Abstract
Background: Ginseng is widely used as herb or food. Different parts of ginseng have diverse usages. However, the comprehensive analysis on the ginsenosides in different parts of ginseng root is scarce. Methods: An ultra-high-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UHPLC-Q-TOF/MS) combined with UNIFI informatics platform and ultra-high-performance liquid chromatography-charged aerosol detection (UHPLC-CAD) were employed to evaluate the different parts of cultivated ginseng root. Results: 105 ginsenosides including 16 new compounds were identified or tentatively characterized. 22 potential chemical markers were identified, 20, 17, and 19 for main root (MR) and fibrous root (FR), main root (MR) and branch root (BR), and main root (MR) and rhizome (RH), respectively. The relative contents of Re, Rb1, 20(R)-Rh1, Rd, and Rf were highest in FR. The relative content of Rg1 was highest in RH. The total relative content of pharmacopoeia indicators Rg1, Re, and Rb1 was highest in FR. Conclusion: The differences among these parts were the compositions and relative contents of ginsenosides. Under our research conditions, the peak area ratio of Rg1 and Re could distinguish the MR and FR samples. Fibrous roots showed rich ingredients and high ginsenosides contents which should be further utilized.
1. Introduction
Ginseng, Panax ginseng Meyer of the Araliaceae family, is the king of herbs in the Orient, and its root has been widely used as a constituent of traditional medicine in China and Korea [1]. The major constituents of ginseng root are ginsenosides with various pharmacological properties, such as antitumor, enhanced immune system, antidiabetes, antifatigue, anti-oxidative, and anti-aging effects [2,3,4,5,6]. At present, ginseng is used not only as a therapeutic medicinal herb but also as a health supplement available on the market to promote longevity and adjust the balance of the human body. Public use of ginseng in the food field continues to grow. The quality of ginseng roots in the food industry and herbal markets must be evaluated and controlled. Nowadays, wild ginseng is rarely available, and the types of ginseng on the market are mostly collected from farms cultivating ginseng in fields. Ginseng has been developed as a valuable industrial crop and is now widely used worldwide [7]. The quality of ginseng root varies from different cultivation environments/areas, cultivation ages and “paozhi”. For example, ginsenosides Rg5, Rh4, Rk1, Rs4, F4, and 20(S)-Rg3 were found to be the quality control markers in different processing methods in distinguishing red ginseng from white ginseng [8]. The metabolic profiles of root, leaf, flower bud, berry, and seed of ginseng were also investigated [9,10]. However, the comprehensive analysis and research on the chemical constituents of different parts of ginseng root are scarce.
Ginseng root is further divided into main root (MR), branch root (BR), rhizome (RH), and fibrous root (FR). The different parts of P. ginseng root have diverse tradition medicine uses. The main root is generally used in Chinese medical clinics and traditional Chinese proprietary medicines and it was used as slice. The rhizome may cause the vomits, so it will be cut off before make slice. The fibrous root and branch root of ginseng root are the residual products of ginseng slice and it is used for the manufacturing of ginsenosides and related preparations, whereas the fibrous and branch roots are often ground to yield powder as health food. Several reports showed that the contents of total saponins in ginseng root were ranked as follows: rhizome > branch root > main root [11,12]. Few reports are available regarding the chemical constituents of different parts of the ginseng root. Therefore, an in-depth and comprehensive study on the ginsenosides of the different parts of ginseng root must be conducted to investigate. The constituents of such parts will provide the material basis reference for further utilization.
In recent years, ultra-high-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UHPLC-Q-TOF/MS) has become a powerful tool for the rapid separation and identification of active components in traditional Chinese medicines due to its advantages of high resolution and sensitivity. In addition, UNIFI, the automated data processing software, is an integrated informatics platform that can incorporate scientific library into a streamlined workflow to identify chemical components from complex raw data [13]. The combination of UHPLC separation, Q-TOF/MS detection, and UNIFI platform has been frequently applied in the characterization of chemical constituents of herbs [14,15]. Charged aerosol detector (CAD) has become a valuable tool for detecting substances with no ultraviolet absorption or only end absorption, such as saponins. This detector has some advantages, such as broad linearity response range, high sensitivity and reproducibility, the signal response consistency is independent of chemical structures, and simple operation [16,17]. The CAD detector coupled with UHPLC for a semiquantitative or quantitative analysis has certain advantages.
A comprehensive analysis on the chemical constituents of ginseng root based on UHPLC-Q-TOF/MS and UHPLC-CAD was performed in this study to evaluate the differences of ginsenosides among MR, BR, RH, and FR of cultivated ginseng root. The UHPLC-Q-TOF/MS combined with the UNIFI informatics platform was used to develop a multicomponent identification workflow for the analysis of extracts from four parts of cultivated ginseng root. Principal component analysis (PCA) and orthogonal partial least squares discrimination analysis (OPLS-DA) was used to profile diverse classes of metabolites of four parts from ginseng root. The UHPLC-CAD data were processed by histogram to intuitively see the differences of ginsenoside compounds in different parts of cultivated ginseng root. The study in this research comparatively analyzes the phytochemicals of different parts of ginseng root for the first time and finds out the similarities and differences among them. These results will support the further research and exploration of potential applications of ginseng root.
2. Results and Discussion
2.1. Identification of Components from Four Different Parts of the Ginseng Root Based on the UNIFI Platform
According to the literature [18], the ginsenosides in P. ginseng are generally divided into several types according to the aglycone moieties: the protopanaxadiol (PPD) type with sugar moieties attached to C-3 and/or C-20, the protopanaxatriol (PPT) type with sugar moieties at C-3, C-6, and/or C-20, the oleanolic acid type (OA), and other PPD and PPT derivatives. In combination with the fragmentation behavior of the reference standards and some studies [7,8,19,20,21,22], we deduced the fragmentation rule of the ginsenoside in ginseng; we also proposed a strategy for characterization and identification of compounds in P. ginseng by using the workflow of UHPLC-Q-TOF/MS combined with UNIFI informatics platform (Figure 1).
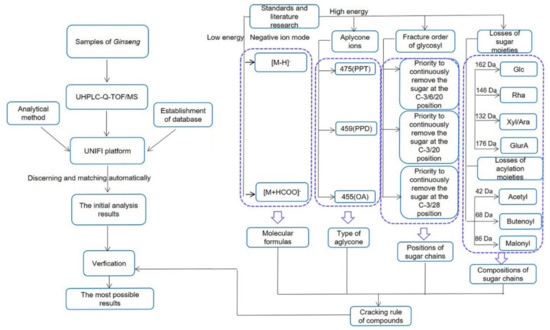
Figure 1.
Workflow of the rapid characterization of ginsenoside in P. ginseng by UHPLC-Q-TOF/MS combined with the UNIFI informatics platform.
The fragmentation rule of ginsenoside in P. ginseng is deducted as follows: for low CE in the ESI− mode, the common adduct ions, such as [M + HCOO]− and deprotonated ions [M − H]−, were often observed, which would allow us to determine the molecular mass and formula of the compounds. The backbone of a compound could be rapidly assigned by the abundant aglycone ions in the high CE channel of the ESI−. The characteristic fragment ion at m/z 475, 459, and 455 correspond to the PPT, PPD, and OA type aglycones, respectively. The positions of the glycosyl chains could be readily determined by the characteristic fragment ions in the negative ion mode.
The characteristic sugar fragments can be found by sequential losses of 162 Da (−Glc), and/or 146 Da (−Rha), and/or 132 Da (−Xyl/Ara), and/or 176 Da (−GlurA). Fragmentation can be conducted in three possible ways when the hydroxy groups on the sugar were replaced by acetyl (Ace), butenoyl (But), or malonyl (Mal) groups, which would be accompanied by the losses of 42 Da (−Ace), 68 Da (−But), 44 Da (−CO2), and 86 Da (−Mal). Sugar chains first loss the acylation moieties and then the glycosyls. After data processing, all discovered components using UNIFI were further verified.
Accordingly, 105 ginsenosides were identified or tentatively characterized in the ESI− modes from MR, BR, FR, and RH (Table 1). The base peak ion (BPI) chromatograms are shown in Figure 2. In the four different parts of the ginseng root, these compounds almost shared constituents, including 57 PPD type saponins, 31 PPT type saponins, 4 OA type saponins, and 13 other compounds. The chemical structures of these compounds were summarized in Table S1. Among them, 83, 101, 99, and 96 ginsenosides were tentatively characterized in the MR, FR, BR, and RH, respectively. This result clearly suggests that these parts could be used as raw materials for the manufacture of ginsenoside-based products.

Table 1.
Identification results of chemical constituents from different parts of P. ginseng root by UHPLC-Q-TOF/MS.
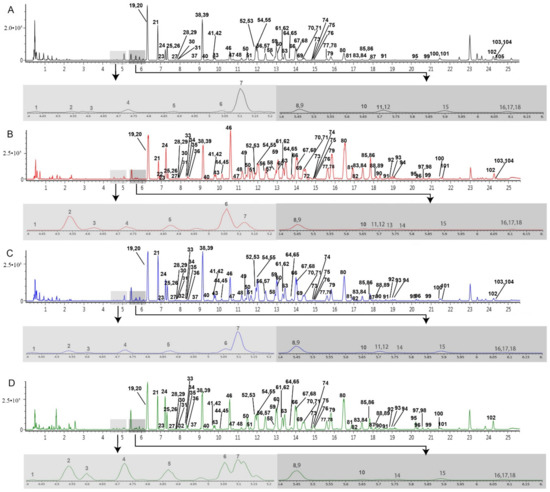
Figure 2.
BPI chromatograms of the different parts of the P. ginseng root in the negative mode analyzed by UHPLC-Q-TOF/MSE. (A) main root (MR), (B) fibrous root (FR), (C) branch root (BR), (D) rhizome (RH). Light gray represents a partial enlarged view of the chromatogram with the retention time from 4.4 to 5.2 min, and dark gray represents a partial enlarged view of the chromatogram with the retention time from 5.4 to 6.2 min.
The compounds were tentatively assigned by matching the molecular formulas and diagnostic fragment ions with those of the published known ginsenosides and the reference standards on the basis of UNIFI.
For example, peaks 5, 7–15, 17–29, 31–33, 36–37, 39–41, 43, 46, and 47 exhibited fragment ions at m/z 475 in the high energy of ESI− corresponding to the PPT aglycone moiety, thereby suggesting that they were the PPT-type ginsenosides. Peak 8, 10, and 14 had the same deprotonated molecular ion [M − H]− at m/z 931 and an adduct ion [M + HCOO]− at m/z 977 in the low energy of ESI− and fragment ions at m/z 799 [M − H − 132]− and 637 [M − H − 132 − Glc]− and 475 [M − H − 132 − 2Glc]− in the high energy of ESI−, thereby suggesting that they were a pair of isomers. In comparison with the reference standard, peak 14 was assigned to be Notoginsenoside R1. Meanwhile, in comparison with literature [7,19], peaks 8 and 10 were tentatively assigned to be Ginsenoside Re4 or its isomers. Peaks 7, 9, 11, 15, 18, 27, 33, 36, and 37 should be in each pair of isomers. Peaks 7, 11, and 15 were tentatively identified as Ginsenoside Re1/Ginsenoside Re2/Ginsenoside Re3 or their isomers. Peak 9 was identified as 20-O-Glucosylginsenoside Rf. Peaks 18, 27, 33, 36, and 37 were tentatively identified as Vinaginsenoside R4/Notoginsenoside R3/Notoginsenoside R6/Notoginsenoside M/Notoginsenoside N or its isomers compared with literature [7,19,23]. This study matched the accurate masses and the fragment ions with those of previous studies [7,19,25,27]; peaks 21, 23, 25, 32, and 41 were tentatively assigned to be 6′-O-Acetyl-ginsenoside Rg1, Yesanchinoside D, or its isomer; peaks 22 and 24 were tentatively assigned to be 6′′′-O-Acetyl-ginsenoside Re or its isomer; peak 29 was assigned to be Ginsenoside Rs4 or Ginsenoside Rs5; they all produced fragment ions [M − H − Ace]− and were acetylated ginsenosides.
Peaks 38, 42, 44–45, 48–52, 54–74, 76–94, 96–101, and 103–104 exhibited the fragment ions at m/z 459 in the high energy of ESI− corresponding to the PPD aglycone moiety, thereby suggesting that they were the PPD-type ginsenosides. This study takes peaks 79 and 85 as examples. Both peaks had the same protonated ion [M + HCOO]− at m/z 991 and the same fragment ions at m/z 783 [M − H − Glc]−, 621 [M − H − 2Glc]−, and 459 [M − H − 3Glc]− in ESI−. As a pair of isomers, peak 79 was identified as ginsenoside Rd compared with the standard; peak 85 was tentatively identified as Gypenoside XVII because it was matched with the characteristic MS fragmentation pattern of Gypenoside XVII reported in literature [20]. Peaks 38, 45, 55, 60, 68, 78, 84, and 88 all produced fragment ions [M − H − Mal]− and were malonylated ginsenosides by matching the accurate masses and the fragment ions with those of previous studies; peaks 96, 91, 94, 98, and 87 produced fragment ions [M − H − But]− and were butenoylated ginsenosides; peaks 44, 51, 52, 54, 58–59, 61, 64, 66–67, 69–74, 76–77, 80–83, 86, 89–90, 93, 99, and 101 produced fragment ions [M − H − Ace]− and were acetylated ginsenosides.
Peaks 30, 53, 75, and 95 exhibited fragment ions at m/z 455 in the high energy of ESI− corresponding to the OA aglycone moiety, thereby suggesting that they were the OA-type ginsenosides. Peak 30 showed a deprotonated molecular ion [M − H]− at m/z 925 in the low energy of ESI−, thereby suggesting that the molecular formula was C47H74O18. In the high energy of ESI−, the fragment ions [M − H − Glc]− at m/z 763, [M − H − Glc − Xyl]− at m/z 631, and [M − H − Glc − Xyl − GlurA]− at m/z 455 could be attributed to the successive loss of the Glc, Xyl, and GlurA groups. Finally, peak 30 was assigned to be Pseudoginsenoside Rt1 [7,28]. Peaks 75 and 95 exhibited the same deprotonated molecular ion [M − H]− at m/z 793 in the low energy of ESI−, thereby suggesting that the molecular formula was C42H66O14 and the same fragment ions [M − H − Glc]− at m/z 631 and [M − H − Glc − GlurA]− at m/z 455 in the high energy of ESI−. The fragment ions could be attributed to the loss of the Glc and GlurA groups. Finally, peaks 75 and 95 were tentatively assigned to be Chikusetsusaponin Iva or its isomer [26].
In this study, we have tentatively identified 16 new compounds (peaks 2, 6, 64, 66, 70–74, 76–77, 82–83, 90, 93, and 95) from MR, BR, FR, and RH on the basis of the result of UNIFI and literatures. For example, peaks 51, 58, 66, 70, 72, and 73 showed the same molecular formula (C60H100O27) and fragment ions at m/z 1209 [M − H − Ace]−, m/z 1077 [M − H − Ace − Xyl]−, m/z 945 [M − H − Ace − 2Xyl]−, m/z 783 [M − H – Ace − 2Xyl − glc]−, m/z 621 [M − H − Ace − 2Xyl − 2glc]−, and m/z 459 [M − H − Ace − 2Xyl − 3glc]− in the high energy of ESI− and they were a pair of isomers. Based on the literatures, we found that only two compounds are present, namely, Ginsenoside Ra5 and (3β,12β)-3-[[2-O-(6-O-Acetyl-β-D-glucopyranosyl)-β-D-glucopyranosyl]oxy]-12-hydroxydammar-24-en-20-yl O-β-D-xylopyranosyl-(1→2)-O-α-L-arabinopyranosyl-(1→6)-β-D-glucopyranoside in accordance with the mass spectral fragmentation rule of these peaks in this experiment; thus, the other four compounds were tentatively new. In the same way, two new compounds were tentatively identified from the isomers of peaks 54, 61, 64, and 74. Two new compounds were tentatively identified from the isomers of peaks 59, 67, 69, 71, and 76. One new compound was tentatively identified from the isomers of peaks 75 and 95. One new compound was tentatively identified from the isomers of peaks 80, 81, 86, 89, and 93. Peaks 83 and 90 were new compounds that have not been searched in the literature and databases. The high collision energy ESI-MS spectra of some representative compounds in each pair of isomers, such as peaks 51, 54, 59, 75, 80, and 83, are shown in Figure S1.
This research is the first to study the comprehensive screening analysis of the different parts of the cultivated ginseng root by using UHPLC-Q-TOF/MS combined with the UNIFI platform. This comprehensive and unique phytochemical profile study revealed the structural diversity of secondary metabolites and the similar patterns in the different parts of ginseng root. Moreover, this study could provide systematic data for clarifying the chemical composition of ginseng root.
2.2. Discrimination of Different Parts of P. ginseng Root by PCA and OPLS-DA Analysis
Multivariate statistical methods are applied to the analysis of metabolite data to discriminate and classify the different parts of ginseng root and identify marker compounds. First, the obtained multivariate dataset of 144 batches of samples that contained the same amount of MR, BR, RH, and FR was analyzed by PCA. The PCA 2D plots of the samples from four parts of ginseng root groups were easily classified into two clusters according to their common spectral characteristics (Figure 3A). An obvious difference can be observed between FR and other parts, and the FR groups were separated. Figure 3A (right to left) shows a transition trend of FR, RH, BR, and MR.
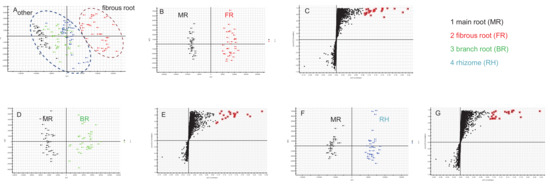
Figure 3.
PCA of four different parts of P. ginseng root (A), OPLS-DA/S plot of MR and FR (B,C), MR and BR (D,E), and MR and RH (F,G) samples in ESI− mode.
Aiming at evaluating the differences of the different parts (MR and FR, MR and BR, and MR and RH) of P. ginseng root, OPLS-DA score plot and S plot were obtained to understand which variables are responsible for this sample separation. The OPLS-DA plotting achieved maximum separation between different groups (panels B, D and F of Figure 3). S plots were then created to explore the potential chemical markers that contributed to the differences (panels C, E and G of Figure 3). On the basis of the VIP values (VIP > 8.5) from univariate statistical analysis, a total of 22 robust known chemical markers between MR and FR, MR and BR, and MR and RH groups of P. ginseng root were marked and listed (Table 2) and there were significant differences in the contents of them. (1) For MR and FR samples, there were 20 potential chemical markers, including 4 PPT-type and 16 PPD-type saponins, with higher contents in FR samples. (2) For MR and BR samples, there were 17 potential chemical markers, including 4 PPT-type and 13 PPD-type saponins, with higher contents in BR samples. (3) For MR and RH samples, there were 19 potential chemical markers, including 4 PPT-type, 13 PPD-type saponins and 2 OA-type (ginsenoside Ro and chikusetsusaponin Iva), with higher contents in RH samples.

Table 2.
The screened marker compounds for the discrimination of the different parts of P. ginseng root.
According to the reports [34,40], ginsenosides in FR were different from MR and BR in America ginseng and notoginseng. From this point of view, the results in this study are similar to America ginseng and notoginseng. A clear separation from the other parts appeared for FR. Therefore, FR might be differently used from the other parts. In addition, many differential monomeric ginsenosides exist in FR. In future research, some studies could be performed on the pharmacological activities and the relationship between the potential markers, and the effects should be established.
2.3. Distribution of Ginsenosides from the Different Parts of P. ginseng Root
The semiquantitative analysis of ginsenosides in different parts of cultivated P. ginseng root is still obscured to date. A poor correlation exists between mass spectral response and chemical composition. Accordingly, the data from UHPLC-CAD were used to communicate additional penetrating understanding of the distribution of compounds with a high content in different parts of cultivated P. ginseng root, especially pharmacopoeia indicators, such as ginsenosides Rg1, Re, and Rb1. The separation effects on the ginseng samples of Waters ACQUITY™ HSS T3, Acclaim RSLC PolarAdvantage, and Phenomenex C18 were compared. Rg1 and Re could not be separated by Waters ACQUITY™ HSS T3. Although Rg1 and Re could be separated by Acclaim RSLC PolarAdvantage column, the separation effect of the other components was worse than that of Phenomenex C18 column. Therefore, the Phenomenex C18 column can be used to separate the components of ginseng root in the Thermo vanquish UHPLC-CAD system.
Compounds with representative structure, different polarity, high relative content and good pharmacological activity and resolution were selected as the object of relative content analysis in this study. Six main compounds, namely, Rg1, Re, Rd, Rb1, Rf, and (20R)-Rh1, were compared with the existing reference standards. The area percentage method was used to calculate the percentage of the peak area of each compound in the sum of all peak areas, as the evaluation index of relative content. Figure 4A shows the liquid chromatograms of different parts of ginseng root. The FR had more abundant components compared with the other three parts. The contents of most saponins in BR, RH, and FR were higher than those in MR.
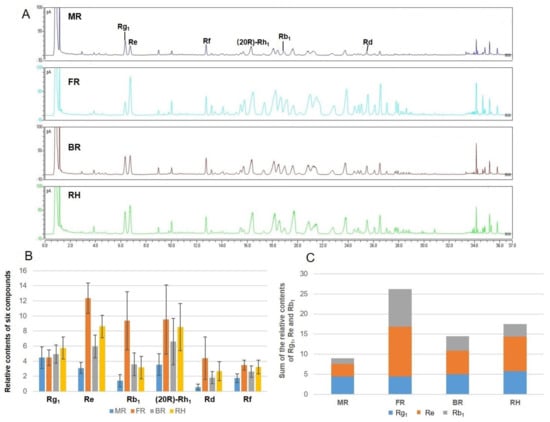
Figure 4.
Chromatograms of the different parts of ginseng root (MR main root, FR fibrous root, BR branch root, and RH rhizome) analyzed by UHPLC-CAD and column chart of relative contents of main components. (A) liquid chromatograms of different parts of ginseng root; (B) relative contents of six components of the different parts of ginseng root; and (C) sum of the peak areas of pharmacopoeia indicators Rg1, Re, and Rb1.
The relative contents of the six components in the different parts of 144 ginseng root samples were analyzed (Figure 4B). The results indicated that except for Rg1, the relative contents of Re, Rb1, (20R)-Rh1, Rd, and Rf were the highest in FR, followed by RH, BR, and MR. The peak areas of Re, Rb1, (20R)-Rh1, Rd, and Rf in FR were 1.43–4, 2.62–6.65, 1.12–2.69, 1.64–7.67, and 1.07–1.98 times than those of the other parts, respectively. The relative content of Rg1 in RH was the highest, followed by BR and FR. The peak area of Rg1 in RH was 1.16–1.29 times than those of the other parts. The sum of the peak areas of pharmacopoeia indicators Rg1, Re, and Rb1 in FR was 1.53–3.25 times than those of the other parts (Figure 4C). Ginseng root has two types, with fibrous root or no fibrous root. Our results explain the reason that the content of ginseng with fibrous root is qualified in the market. Under our research conditions, we also found some interesting results. The range of Rg1/Re was from 0.19 to 0.64 in FR samples, while the range of Rg1/Re was from 0.75 to 2.00 in MR samples. The peak area ratio of Rg1 and Re might be the marker of MR and FR samples. This idea laid a theoretical foundation for strengthening the comprehensive utilization of various parts of the roots of garden ginseng in the future.
3. Materials and Methods
3.1. Materials and Reagents
Thirty-six batches of representative cultivated ginseng root were collected or purchased from different cultivation areas in Jilin, Liaoning, and Heilongjiang Provinces, the main source of ginseng in China. A detailed sample list is provided in Table S2. The identity of all samples was confirmed by Prof. Bao-lin Guo of the Institute of Medicinal Plant Development, Beijing, China. Each whole ginseng root was divided into four parts: main root (MR), branch root (BR), rhizome (RH), and fibrous root (FR). One hundred forty-four different parts samples of ginseng root are present.
Ginsenosides Rb1, Re, Rg1, Rd, Rc, Rf, Ro, Rg2, 20(R)-Rh1, 20(S)-Rh2, 20(R)-Rh2, 20(S)-Rg3, and 20(R)-Rg3 and notoginsenosides R1 and R2 were isolated in our laboratory and identified by spectroscopic data. The purities of these standards were better than 98% by the HPLC analysis. All samples were stored at 4 °C before use.
Acetonitrile (HPLC grade) was purchased from Fisher Scientific Co. (Loughborough, UK). Distilled water was purchased from Watsons (Guangzhou, China). Formic acid (MS grade) was purchased from Thermo Fisher Scientific Co. Ltd. (Waltham, MA, USA). The other reagents were commercially obtained in analytical purity (Beijing, China).
3.2. Sample Preparation and Extraction
All the samples were separately dried, ground, and sieved (Chinese National Standard Sieve 3, R40/3 Series) to obtain the homogeneous powder. An aliquot of 0.2 g accurately weighed fine powder (<40 mesh) of each sample was soaked in a 10 mL centrifuge tube containing 3 mL of 70% (v/v) methanol, tightly plugged, shaken, weighed. After sonication for 30 min, the solutions were cooled to room temperature and made up for weight loss with 70% aqueous MeOH. All the solutions were filtered through a 0.22 μm filter membrane before analysis.
3.3. UHPLC-Q-TOF/MS and UHPLC-CAD Analysis
A UHPLC-Q-TOF/MS analysis was performed on a Waters ACQUITY I-Class system (Waters Corporation, Milford, MA, USA) coupled with a VION-IMS-QTOF system (Waters Corporation, Wilmslow, UK). A Waters ACQUITY™ UPLC HSS T3 column (100 × 2.1 mm, 1.8 μm) was used with the column temperature at 40 °C. The mobile phases were water with 0.1% formic acid (A) and acetonitrile (B). The gradient used was as follows: (0–1) min, 5%→15% B; (1–8) min, 15%→31% B; (8–16) min, 31%→35% B; (16–21) min, 35%→49% B; (21–24) min, 49%→60% B; (24–26) min, 60% B; (26–27) min, 60%→95% B; (27–28) min, 95% B; (28–29) min, 95%→5% B; and (29–31) min, 5% B. The flow rate was 0.5 mL/min. The injection volume of the sample was 1 μL. The data acquisition mode was MSE. Each sample was injected for ESI− analyses, and data were acquired from 50 Da to 1600 Da. A QC sample containing pooled different parts was regularly injected to monitor the system stability and minimize the analytical variation.
For MS conditions: the source temperature was 110 °C, and the desolvation temperature was 550 °C, and the desolvation gas flow was 1000 L/h. The capillary voltage was 2.5 kV (ESI−). At a low CE scan, the cone voltage was 50 V, and the collision energy was 4 eV. At a high CE scan, the cone voltage was 50 V, and the collision energy was ramping 30–50 eV. Leucine-enkephalin was used as lock mass. The instrument was controlled by UNIFI software (version 1.9.4, Waters Corp., Milford, MA, USA).
A UHPLC-CAD analysis was performed on the Thermo Vanquish UHPLC system (ThermoFisher Scientific, Germering, Bavaria, Germany). A Phenomenex C18 column (100 × 4.6 mm, 2.6 μm) was used with a column temperature at 25 °C. The mobile phases were water with 0.1% formic acid (A) and acetonitrile (B). The gradient used was as follows: (−5–0) min, 21% B; (0–6) min, 21% B; (6–10) min, 21%→29% B; (10–21) min, 29% B; (21–26) min, 29%→35% B; (26–32) min, 35%→53% B; (32–33) min, 53%→95% B; (33–36) min, 95% B; and (36–37) min, 95%→21% B. The flow rate was 1.0 mL/min. The injection volume of the sample was 10 μL.
3.4. Data Analysis by the UNIFI Informatics Platform
A database of the total chemical ingredients of the ginseng (304 compounds) was created for UNIFI 1.9.4 (Waters Corporation, Milford, MA, USA) on the basis of the result of the literature and some online databases or internet search engines, such as PubMed, Full-Text Database (CNKI), SciFinder, ChemSpider, Web of Science, and Medline [19,29,38,41,42]. The database included the compound names, molecular formulas, chemical structures, and fragment ions. Data analysis was accomplished by UNIFI 1.9.4., and the parameter setting is as follows: the maximum allowed number of peak detections was 1000 for 2D peak detection, the peak intensity threshold was 25 counts for high energy and 200 counts for low energy in the 3D peak detection. The mass and fragment errors were set to be 10 mDa for chemical identification, which would be the exactly predicted fragments from the structure. We also selected +HCOO−, −H, and −H + 2H2O as adducts in ESI− mode. Leucine-enkephalin was used as the reference compound to ensure the mass accuracy, and [M − H]− 554.2620 was used in the negative ion.
3.5. Multivariate Statistical Analysis
All data acquisition in MSE mode was in the continuum mode, and the raw data were processed by UNIFI 1.9.4. The data analysis included deconvolution, alignment, and data reduction to provide a list of mass and retention time pairs along with the corresponding peak areas for all detected peaks from each file in the data set. The processed data list was then imported by the PCA and OPLS-DA. All the test groups were discriminated in the PCA to investigate whether different groups could be separated. The parameters used in the analysis were 0–27 min for the retention time range, 100–1500 Da for the mass range, 0.02 Da for the mass tolerance, and 0.10 min for the retention time tolerance. The isotopic peaks were excluded for analysis. Then, OPLS-DA was carried out to discriminate ions contributing to the classification among the samples. The results were visualized in a score plot to show the group clusters and an S plot to show the variables that contribute to the classification.
4. Conclusions
A strategy based on UHPLC-Q-TOF/MS coupled with the UNIFI informatics platform was developed to effectively profile and characterize of ginsenosides in four parts of P. ginseng root. One hundred and five ginsenosides including 16 new compounds were identified or tentatively characterized. Among them, 83, 101, 99, and 96 ginsenosides were tentatively characterized in the MR, FR, BR, and RH, respectively. UHPLC-Q-TOF/MS analysis combined with multivariate statistical analysis showed an obvious difference between fibrous root and other parts. A total of 22 (20, 17, and 19 markers for MR and FR, MR and BR, and MR and RH, respectively) robust known chemical markers were identified and they were with lower contents in MR samples than other parts. The markers between MR and FR, MR and BR, and MR and RH groups included PPT-types and PPD-types. In addition, two OA-type ginsenosides (ginsenoside Ro and chikusetsusaponin Iva) were higher in RH samples than MR samples. Finally, the UHPLC-CAD semiquantitative results showed that the relative contents of Re, Rb1, 20(R)-Rh1, Rd, and Rf were highest in FR, followed by RH, BR, and MR. The relative content of Rg1 was highest in RH and the total content of pharmacopoeia indicators Rg1, Re, and Rb1 was highest in FR. Under our research conditions, the range of Rg1/Re was from 0.19 to 0.64 in FR samples, while the range of Rg1/Re was from 0.75 to 2.00 in MR samples. The peak area ratio of Rg1 and Re might be the marker of MR and FR samples.
This study systematically revealed the differences of ginsenoside components in the different parts of cultivated ginseng root. These parts were all rich in ginsenosides and contained similar structural types. The differences among them were in the compositions and relative contents of ginsenosides. Fibrous roots showed rich ingredients and high ginsenosides contents which should be further utilized. The research results provided a basis for the rational development and utilization of ginseng root.
Supplementary Materials
The following are available online, Figure S1: High collision energy ESI-MS spectra of peaks 51(A), 54(B), 59(C), 75(D), 80(E), and 83(F), Table S1: Chemical structures of the detected compounds in different parts of P. ginseng root, Table S2: Detailed information of the tested P. ginseng whole root samples.
Author Contributions
Conceptualization, B.G. and B.M.; resources, J.P. and B.G.; data curation, J.P.; writing—original draft preparation, J.P.; writing—review and editing, B.M., B.G., W.Z., X.P., J.Z., X.C., M.Y., and K.Y.; project administration, B.M.; funding acquisition, B.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science and Technology Major Project (2017ZX09301072) and the National Natural Science Foundation of China (82074008).
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to the protection for the former research of this study.
Conflicts of Interest
The authors declare no conflict of interest concerning this article.
Sample Availability
Samples of the compounds are not available from the authors.
References
- Yun, T.K. Brief introduction of Panax ginseng CA Meyer. J. Korean Med. Sci. 2001, 16, S3–S5. [Google Scholar] [CrossRef]
- Choi, K.-T. Botanical characteristics, pharmacological effects and medicinal components of Korean Panax ginseng CA Meyer. Acta Pharmacol. Sin. 2008, 29, 1109–1118. [Google Scholar] [CrossRef]
- Yue, P.Y.K.; Mak, N.K.; Cheng, Y.K.; Leung, K.W.; Ng, T.B.; Fan, D.T.P.; Yeung, H.W.; Wong, R.N.S. Pharmacogenomics and the Yin/Yang actions of ginseng: Anti-tumor, angiomodulating and steroid-like activities of ginsenosides. Chin. Med. 2007, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Kenarova, B.; Neychev, H.; Hadjiivanova, C.; Petkov, V.D. Immunomodulating activity of ginsenoside Rg1 from Panax ginseng. Jpn. J. Pharmcol. 1990, 54, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Qu, S.; Sui, D.; Yu, X.; Lv, Z. Effects of ginsenoside-Rb on blood lipid metabolism and anti-oxidation in hyperlipidemia rats. Zhongguo Zhong Yao Za Zhi 2004, 29, 1085–1088. (In Chinese) [Google Scholar]
- Cheng, Y.; Shen, L.-H.; Zhang, J.-T. Anti-amnestic and anti-aging effects of ginsenoside Rg1 and Rb1 and its mechanism of action. Acta Pharmacol. Sin. 2005, 26, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.P.; Zhang, Y.B.; Yang, X.W.; Yang, X.B.; Xu, W.; Xu, F.; Cai, S.Q.; Wang, Y.P.; Xu, Y.H.; Zhang, L.X. High-performance liquid chromatography with diode array detector and electrospray ionization ion trap time-of-flight tandem mass spectrometry to evaluate ginseng roots and rhizomes from different regions. Molecules 2016, 21, 603. [Google Scholar] [CrossRef]
- Zhou, Q.L.; Zhu, D.N.; Yang, X.W.; Xu, W.; Wang, Y.P. Development and validation of a UFLC-MS/MS method for simultaneous quantification of sixty-six saponins and their six aglycones: Application to comparative analysis of red ginseng and white ginseng. J. Pharm. Biomed. Anal. 2018, 159, 153–165. [Google Scholar] [CrossRef]
- Qiu, S.; Yang, W.Z.; Yao, C.L.; Qiu, Z.D.; Shi, X.J.; Zhang, J.X.; Hou, J.J.; Wang, Q.R.; Wu, W.Y.; Guo, D.A. Nontargeted metabolomic analysis and “commercial-homophyletic” comparison-induced biomarkers verification for the systematic chemical differentiation of five different parts of Panax ginseng. J. Chromatogr. A 2016, 1453, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.W.; Li, D.K.; Wang, T.; Wu, Y.C.; Zhao, Y.; Zhou, D.Z.; Zhang, T.; Ye, Z.L. Application of metabolomics approach to study of different parts of Mountain Cultivated Ginseng using UHPLC-QTOF/MS. Acta Pharm. Sin. B 2016, 51, 1609–1615. [Google Scholar]
- Chen, Y.; Zhao, Z.; Chen, H.; Brand, E.; Yi, T.; Qin, M.; Liang, Z. Determination of ginsenosides in Asian and American ginsengs by liquid chromatography-quadrupole/time-of-flight MS: Assessing variations based on morphological characteristics. J. Ginseng Res. 2017, 41, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Chen, Y.; Xu, L.; Qin, M.; Yi, T.; Chen, H.; Zhao, Z. Localization of ginsenosides in the rhizome and root of Panax ginseng by laser microdissection and liquid chromatography-quadrupole/time of flight-mass spectrometry. J. Pharm. Biomed. Anal. 2015, 105, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.X.; Li, M.; Qiao, L.R.; Yao, Z.H.; Li, C.; Shen, X.Y.; Wang, Y.; Yu, K.; Yao, X.S.; Dai, Y. Rapid characterization of Ziziphi Spinosae Semen by UPLC/Qtof MS with novel informatics platform and its application in evaluation of two seeds from Ziziphus species. J. Pharm. Biomed. Anal. 2016, 122, 59–80. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, N.; Wang, Z.; Qi, Z.; Zhu, H.; Zheng, B.; Li, P.; Liu, J. Nontargeted metabolomic analysis of four different parts of Platycodon grandiflorum GROWN in Northeast China. Molecules 2017, 22, 1280. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, C.; Lin, H.; Liu, Y.; Li, Y.; Zhao, Y.; Li, P.; Liu, J. Discovery of the potential biomarkers for discrimination between Hedyotis diffusa and Hedyotis corymbosa by UPLC-QTOF/MS metabolome analysis. Molecules 2018, 23, 1525. [Google Scholar] [CrossRef]
- Schilling, K.; Holzgrabe, U. Recent applications of the Charged Aerosol Detector for liquid chromatography in drug quality control. J. Chromatogr. A 2020, 1619, 460911. [Google Scholar] [CrossRef]
- Almeling, S.; Ilko, D.; Holzgrabe, U. Charged aerosol detection in pharmaceutical analysis. J. Pharm. Biomed. Anal. 2012, 69, 50–63. [Google Scholar] [CrossRef]
- Kim, N.; Kim, K.; Lee, D.; Shin, Y.S.; Bang, K.H.; Cha, S.W.; Lee, J.W.; Choi, H.K.; Hwang, B.Y.; Lee, D. Nontargeted metabolomics approach for age differentiation and structure interpretation of age-dependent key constituents in hairy roots of Panax ginseng. J. Nat. Prod. 2012, 75, 1777–1784. [Google Scholar] [CrossRef]
- Qiu, S.; Yang, W.Z.; Shi, X.J.; Yao, C.L.; Yang, M.; Liu, X.; Jiang, B.H.; Wu, W.Y.; Guo, D.A. A green protocol for efficient discovery of novel natural compounds: Characterization of new ginsenosides from the stems and leaves of Panax ginseng as a case study. Anal. Chim. Acta 2015, 893, 65–76. [Google Scholar] [CrossRef]
- Zhu, H.; Lin, H.; Tan, J.; Wang, C.; Wang, H.; Wu, F.; Dong, Q.; Liu, Y.; Li, P.; Liu, J. UPLC-QTOF/MS-Based nontargeted metabolomic analysis of Mountain- and Garden-Cultivated ginseng of different ages in Northeast China. Molecules 2018, 24, 33. [Google Scholar] [CrossRef]
- Lee, J.W.; Choi, B.R.; Kim, Y.C.; Choi, D.J.; Lee, Y.S.; Kim, G.S.; Baek, N.I.; Kim, S.Y.; Lee, D.Y. Comprehensive profiling and quantification of ginsenosides in the root, stem, leaf, and berry of Panax ginseng by UPLC-QTOF/MS. Molecules 2017, 22, 2147. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Ji, S.H.; Lee, Y.S.; Choi, D.J.; Choi, B.R.; Kim, G.S.; Baek, N.I.; Lee, D.Y. Mass spectrometry based profiling and imaging of various ginsenosides from Panax ginseng roots at different ages. Int. J. Mol. Sci. 2017, 18, 1114. [Google Scholar] [CrossRef]
- Dan, M.; Su, M.; Gao, X.; Zhao, T.; Zhao, A.; Xie, G.; Qiu, Y.; Zhou, M.; Liu, Z.; Jia, W. Metabolite profiling of Panax notoginseng using UPLC-ESI-MS. Phytochemistry 2008, 69, 2237–2244. [Google Scholar] [CrossRef]
- Wang, L.L.; Han, L.F.; Yu, H.S.; Sang, M.M.; Liu, E.W.; Zhang, Y.; Fang, S.M.; Wang, T.; Gao, X.M. Analysis of the constituents in “Zhu She Yong Xue Shuan Tong” by ultra high performance liquid chromatography with quadrupole time-of-flight mass spectrometry combined with preparative high performance liquid chromatography. Molecules 2015, 20, 20518–20537. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Li, J.; Zhang, X.; Pei, J.; Huang, L. An integrated LC-MS-Based strategy for the quality assessment and discrimination of three Panax species. Molecules 2018, 23, 2988. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Chen, Y.; Liang, J.; Wang, C.Z.; Liu, X.; Yan, Z.; Tang, Y.; Li, J.; Yuan, C.S. Component analysis and target cell-based neuroactivity screening of Panax ginseng by ultra-performance liquid chromatography coupled with quadrupole-time-of-flight mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2016, 1038, 1–11. [Google Scholar] [CrossRef]
- Wu, W.; Sun, L.; Zhang, Z.; Guo, Y.; Liu, S. Profiling and multivariate statistical analysis of Panax ginseng based on ultra-high-performance liquid chromatography coupled with quadrupole-time-of-flight mass spectrometry. J. Pharm. Biomed. Anal. 2015, 107, 141–150. [Google Scholar] [CrossRef]
- Liang, C.; Ding, Y.; Nguyen, H.T.; Kim, J.A.; Boo, H.J.; Kang, H.K.; Nguyen, M.C.; Kim, Y.H. Oleanane-type triterpenoids from Panax stipuleanatus and their anticancer activities. Bioorganic Med. Chem. Lett. 2010, 20, 7110–7115. [Google Scholar] [CrossRef]
- Yang, W.Z.; Hu, Y.; Wu, W.Y.; Ye, M.; Guo, D.A. Saponins in the genus Panax L. (Araliaceae): A systematic review of their chemical diversity. Phytochemistry 2014, 106, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Yang, W.Z.; Yao, C.L.; Shi, X.J.; Li, J.Y.; Lou, Y.; Duan, Y.N.; Wu, W.Y.; Guo, D.A. Malonylginsenosides with Potential antidiabetic activities from the flower buds of Panax ginseng. J. Nat. Prod. 2017, 80, 899–908. [Google Scholar] [CrossRef]
- Zhu, G.Y.; Li, Y.W.; Hau, D.K.; Jiang, Z.H.; Yu, Z.L.; Fong, W.F. Protopanaxatriol-type ginsenosides from the root of Panax ginseng. J. Agric. Food Chem. 2011, 59, 200–205. [Google Scholar] [CrossRef]
- Yang, W.Z.; Ye, M.; Qiao, X.; Liu, C.F.; Miao, W.J.; Bo, T.; Tao, H.Y.; Guo, D.A. A strategy for efficient discovery of new natural compounds by integrating orthogonal column chromatography and liquid chromatography/mass spectrometry analysis: Its application in Panax ginseng, Panax quinquefolium and Panax notoginseng to characterize 437 potential new ginsenosides. Anal. Chim. Acta 2012, 739, 56–66. [Google Scholar] [PubMed]
- Mao, Q.; Yang, J.; Cui, X.M.; Li, J.J.; Qi, Y.T.; Zhang, P.H.; Wang, Q. Target separation of a new anti-tumor saponin and metabolic profiling of leaves of Panax notoginseng by liquid chromatography with eletrospray ionization quadrupole time-of-flight mass spectrometry. J. Pharm. Biomed. Anal. 2012, 59, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Zhu, H.; Tan, J.; Wang, C.; Dong, Q.; Wu, F.; Wang, H.; Liu, J.; Li, P.; Liu, J. Comprehensive Investigation on metabolites of wild-simulated American Ginseng root based on ultra-high-performance liquid chromatography-quadrupole time-of-flight mass spectrometry. J. Agric. Food Chem. 2019, 67, 5801–5819. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Sugimoto, S.; Matsuda, H.; Yoshikawa, M. Medicinal flowers. XVII. New dammarane-type triterpene glycosides from flower buds of American ginseng, Panax quinquefolium L. Chem. Pharm. Bull. 2007, 55, 1342–1348. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Murakami, T.; Yashiro, K.; Yamahara, J.; Matsuda, H.; Saijoh, R.; Tanaka, O. Bioactive saponins and glycosides. XI. Structures of new dammarane-type triterpene oligoglycosides, quinquenosides I, II, III, IV, and V, from American ginseng, the roots of Panax quinquefolium L. Chem. Pharm. Bull. 1998, 46, 647–654. [Google Scholar] [CrossRef]
- Zhu, G.Y.; Li, Y.W.; Hau, D.K.; Jiang, Z.H.; Yu, Z.L.; Fong, W.F. Acylated protopanaxadiol-type ginsenosides from the root of Panax ginseng. Chem. Biodivers. 2011, 8, 1853–1863. [Google Scholar] [CrossRef]
- Wu, W.; Jiao, C.; Li, H.; Ma, Y.; Jiao, L.; Liu, S. LC-MS based metabolic and metabonomic studies of Panax ginseng. Phytochem. Anal. 2018, 29, 331–340. [Google Scholar] [CrossRef]
- Lee, S.M.; Shon, H.J.; Choi, C.S.; Hung, T.M.; Min, B.S.; Bae, K. Ginsenosides from heat processed ginseng. Chem. Pharm. Bull. 2009, 57, 92–94. [Google Scholar] [CrossRef]
- Wang, J.R.; Yau, L.F.; Gao, W.N.; Liu, Y.; Yick, P.W.; Liu, L.; Jiang, Z.H. Quantitative comparison and metabolite profiling of saponins in different parts of the root of Panax notoginseng. J. Agric. Food Chem. 2014, 62, 9024–9034. [Google Scholar] [CrossRef]
- Wang, Y.; Choi, H.K.; Brinckmann, J.A.; Jiang, X.; Huang, L. Chemical analysis of Panax quinquefolius (North American ginseng): A review. J. Chromatogr. A 2015, 1426, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.L.; Pan, H.Q.; Wang, H.; Yao, S.; Yang, W.Z.; Hou, J.J.; Jin, Q.H.; Wu, W.Y.; Guo, D.A. Global profiling combined with predicted metabolites screening for discovery of natural compounds: Characterization of ginsenosides in the leaves of Panax notoginseng as a case study. J. Chromatogr. A 2018, 1538, 34–44. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).