Zymography for Picogram Detection of Lipase and Esterase Activities
Abstract
1. Introduction
2. Results
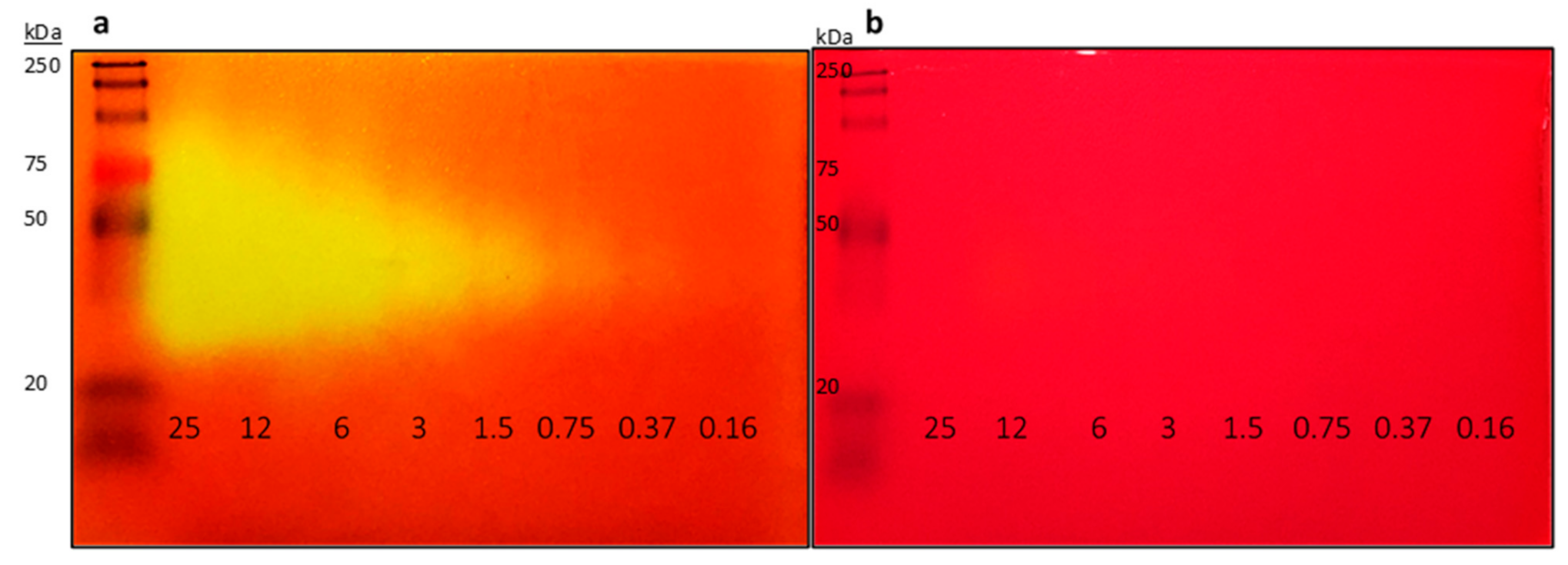
2.1. Improvement of Zymogram Detection Limit to Enable Picogram Detection of Lipases
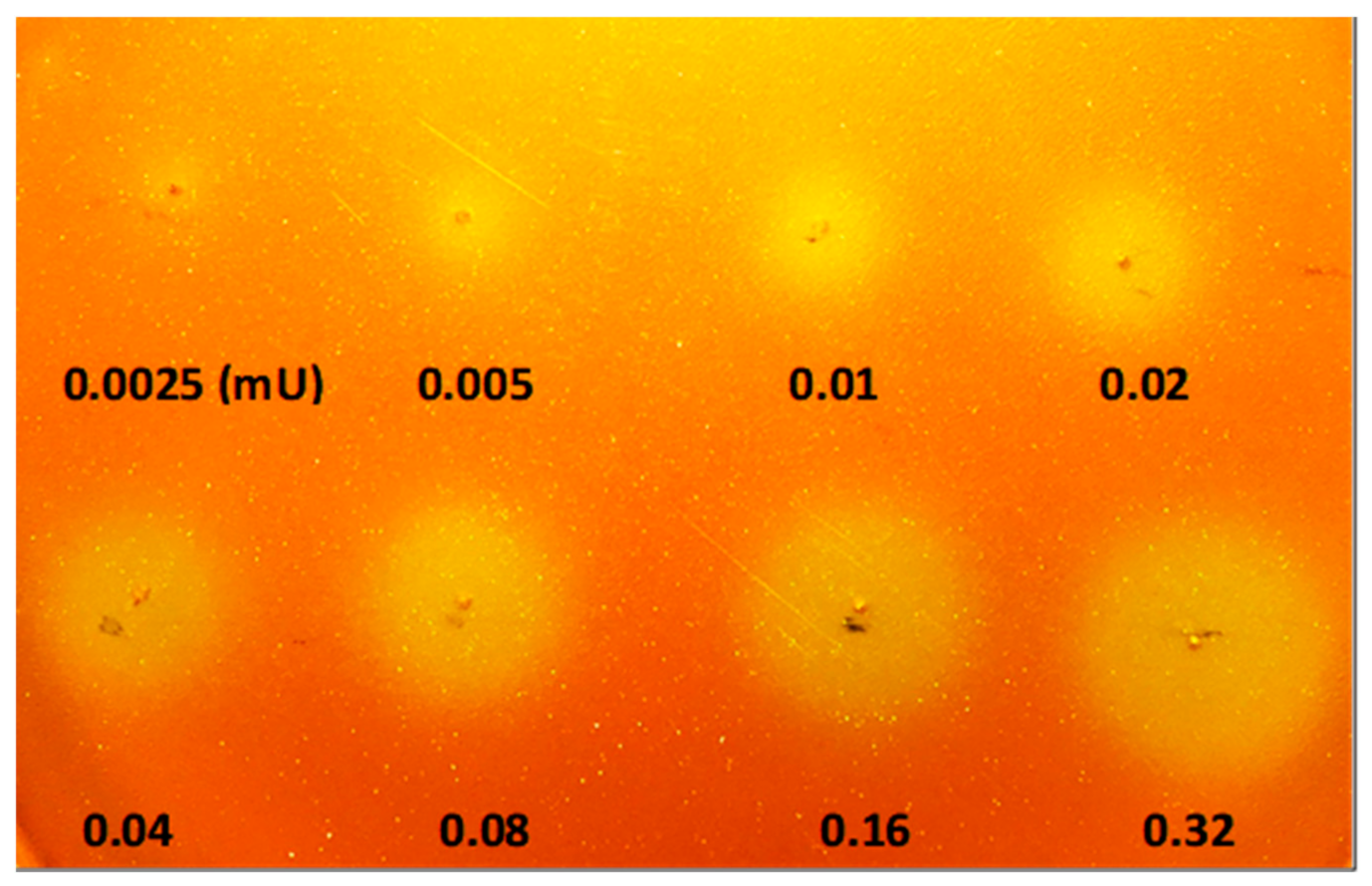
2.2. Calibration of Lipolytic Activity Detection Limit on Chromogenic Agar Plate
2.3. Optimisation of Buffer Strength Effect on the Detection Sensitivity
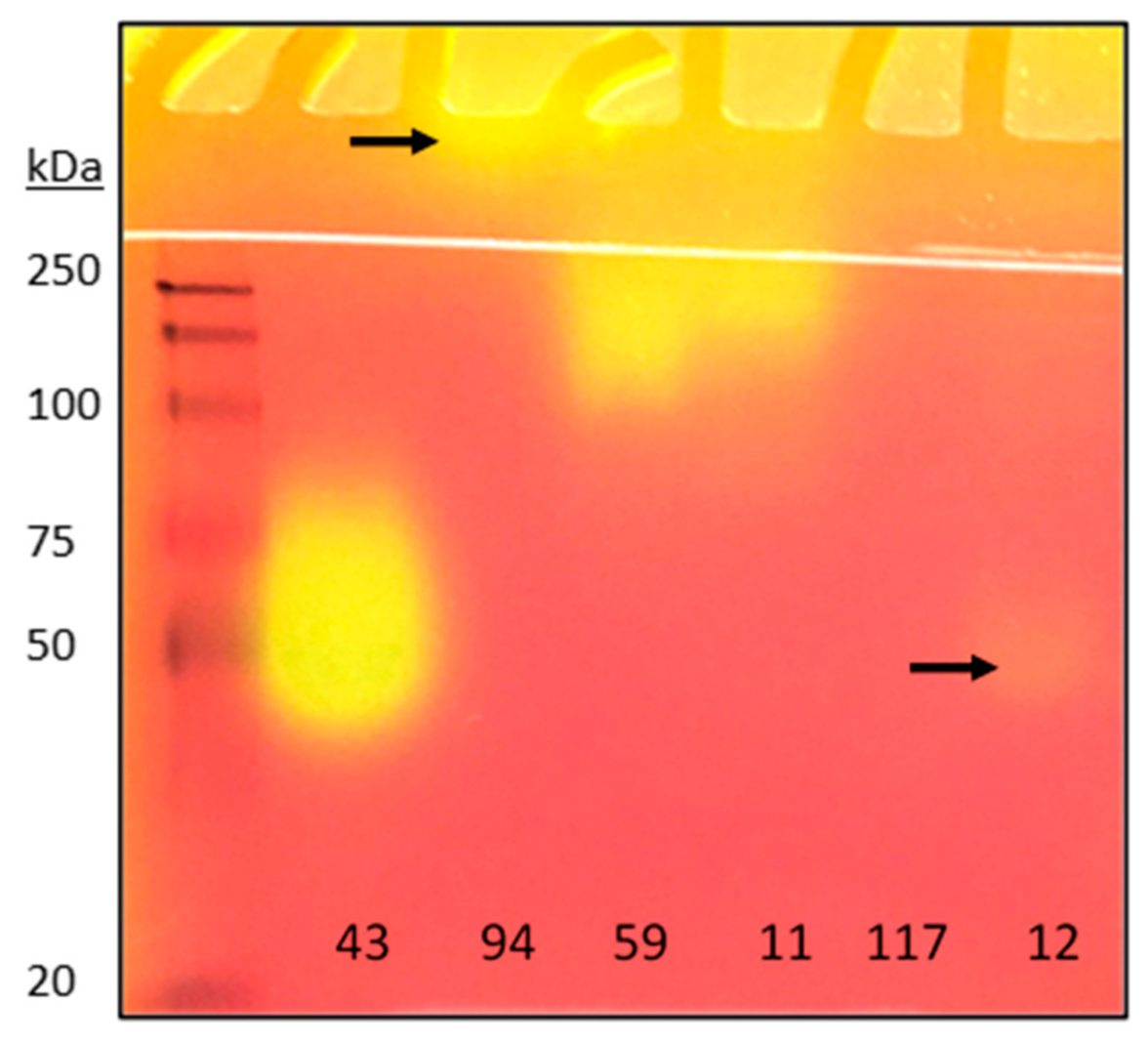
2.4. Detection of Lipases from Crude Microbial Cultures
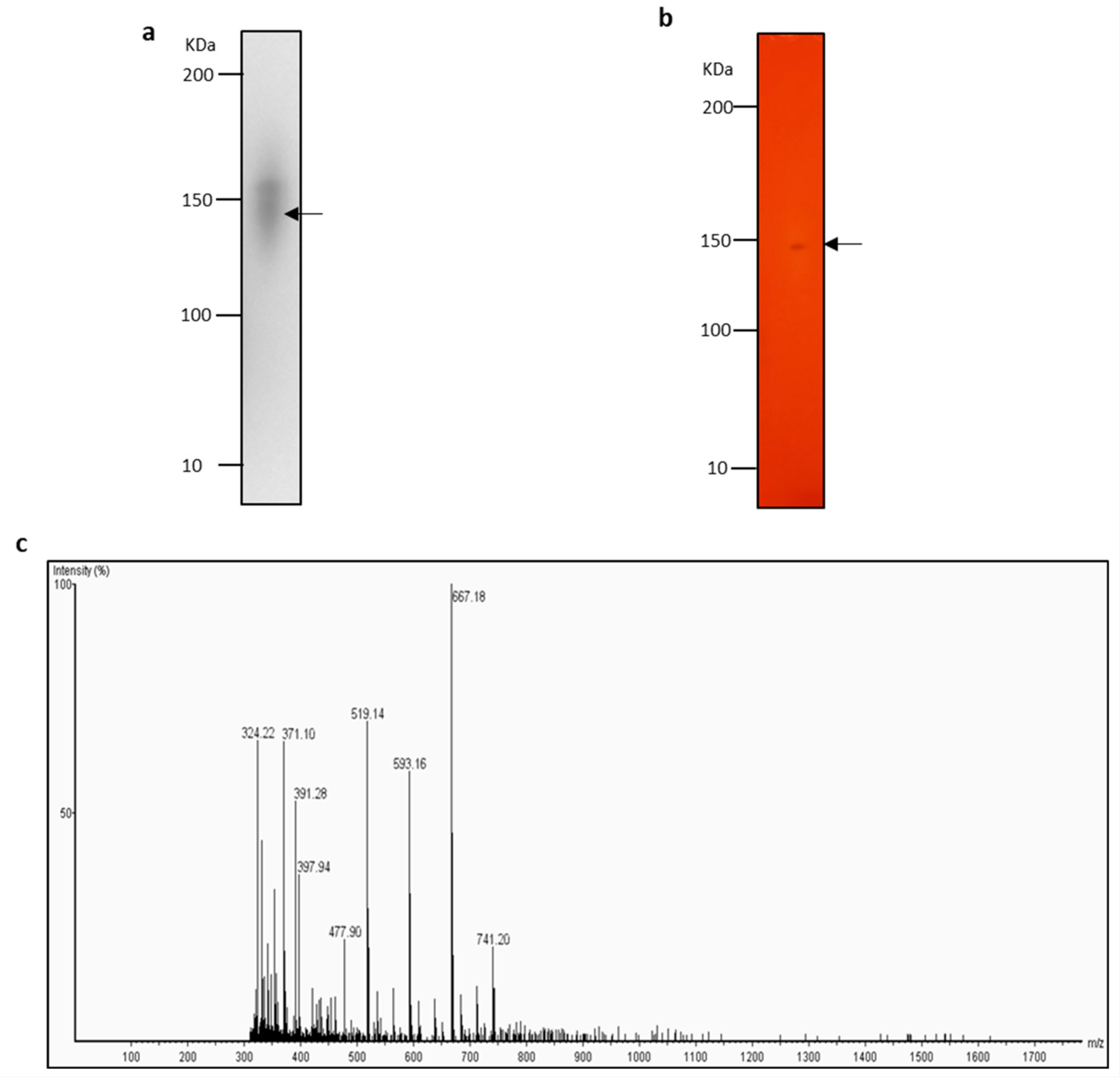
2.5. Identification of Lipase with Zymogram-LC-MS
3. Discussion
4. Materials and Methods
4.1. Isolation and Cultivation of Microbial Strains
4.2. Detection of Lipase Activity via Chromogenic Agar Plates
4.3. Detection of Lipase Activity by Zymography
4.4. Peptide Mass Fingerprinting with LC-MS
4.5. Data Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bolsoni-Lopes, A.; Alonso-Vale, M.I. Lipolysis and lipases in white adipose tissue-An update. Arch. Endocrinol. Metab. 2015, 59, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Krajmalnik-Brown, R.; Ilhan, Z.E.; Kang, D.W.; DiBaise, J.K. Effects of gut microbes on nutrient absorption and energy regulation. Nutr. Clin. Pract. 2012, 27, 201–214. [Google Scholar] [CrossRef]
- Gacser, A.; Stehr, F.; Kroger, C.; Kredics, L.; Schafer, W.; Nosanchuk, J.D. Lipase 8 affects the pathogenesis of Candida albicans. Infect. Immun. 2007, 75, 4710–4718. [Google Scholar] [CrossRef] [PubMed]
- Karigar, C.S.; Rao, S.S. Role of microbial enzymes in the bioremediation of pollutants: A review. Enzyme Res. 2011, 2011, 805187. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Uyama, H.; Takamoto, T. Lipase-catalyzed degradation of polyesters in organic solvents. A new methodology of polymer recycling using enzyme as catalyst. Biomacromolecules 2000, 1, 3–5. [Google Scholar] [CrossRef]
- Zhang, K.P.; Lai, J.Q.; Huang, Z.L.; Yang, Z. Penicillium expansum lipase-catalyzed production of biodiesel in ionic liquids. Bioresour. Technol. 2011, 102, 2767–2772. [Google Scholar] [CrossRef]
- Zechner, R.; Zimmermann, R.; Eichmann, T.O.; Kohlwein, S.D.; Haemmerle, G.; Lass, A.; Madeo, F. FAT SIGNALS--lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012, 15, 279–291. [Google Scholar] [CrossRef]
- Fojan, P.; Jonson, P.H.; Petersen, M.T.; Petersen, S.B. What distinguishes an esterase from a lipase: A novel structural approach. Biochimie 2000, 82, 1033–1041. [Google Scholar] [CrossRef]
- Hasan, F.; Shah, A.A.; Hameed, A. Industrial applications of microbial lipases. Enzyme Microb. Technol. 2006, 39, 235–251. [Google Scholar] [CrossRef]
- Houde, A.; Kademi, A.; Leblanc, D. Lipases and their industrial applications: An overview. Appl. Biochem. Biotechnol. 2004, 118, 155–170. [Google Scholar] [CrossRef]
- Kumar, A.; Dhar, K.; Kanwar, S.S.; Arora, P.K. Lipase catalysis in organic solvents: Advantages and applications. Biol. Proced. Online 2016, 18, 2. [Google Scholar] [CrossRef] [PubMed]
- Jette, J.F.; Ziomek, E. Determination of lipase activity by a rhodamine-triglyceride-agarose assay. Anal. Biochem. 1994, 219, 256–260. [Google Scholar] [CrossRef]
- Kouker, G.; Jaeger, K.E. Specific and sensitive plate assay for bacterial lipases. Appl. Environ. Microbiol. 1987, 53, 211–213. [Google Scholar] [CrossRef]
- Farnet, A.M.; Qasemian, L.; Goujard, L.; Gil, G.; Guiral, D.; Ruaudel, F.; Ferre, E. A modified method based on p-nitrophenol assay to quantify hydrolysis activities of lipases in litters. Soil Biol. Biochem. 2010, 42, 386–389. [Google Scholar] [CrossRef]
- Gupta, N.; Rathi, P.; Gupta, R. Simplified para-nitrophenyl palmitate assay for lipases and esterases. Anal. Biochem. 2002, 311, 98–99. [Google Scholar] [CrossRef]
- Jacks, T.J.; Kircher, H.W. Fluorometric assay for the hydrolytic activity of lipase using fatty acyl esters of 4-methylumbelliferone. Anal. Biochem. 1967, 21, 279–285. [Google Scholar] [CrossRef]
- Margesin, R.; Feller, G.; Hämmerle, M.; Stegner, U.; Schinner, F. A colorimetric method for the determination of lipase activity in soil. Biotechnol. Lett. 2002, 24, 27–33. [Google Scholar] [CrossRef]
- Roberts, I.M. Hydrolysis of 4-methylumbelliferyl butyrate: A convenient and sensitive fluorescent assay for lipase activity. Lipids 1985, 20, 243–247. [Google Scholar] [CrossRef]
- Roy, R.N. Fluorimetric assay of the activity of extracellular lipases of Pseudomonas fluorescens and Serratia marcescens. J. Appl. Bacteriol. 1980, 49, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Schultz, N.; Hobley, T.J.; Syldatk, C. Spectrophotometric assay for online measurement of the activity of lipase immobilised on micro-magnetic particles. Biotechnol. Lett. 2007, 29, 365–371. [Google Scholar] [CrossRef]
- Singh, R.; Gupta, N.; Goswami, V.K.; Gupta, R. A simple activity staining protocol for lipases and esterases. Appl. Microbiol. Biotechnol. 2006, 70, 679–682. [Google Scholar] [CrossRef]
- Yadav, R.P.; Saxena, R.K.; Gupta, R.; Davidson, W.S. Rapid zymogram for lipase. Biotechniques 1998, 24, 754–756. [Google Scholar] [CrossRef]
- Bornscheuer, U.T.; Altenbuchner, J.; Meyer, H.H. Directed evolution of an esterase: Screening of enzyme libraries based on pH-indicators and a growth assay. Bioorg. Med. Chem. 1999, 7, 2169–2173. [Google Scholar] [CrossRef]
- Snellman, E.A.; Sullivan, E.R.; Colwell, R.R. Purification and properties of the extracellular lipase, LipA, of Acinetobacter sp. RAG-1. Eur. J. Biochem. 2002, 269, 5771–5779. [Google Scholar] [CrossRef]
- Welshons, W.V.; Wolf, M.F.; Murphy, C.S.; Jordan, V.C. Estrogenic activity of phenol red. Mol. Cell Endocrinol. 1988, 57, 169–178. [Google Scholar] [CrossRef]
- Chahinian, H.; Nini, L.; Boitard, E.; Dubes, J.P.; Comeau, L.C.; Sarda, L. Distinction between esterases and lipases: A kinetic study with vinyl esters and TAG. Lipids 2002, 37, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Duarte, J.G.; Leone-Ignacio, K.; da Silva, J.A.C.; Fernandez-Lafuente, R.; Freire, D.M.G. Rapid determination of the synthetic activity of lipases/esterases via transesterification and esterification zymography. Fuel 2016, 177, 123–129. [Google Scholar] [CrossRef]
- Kwon, M.A.; Kim, H.S.; Hahm, D.H.; Song, J.K. Synthesis activity-based zymography for detection of lipases and esterases. Biotechnol. Lett. 2011, 33, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.P.; Saxena, R.K.; Gupta, R.; Davidson, S. Lipase production by Aspergillus and Penicillium species. Folia Microbiol. 1998, 43, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Ko, M.K.; Jeon, W.B.; Kim, K.S.; Lee, H.H.; Seo, H.H.; Kim, Y.S.; Oh, B.J. A Colletotrichum gloeosporioides-induced esterase gene of nonclimacteric pepper (Capsicum annuum) fruit during ripening plays a role in resistance against fungal infection. Plant. Mol. Biol 2005, 58, 529–541. [Google Scholar] [CrossRef]
- Venkatesagowda, B.; Ebenezer, P. Optimization of extracellular lipase production in Colletotrichum gloeosporioides by solid state fermentation. Ind. J. Sci. Technol. 2008, 1, 1–8. [Google Scholar]
- Denise, S.; Livia, T.A.S.; Jamil, S.O.; Marcelo, M.S.; Inayara, C.A.L.; Gecernir, C.; Jacqueline, A.T. Colletotrichum gloeosporioides lipase: Characterization and use in hydrolysis and esterifications. Afr. J. Microbiol. Res. 2015, 9, 1322–1330. [Google Scholar] [CrossRef]
- Bandmann, N.; Collet, E.; Leijen, J.; Uhlen, M.; Veide, A.; Nygren, P.A. Genetic engineering of the Fusarium solani pisi lipase cutinase for enhanced partitioning in PEG-phosphate aqueous two-phase systems. J. Biotechnol. 2000, 79, 161–172. [Google Scholar] [CrossRef]
- Jallouli, R.; Parsiegla, G.; Carriere, F.; Gargouri, Y.; Bezzine, S. Efficient heterologous expression of Fusarium solani lipase, FSL2, in Pichia pastoris, functional characterization of the recombinant enzyme and molecular modeling. Int. J. Biol. Macromol. 2017, 94, 61–71. [Google Scholar] [CrossRef]
- Maia, M.M.; Heasley, A.; Camargo de Morais, M.M.; Melo, E.H.; Morais, M.A., Jr.; Ledingham, W.M.; Lima Filho, J.L. Effect of culture conditions on lipase production by Fusarium solani in batch fermentation. Bioresour. Technol. 2001, 76, 23–27. [Google Scholar] [CrossRef]
- Martinez, C.; De Geus, P.; Lauwereys, M.; Matthyssens, G.; Cambillau, C. Fusarium solani cutinase is a lipolytic enzyme with a catalytic serine accessible to solvent. Nature 1992, 356, 615–618. [Google Scholar] [CrossRef] [PubMed]
- Wongwatanapaiboon, J.; Malilas, W.; Ruangchainikom, C.; Thummadetsak, G.; Chulalaksananukul, S.; Marty, A.; Chulalaksananukul, W. Overexpression of Fusarium solani lipase in Pichia pastoris and its application in lipid degradation. Biotechnol. Biotechnol. Equip. 2016, 30, 885–893. [Google Scholar] [CrossRef]
- Egmond, M.R.; de Vlieg, J. Fusarium solani pisi cutinase. Biochimie 2000, 82, 1015–1021. [Google Scholar] [CrossRef]
- Dai, D.; Xia, L. Enhanced production of Penicillium expansum PED-03 lipase through control of culture conditions and application of the crude enzyme in kinetic resolution of racemic Allethrolone. Biotechnol. Prog. 2005, 21, 1165–1168. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.-Q.; Hu, Z.-L.; Sheldon, R.A.; Yang, Z. Catalytic performance of cross-linked enzyme aggregates of Penicillium expansum lipase and their use as catalyst for biodiesel production. Process. Biochemistry 2012, 47, 2058–2063. [Google Scholar] [CrossRef]
- Li, N.-W.; Zong, M.-H.; Wu, H. Highly efficient transformation of waste oil to biodiesel by immobilized lipase from Penicillium expansum. Process. Biochemistry 2009, 44, 685–688. [Google Scholar] [CrossRef]
- Stocklein, W.; Sztajer, H.; Menge, U.; Schmid, R.D. Purification and properties of a lipase from Penicillium expansum. Biochim. Biophys. Acta 1993, 1168, 181–189. [Google Scholar] [CrossRef]
- Coradi, G.V.; da Visitação, V.L.; de Lima, E.A.; Saito, L.Y.T.; Palmieri, D.A.; Takita, M.A.; de Oliva Neto, P.; de Lima, V.M.G. Comparing submerged and solid-state fermentation of agro-industrial residues for the production and characterization of lipase by Trichoderma harzianum. Ann. Microbiol. 2012, 63, 533–540. [Google Scholar] [CrossRef]
- Toscano, L.; Montero, G.; Cervantes, L.; Stoytcheva, M.; Gochev, V.; Beltrán, M. Production and Partial Characterization of Extracellular Lipase fromTrichoderma Harzianumby Solid-State Fermentation. Biotechnol. Biotechnol. Equip. 2014, 27, 3776–3781. [Google Scholar] [CrossRef]
- Ulker, S.; Özel, A.; Çolak, A.; Alpay Karaoğlu, Ş. Isolation, production, and characterization of an extracellular lipase from Trichoderma harzianum isolated from soil. Turk. J. Biol. 2011, 35, 543–550. [Google Scholar]
- Elad, Y.; Chet, I.; Henis, Y. Degradation of plant pathogenic fungi by Trichoderma harzianum. Can. J. Microbiol. 1982, 28, 719–725. [Google Scholar] [CrossRef]
- Benhamou, N.; Chet, I. Parasitism of sclerotia of Sclerotium rolfsii by Trichoderma harzianum: Ultrastructural and cytochemical aspects of the interaction. Phytopathology 1996, 86, 405–416. [Google Scholar] [CrossRef]
- Sheludchenko, M.S.; Kolomytseva, M.P.; Travkin, V.M.; Akimov, V.N.; Golovleva, L.A. [Degradation of aniline by Delftia tsuruhatensis 14S in batch and continuous processes]. Prikl. Biokhim. Mikrobiol. 2005, 41, 530–534. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. SDS-Polyacrylamide Gel Electrophoresis of Proteins. CSH Protoc 2006, 2006. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Michalski, A.; Damoc, E.; Hauschild, J.P.; Lange, O.; Wieghaus, A.; Makarov, A.; Nagaraj, N.; Cox, J.; Mann, M.; Horning, S. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell Proteomics 2011, 10, M111.011015. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Zhang, K.; Hendrie, C.; Liang, C.; Li, M.; Doherty-Kirby, A.; Lajoie, G. PEAKS: Powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 2337–2342. [Google Scholar] [CrossRef] [PubMed]
| Lipolase Amount (mU) | MOPS Concentration (mM) | ||||
|---|---|---|---|---|---|
| 5 | 1 | 0.5 | 0.25 | 0.1 | |
| 180 | 7 | 9 | 11.5 | 12.5 | 15 |
| 90 | 6 | 8 | 11 | 11.5 | 14 |
| 45 | 4.5 | 7 | 9.5 | 11 | 13 |
| 22.5 | 5 | 6 | 9 | 10 | 12 |
| 11.2 | 4.5 | 6 | 8.5 | 9 | 11 |
| 5.6 | 3 | 5.5 | 8 | 8.5 | 10 |
| 2.8 | - * | 5 | 7 | 8 | 9.5 |
| 1.4 | - | 4 | 6 | 7 | 8.5 |
| No. | Accession (UniProt) | Mass | Retention Time | Peptide |
|---|---|---|---|---|
| 1 | O59952|LIP_THELA | 760.4079 | 8.33 | SVADTLR |
| 2 | 1164.625 | 8.67 | ITHTNDIVPR | |
| 3 | 928.5342 | 9.61 | SGTLVPVTR | |
| 4 | 2192.069 | 11.23 | PPREFGYSHSSPEYWIK | |
| 5 | 1630.7263 | 12.8 | GN(+98)GYDIDVFSYGAPR | |
| 6 | 1661.841 | 14.57 | SIENWIGNLNFDLK |
| No. | Accession (UniProt) | Coverage (%) | No. of Peptides | No. of Unique Peptides | Protein Description |
|---|---|---|---|---|---|
| 1 | P00761|TRYP_PIG | 44 | 20 | 16 | Trypsin |
| 2 | A5A6M6|K2C1_PANTR | 15 | 7 | 5 | Keratin type II cytoskeletal 1 |
| 3 | P04264|K2C1_HUMAN | 15 | 7 | 5 | Keratin type II cytoskeletal 1 |
| 4 | O59952|LIP_THELA | 25 | 6 | 6 | Lipase |
| 5 | Q6XQI4|L_JUNIN | 1 | 2 | 2 | RNA-directed RNA polymerase L |
| 6 | Q0DKN3|VLN1_ORYSJ | 7 | 2 | 2 | Villin-1 |
| 7 | Q9LG11|HAC4_ARATH | 4 | 2 | 2 | Histone acetyltransferase 4 |
| Detection Method | Detection Principle | Detection Limit | Assay Time (min) | Advantages | Disadvantages | Reference |
|---|---|---|---|---|---|---|
| Zymography-LC-MS | pH changes from hydrolysis of triacylglycerols | 0.375 mU or 180 pg | 5–15 | Low cost Rapid Detect picogram amounts Versatile with fatty substrates Identify protein sequences Adapted for temperature-sensitive proteins | Coupled with LC-MS detection | (This study) |
| Zymography | White precipitate from esterification and transesterification | Not stated | 1440 | Detection of esterification | Long incubation time Low sensitivity | [27] |
| Zymography | White precipitate from esterification of alcohol and fatty acid | Not stated | 30–720 | Detection of esterification Low cost | Long incubation time Low sensitivity | [28] |
| Zymography | pH changes from hydrolysis of triacylglycerols | 0.5–2.0 U or 5–20 µg | 5–15 | Low cost Rapid | Strong buffering capacity Unable to detect picogram amounts Molten agar overlay Denaturation of proteins Low sensitivity | [21] |
| Zymography | pH changes from hydrolysis of triacylglycerols | 0.17–0.66 U | 360–720 | Low cost | Long incubation time Low sensitivity | [22] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ng, A.M.J.; Zhang, H.; Nguyen, G.K.T. Zymography for Picogram Detection of Lipase and Esterase Activities. Molecules 2021, 26, 1542. https://doi.org/10.3390/molecules26061542
Ng AMJ, Zhang H, Nguyen GKT. Zymography for Picogram Detection of Lipase and Esterase Activities. Molecules. 2021; 26(6):1542. https://doi.org/10.3390/molecules26061542
Chicago/Turabian StyleNg, Andre Mong Jie, Hongfang Zhang, and Giang Kien Truc Nguyen. 2021. "Zymography for Picogram Detection of Lipase and Esterase Activities" Molecules 26, no. 6: 1542. https://doi.org/10.3390/molecules26061542
APA StyleNg, A. M. J., Zhang, H., & Nguyen, G. K. T. (2021). Zymography for Picogram Detection of Lipase and Esterase Activities. Molecules, 26(6), 1542. https://doi.org/10.3390/molecules26061542






