chem. Res. 2019, 52, 1915–1927. [Google Scholar] [CrossRef]
- Majdecki, M.; Niedbała, P.; Jurczak, J. Amide-Based Cinchona Alkaloids as Phase-Transfer Catalysts: Synthesis and Potential Application. Org. Lett. 2019, 21, 8085–8090. [Google Scholar] [CrossRef]
- Borissov, A.; Marques, I.; Lim, J.Y.C.; Félix, V.; Smith, M.D.; Beer, P.D. Anion Recognition in Water by Charge-Neutral Halogen and Chalcogen Bonding Foldamer Receptors. J. Am. Chem. Soc. 2019, 141, 4119–4129. [Google Scholar] [CrossRef] [PubMed]
- Bunchuay, T.; Docker, A.; Martinez-Martinez, A.J.; Beer, P.D. A Potent Halogen-Bonding Donor Motif for Anion Recognition and Anion Template Mechanical Bond Synthesis. Angew. Chem. Int. Ed. 2019, 58, 13823–13827. [Google Scholar] [CrossRef] [PubMed]
- Majdecki, M.; Niedbała, P.; Jurczak, J. Synthesis of C2 Hybrid Amide-Based PTC Catalysts and Their Comparison with Saturated Analogues. Chem. Sel. 2020, 5, 6424–6429. [Google Scholar] [CrossRef]
- Lv, S.; Li, X.; Yang, L.; Wang, X.; Zhang, J.; Zhang, G.; Jiang, J. Azopyrazole-Based Photoswitchable Anion Receptor for Dihydrogen Phosphate Transport. J. Phys. Chem. 2020, 124, 9692–9697. [Google Scholar] [CrossRef]
- Wang, F.; Sen, S.; Chen, C.; Bähring, S.; Lei, C.; Duan, Z.; Zhang, Z.; Sessler, J.L.; Jana, A. Self-Assembled Cagelike Receptor That Binds Biologically Relevant Dicarboxylic Acids via Proton-Coupled Anion Recognition. J. Am. Chem. Soc. 2020, 142, 1987–1994. [Google Scholar] [CrossRef] [PubMed]
- Kohnke, F.H. Calixpyrroles: From Anion Ligands to Potential Anticancer Drugs. Eur. J. Org. Chem. 2020, 4261–4272. [Google Scholar] [CrossRef]
- Chmielewski, M.J.; Jurczak, J. A hybrid macrocycle containing benzene and pyridine subunits is a better anion receptor than both its homoaromatic congeners. Tetrahedron Lett. 2005, 46, 3085–3088. [Google Scholar] [CrossRef]
- Zieliński, T.; Kędziorek, M.; Jurczak, J. Bisamides Derived from Azulene-1,3- and -5,7-dicarboxylic Acids as New Building Blocks for Anion Receptors. Chem. Eur. J. 2008, 14, 838–846. [Google Scholar] [CrossRef] [PubMed]
- Zieliński, T.; Dydio, P.; Jurczak, J. Synthesis, structure and the binding properties of the amide-based anion receptors derived from 1H-indole-7-amine. Tetrahedron 2008, 64, 568–574. [Google Scholar] [CrossRef]
- Niedbała, P.; Majdecki, M.; Dąbrowa, K.; Jurczak, J. Selective Carboxylate Recognition Using Urea-Functionalized Unclosed Cryptands: Mild Synthesis and Complexation Studies. J. Org. Chem. 2020, 85, 5058–5064. [Google Scholar] [CrossRef] [PubMed]
- Niedbała, P.; Jurczak, J. One-Pot Parallel Synthesis of Unclosed Cryptands—Searching for Selective Anion Receptors via Static Combinatorial Chemistry Techniques. ACS Omega 2020, 5, 26271–26277. [Google Scholar] [CrossRef]
- Dąbrowa, K.; Niedbała, P.; Pawlak, M.; Lindner, M.; Ignacak, W.; Jurczak, J. Tuning Anion-Binding Properties of 22-Membered Unclosed Cryptands by Structural Modification of the Lariat Arm. ACS Omega 2020, 5, 29601–29608. [Google Scholar] [CrossRef]
- Niedbała, P.; Jurczak, J. Effective synthetic strategy towards highly selective macrocyclic anion receptors based on static combinatorial chemistry. Tetrahedron 2020, 76, 131693. [Google Scholar] [CrossRef]
- Maeda, H.; Haketa, Y.; Nakanishi, T. Aryl-Substituted C3-Bridged Oligopyrroles as Anion Receptors for Formation of Supramolecular Organogels. J. Am. Chem. Soc. 2007, 129, 13661–13764. [Google Scholar] [CrossRef] [PubMed]
- Mazik, M.; Sonnenberg, C. Isopropylamino and Isobutylamino Groups as Recognition Sites for Carbohydrates: Acyclic Receptors with Enhanced Binding Affinity toward β-Galactosides. J. Org. Chem. 2010, 75, 6416–6423. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Hong, J. Molecular Recognition of Pyrophosphate with Extended Bis(Zn(II)-DPA) Derivatives. J. Org. Chem. 2019, 84, 15797–15804. [Google Scholar] [CrossRef]
- Gale, P.A.; Hiscock, J.R.; Moore, S.J.; Caltagirone, C.; Hursthouse, M.B.; Light, M.E. Anion-Anion Proton Transfer in Hydrogen Bonded Complexes. Chem. Asian J. 2010, 5, 555–561. [Google Scholar] [CrossRef]
- Cholewiak, A.; Tycz, A.; Jurczak, J. 8-Propyldithieno[3,2-b:2′,3′-e]pyridine-3,5-diamine (DITIPIRAM) Derivatives as Neutral Receptors Tailored for Binding of Carboxylates. Org. Lett. 2017, 19, 3001–3004. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Barrientos, D.; Rojas-Hernández, A.; Gutiérrez, A.; Moya-Hernández, R.; Gómez-Balderas, R.; Ramírez-Silva, M.T. Determination of pKa values of tenoxicam from 1H-NMR chemical shifts and of oxicams from electrophoretic mobilities (CZE) with the aid of programs SQUAD and HYPNMR. Talanta 2009, 80, 754–762. [Google Scholar] [CrossRef]
- Dodziuk, H. Rigidity versus flexibility. A review of experimental and theoretical studies pertaining to the cyclodextrin nonrigidity. J. Mol. Struct. 2002, 614, 33–45. [Google Scholar] [CrossRef]
- Zhao, Y.; Truhlar, D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar] [CrossRef]
- Kozuch, S.; Martin, J.M.L. Halogen Bonds: Benchmarks and Theoretical Analysis. J. Chem. Theory Comput. 2013, 9, 1918–1931. [Google Scholar] [CrossRef] [PubMed]
- Bauzá, A.; Alkorta, I.; Frontera, A.; Elguero, J. On the Reliability of Pure and Hybrid DFT Methods for the Evaluation of Halogen, Chalcogen, and Pnicogen Bonds Involving Anionic and Neutral Electron Donors. J. Chem. Theory Comput. 2013, 9, 5201–5210. [Google Scholar] [CrossRef]
- Dey, K.R.; Wong, B.M.; Hossain, M.A. Rational design of a macrocycle-based chemosensor for anions. Tetrahedron Lett. 2010, 51, 1329–1332. [Google Scholar] [CrossRef] [PubMed]
- Hohenstein, E.G.; Chill, S.T.; Sherrill, C.D. Assessment of the Performance of the M05−2X and M06−2X Exchange-Correlation Functionals for Noncovalent Interactions in Biomolecules. J. Chem. Theory Comput. 2008, 4, 1996–2000. [Google Scholar] [CrossRef] [PubMed]
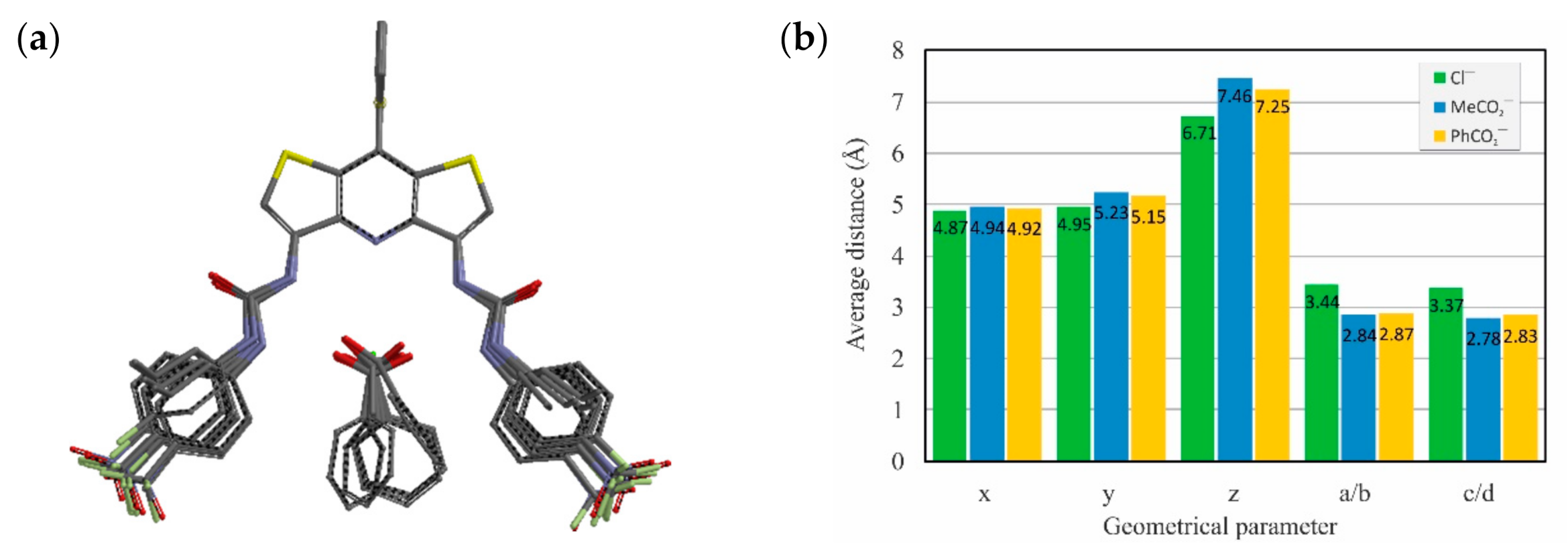
| Receptor | Rb | Solvent System | Cl− | MeCO2− | PhCO2− |
|---|---|---|---|---|---|
| 3 | H | DMSO-d6 + 0.5% H2O | 3350 ± 60 | >10,000 c | >10,000 c |
| DMSO-d6 + 10% CD3OH | 950 ± 10 | 4200 ± 90 | 1560 ± 20 | ||
| 5 | CF3 | DMSO-d6 + 0.5% H2O | 3400 ± 80 | >10,000 c | >10,000 c |
| DMSO-d6 + 10% CD3OH | 1040 ± 20 | 6300 ± 300 | 2400 ± 80 | ||
| 6 | F | DMSO-d6 + 0.5% H2O | 4650 ± 250 | >10,000 c | >10,000 c |
| DMSO-d6 + 10% CD3OH | 1250 ± 20 | 6400 ± 300 | 2050 ± 40 | ||
| 7 | Me | DMSO-d6 + 0.5% H2O | 2500 ± 50 | >10,000 c | n.d. d |
| DMSO-d6 + 10% CD3OH | 915 ± 10 | 2950 ± 150 | n.d. d |
| Scheme | Geometrical Descriptors (Å) | Hydrogen Bond Lengths (Å) | |||||
|---|---|---|---|---|---|---|---|
| x | y | z | a | b | c | d | |
| 6∙(DMSO∙H2O) | 4.89 | 5.11 | 7.12 | 3.18 | 3.19 | 2.84 | 2.83 |
| 6∙TBACl a | 4.82 | 4.87 | 6.54 | 3.39 | 3.46 | 3.29 | 3.30 |
| Difference solvate—complex (Å) | 0.07 | 0.24 | 0.58 | −0.21 | −0.27 | −0.45 | −0.47 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niedbała, P.; Dąbrowa, K.; Cholewiak-Janusz, A.; Jurczak, J. Solution and Solid State Studies of Urea Derivatives of DITIPIRAM Acting as Powerful Anion Receptors. Molecules 2021, 26, 1788. https://doi.org/10.3390/molecules26061788
Niedbała P, Dąbrowa K, Cholewiak-Janusz A, Jurczak J. Solution and Solid State Studies of Urea Derivatives of DITIPIRAM Acting as Powerful Anion Receptors. Molecules. 2021; 26(6):1788. https://doi.org/10.3390/molecules26061788
Chicago/Turabian StyleNiedbała, Patryk, Kajetan Dąbrowa, Agnieszka Cholewiak-Janusz, and Janusz Jurczak. 2021. "Solution and Solid State Studies of Urea Derivatives of DITIPIRAM Acting as Powerful Anion Receptors" Molecules 26, no. 6: 1788. https://doi.org/10.3390/molecules26061788
APA StyleNiedbała, P., Dąbrowa, K., Cholewiak-Janusz, A., & Jurczak, J. (2021). Solution and Solid State Studies of Urea Derivatives of DITIPIRAM Acting as Powerful Anion Receptors. Molecules, 26(6), 1788. https://doi.org/10.3390/molecules26061788








