Androgen Receptor Binding Category Prediction with Deep Neural Networks and Structure-, Ligand-, and Statistically Based Features
Abstract
1. Introduction
2. Results
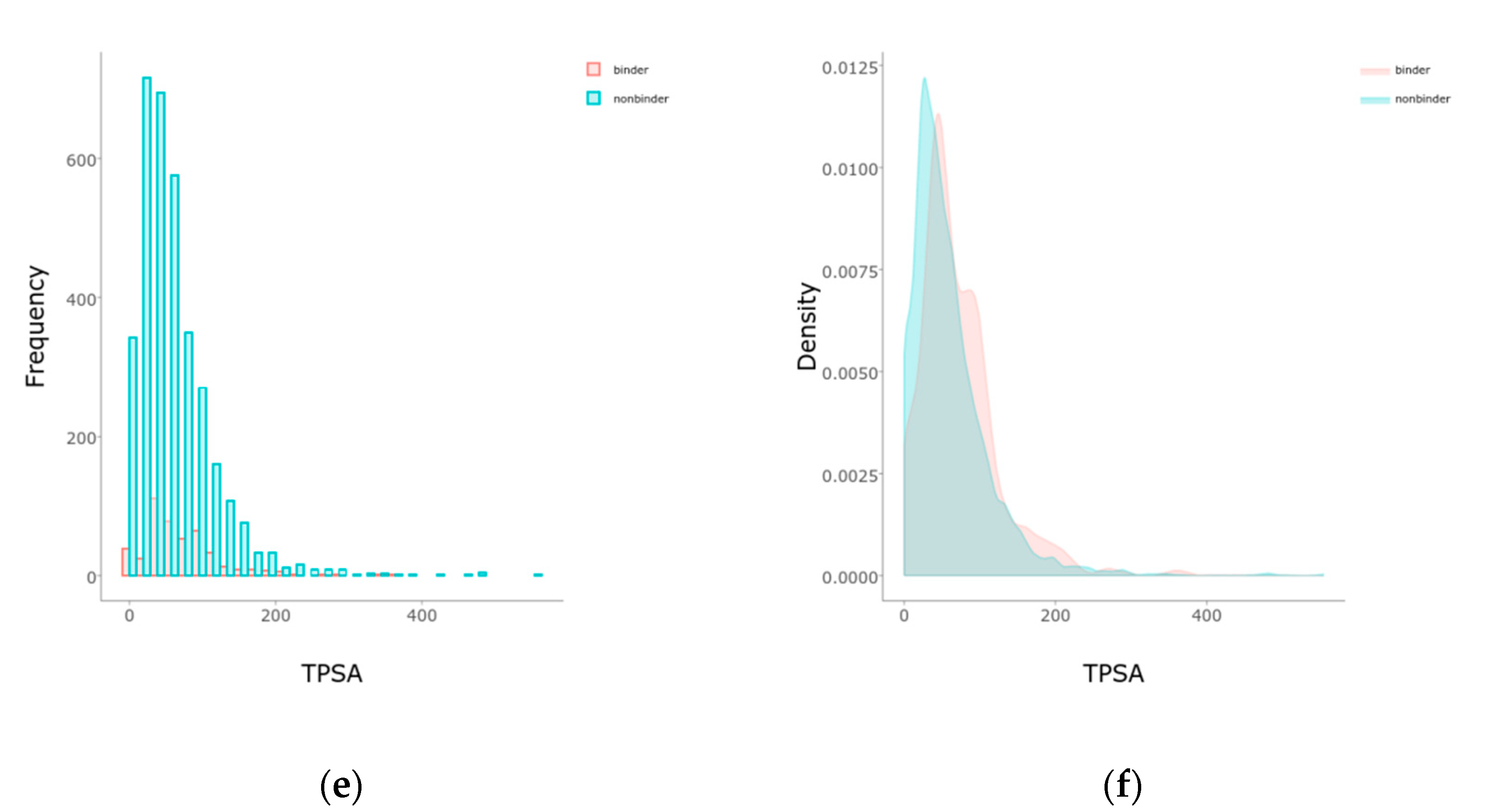
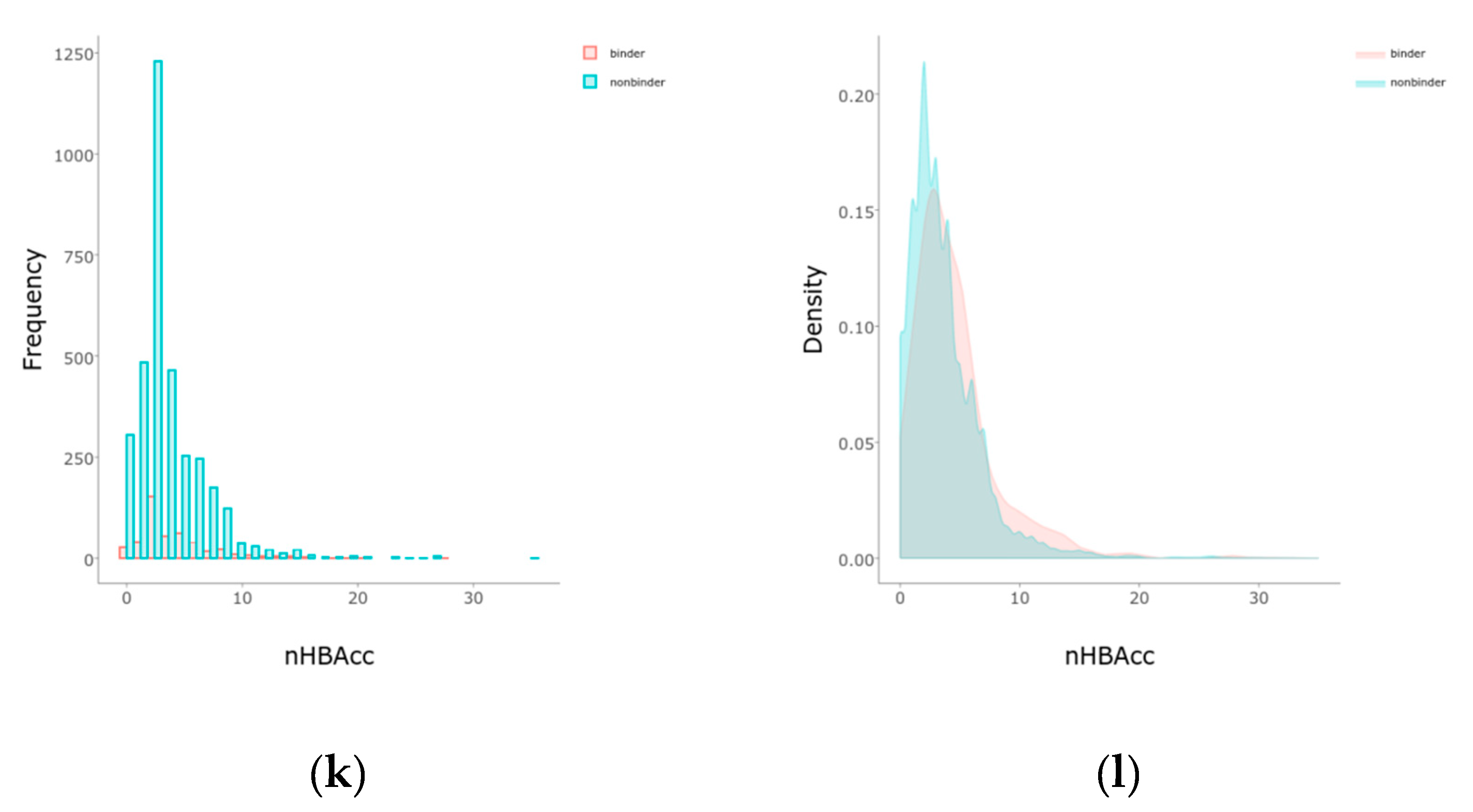
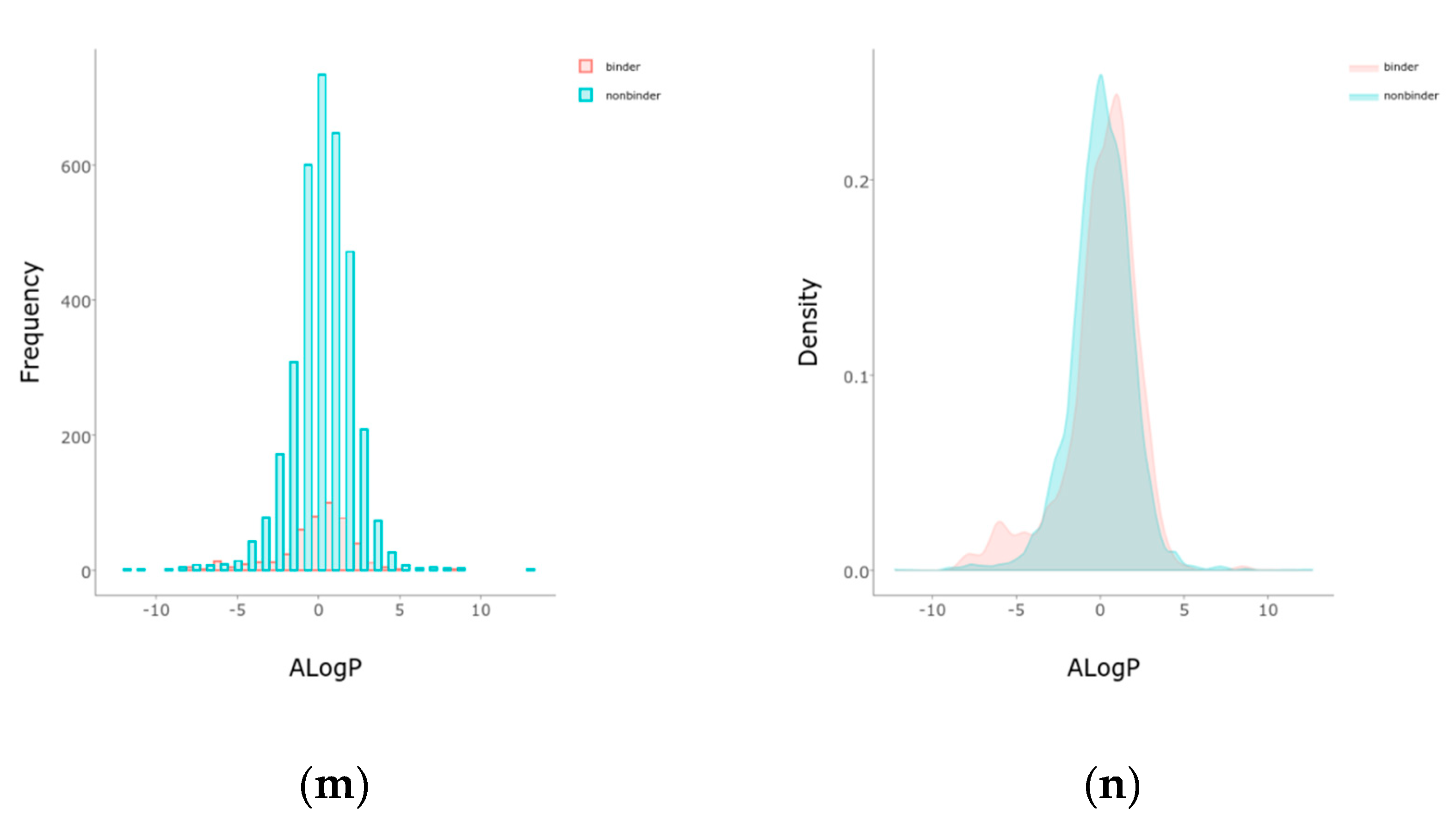
2.1. Balancing and Initial Separations
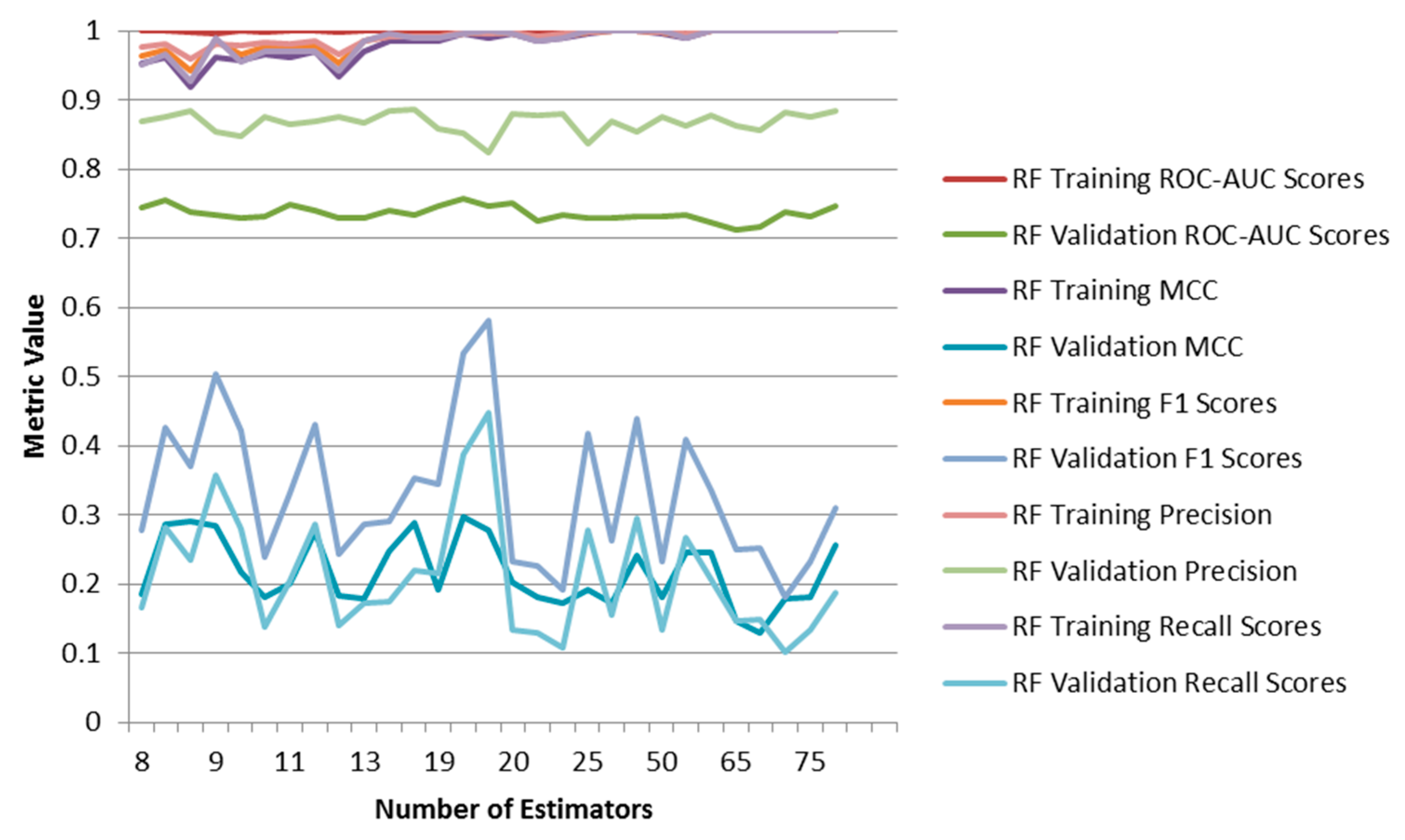
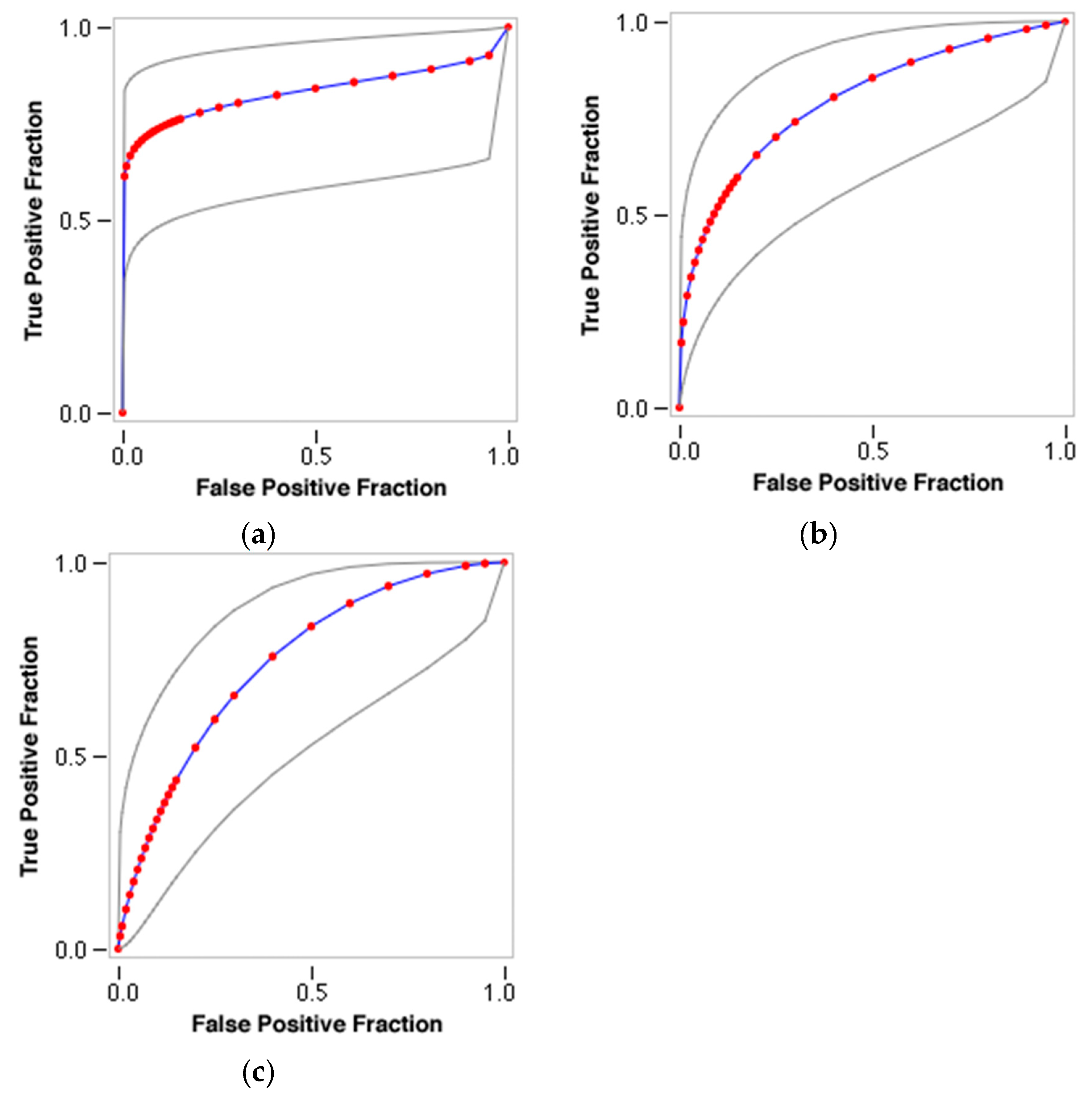
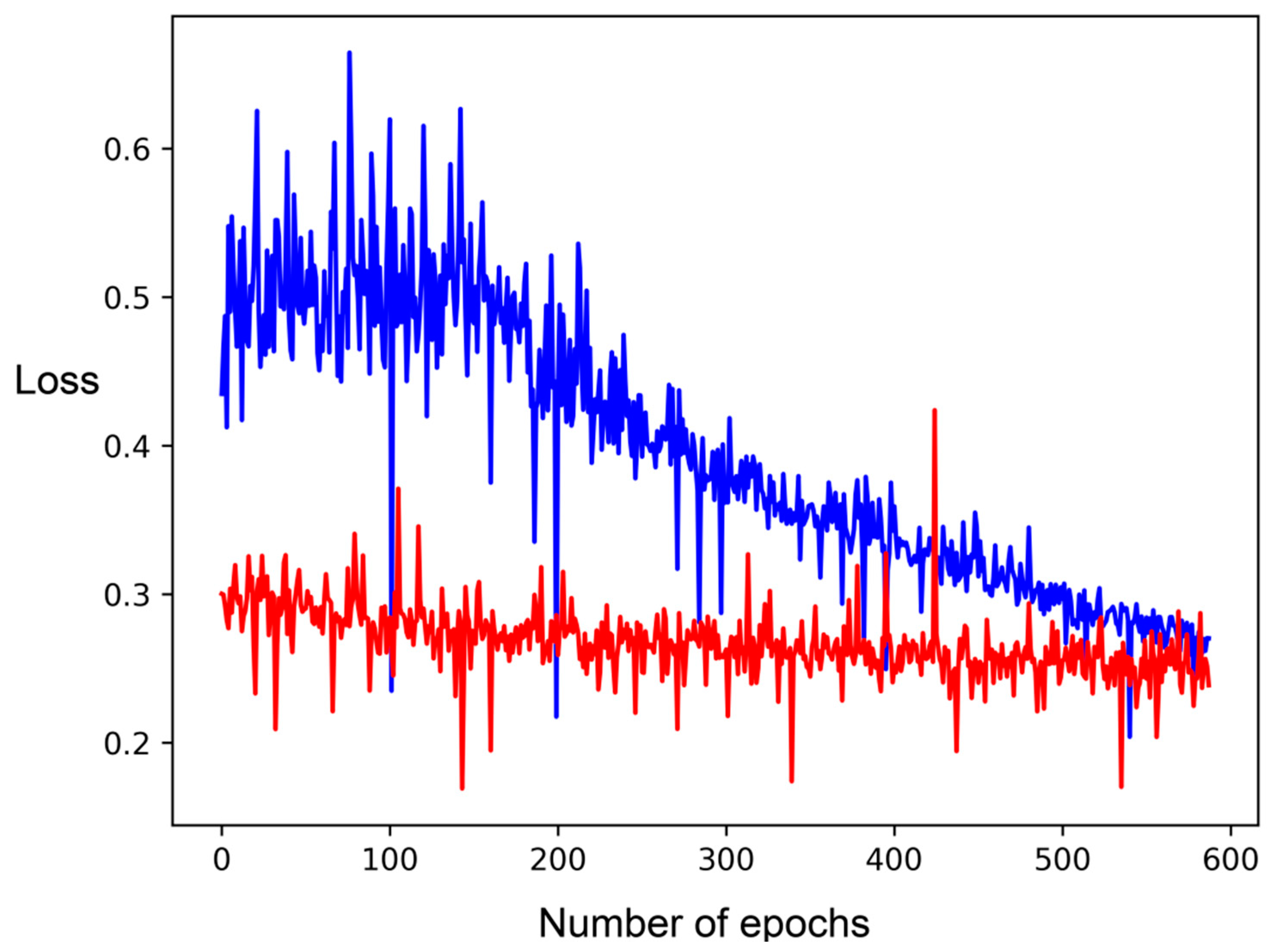
2.2. Comparison to RF and CNN
2.3. DNN with Different Featurizations
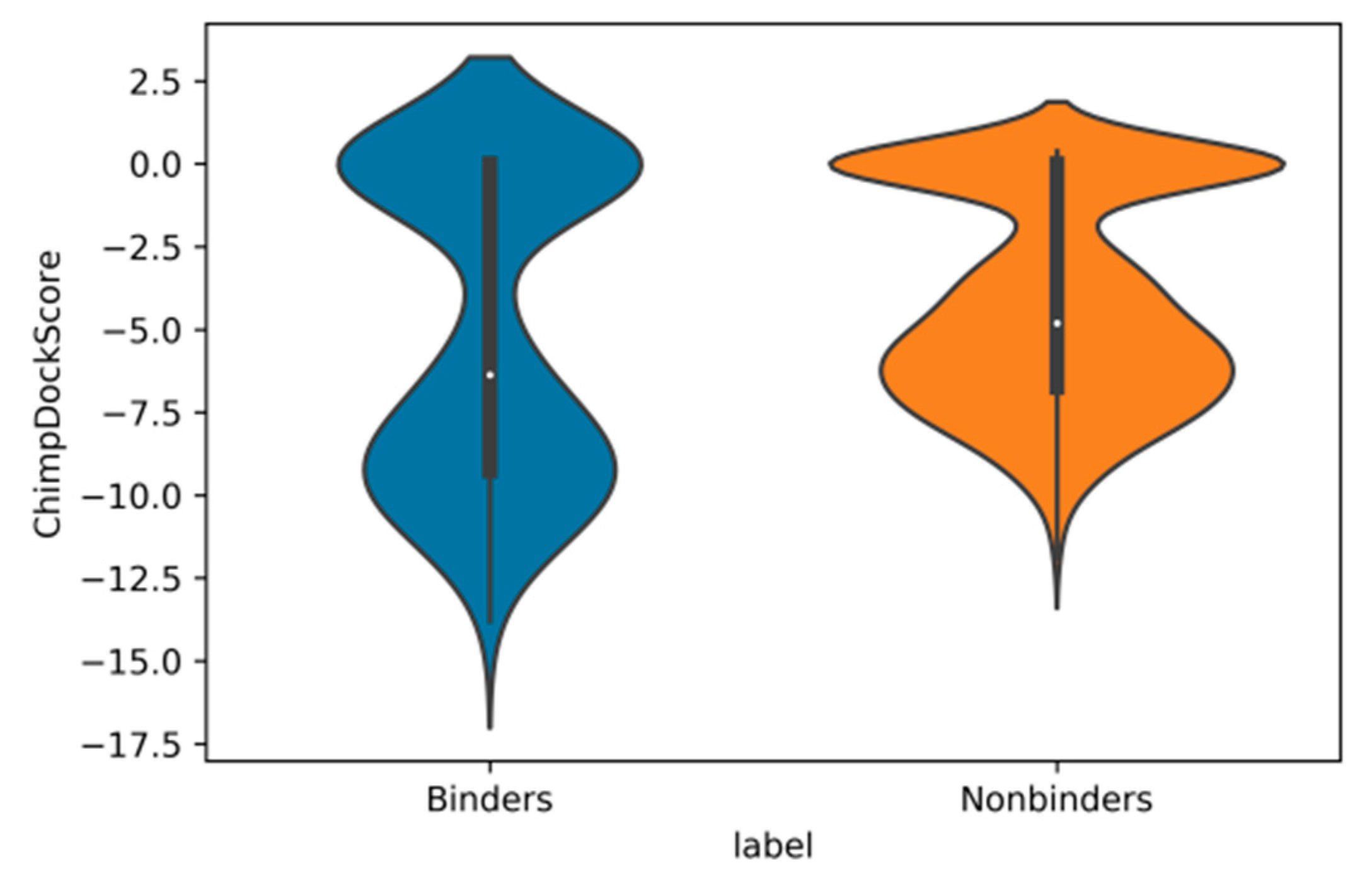
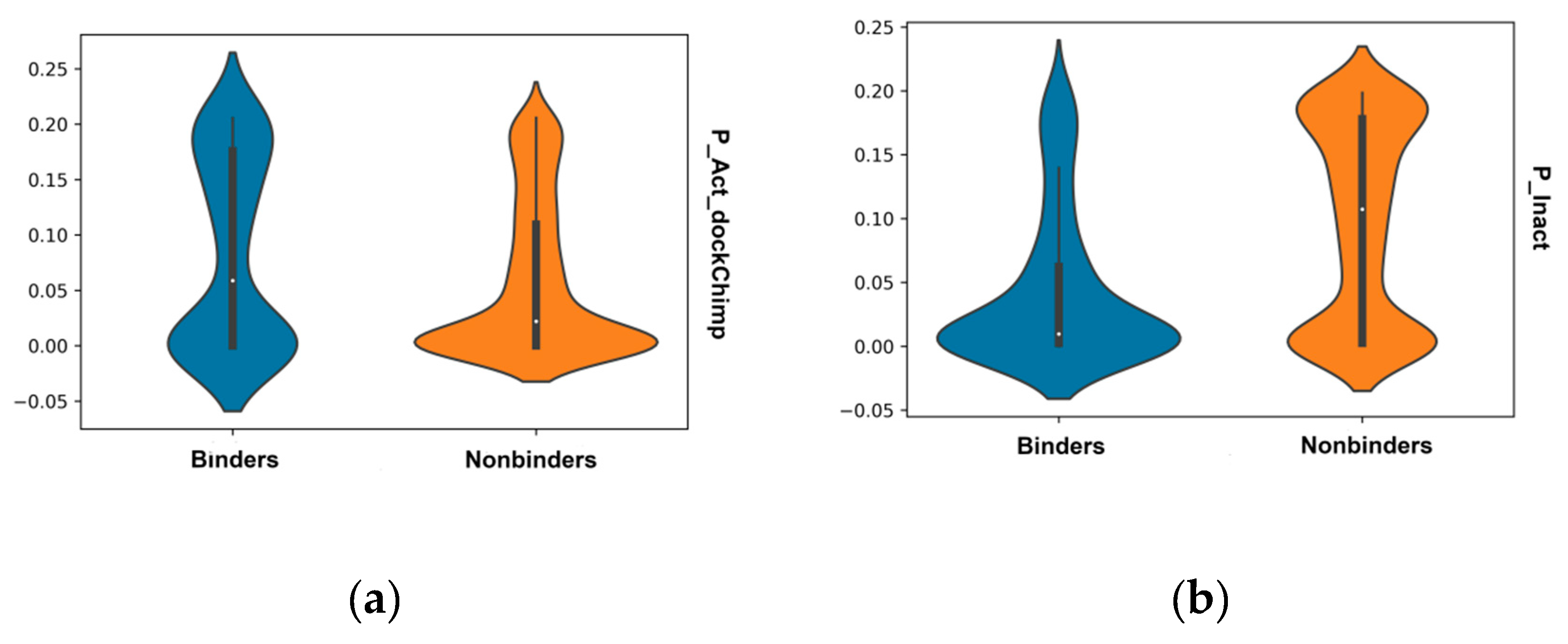
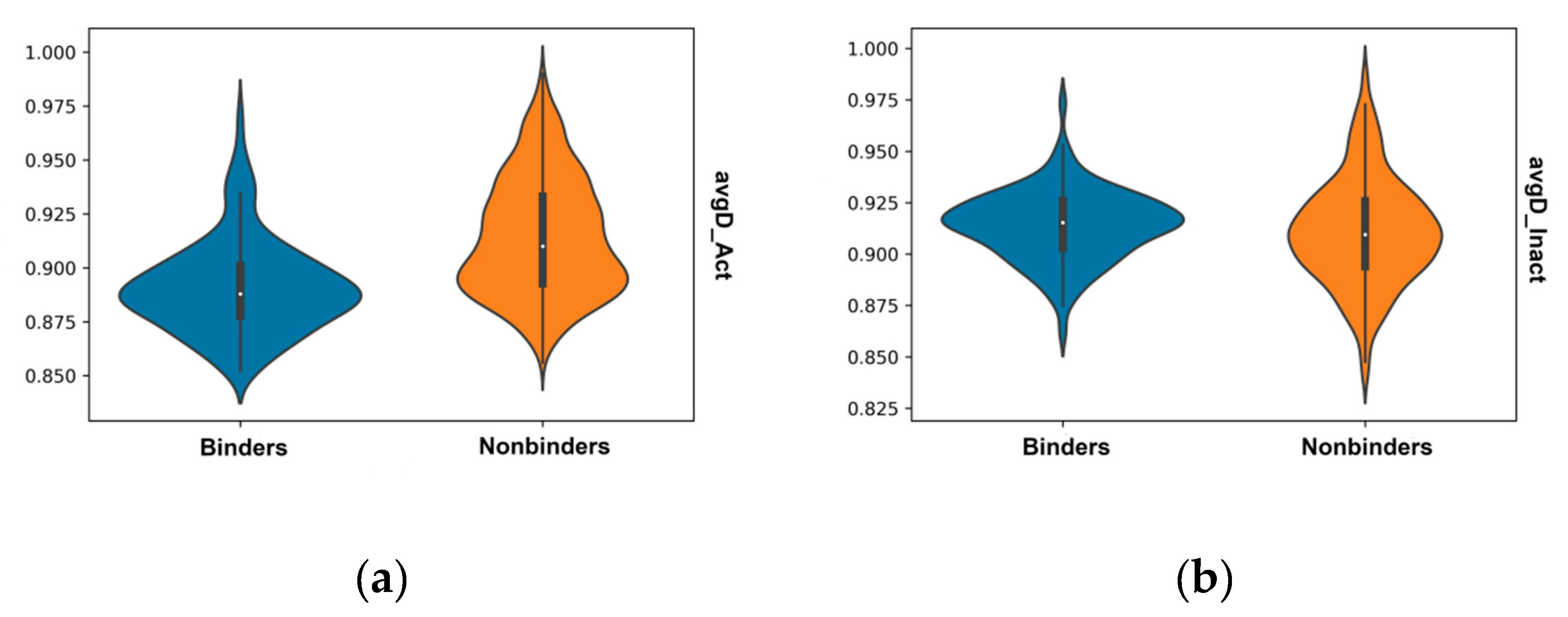
2.4. Validation of Docking and Comparison to Other Methods
3. Discussion
- Strong: Activity concentration < 0.09 μM
- Moderate: Activity concentration 0.09–0.18 μM
- Weak: Activity concentration 0.18–20 μM
- Very Weak: Activity concentration 20–800 μM
- Inactive: Activity concentration > 800 μM
4. Materials and Methods
4.1. Training Set
4.2. Independent Evaluation Set
4.3. Features
4.4. Models
4.5. Metrics
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Sifakis, D.; Androutsopoulos, V.P.; Tsatsakis, A.M.; Spandidos, D.A. Human exposure to endocrine disrupting chemicals: Effects on the male and female reproductive systems. Environ. Toxicol. Pharmacol. 2017, 51, 56–70. [Google Scholar] [CrossRef]
- Cheung, A.; Zajac, J.; Grossmann, M. Muscle and bone effects of androgen deprivation therapy: Current and emerging therapies. Endocr. Relat. Cancer 2014, 21, R371–R394. [Google Scholar] [CrossRef]
- Manolagas, S.C.; O’Brien, C.A.; Almeida, M. The role of estrogen and androgen receptors in bone health and disease. Nat. Rev. Endocrinol. 2013, 9, 699–712. [Google Scholar] [CrossRef]
- Mendelsohn, M.E.; Karas, R.H. Molecular and Cellular Basis of Cardiovascular Gender Differences. Science 2005, 308, 1583–1587. [Google Scholar] [CrossRef]
- Nadal, M.; Prekovic, S.; Gallastegui, N.; Helsen, C.; Abella, M.; Zielinska, K.; Gay, M.; Vilaseca, M.; Taulès, M.; Houtsmuller, A.B.; et al. Structure of the homodimeric androgen receptor ligand-binding domain. Nat. Commun. 2017, 8, 14388. [Google Scholar] [CrossRef]
- Mansouri, K.; Kleinstreuer, N.; Abdelaziz, A.M.; Alberga, D.; Alves, V.M.; Andersson, P.L.; Andrade, C.H.; Bai, F.; Balabin, I.; Ballabio, D.; et al. CoMPARA: Collaborative Modeling Project for Androgen Receptor Activity. Environ. Health Perspect. 2020, 128, 27002. [Google Scholar] [CrossRef]
- Trisciuzzi, D.; Alberga, D.; Mansouri, K.; Judson, R.; Novellino, E.; Mangiatordi, G.F.; Nicolotti, O. Predictive Structure-Based Toxicology Approaches to Assess the Androgenic Potential of Chemicals. J. Chem. Inf. Model. 2017, 57, 2874–2884. [Google Scholar] [CrossRef]
- García-Sosa, A.T.; Maran, U. Combined docking, chemical fingerprints, and Naïve Bayesian classifiers for the androgen receptor binding activity of the CoMPARA data of substances of environmental and health concern. 2021; submitted. [Google Scholar]
- Schneider, P.; Walters, W.P.; Plowright, A.T.; Sieroka, N.; Listgarten, J.; Goodnow, R.A.; Fisher, J.; Jansen, J.M.; Duca, J.S.; Rush, T.S.; et al. Rethinking drug design in the artificial intelligence era. Nat. Rev. Drug Discov. 2020, 19, 353–364. [Google Scholar] [CrossRef]
- Peña-Guerrero, J.; Nguewa, P.A.; García-Sosa, A.T. Machine Learning, Artificial Intelligence, and Data Science Breaking into Drug Design and Neglected Diseases. WIREs Comput. Mol. Sci. 2021, e1513. [Google Scholar] [CrossRef]
- Raj, R.J.S.; Shobana, S.J.; Pustokhina, I.V.; Pustokhin, D.A.; Gupta, D.; Shankar, K. Optimal feature selection-based medical image classification using deep learning model in internet of medical things. IEEE Access 2020, 8, 58006–58017. [Google Scholar] [CrossRef]
- Pustokhina, I.V.; Pustokhin, D.A.; Rodrigues, J.J.P.C.; Gupta, D.; Khanna, A.; Shankar, K.; Seo, C.; Joshi, G.P. Automatic vehicle license plate recognition using optimal k-means with convolutional neural network for intelligent transportation systems. IEEE Access 2020, 8, 92907–92917. [Google Scholar] [CrossRef]
- Khamparia, A.; Pandey, B.; Tiwari, S.; Gupta, D.; Khanna, A.; Rodrigues, J.J.P.C. An integrated hybrid CNN-RNN model for visual description and generation of captions. Circuits Syst. Signal Process. 2020, 39, 776–788. [Google Scholar] [CrossRef]
- Varela-Santos, S.; Melin, P. A new approach for classifying coronavirus COVID-19 based on its manifestation on chest X-rays using texture features and neural networks. Inf. Sci. 2021, 545, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Yosipof, A.; Guedes, R.C.; García-Sosa, A.T. Data Mining and Machine Learning Models for Predicting Drug Likeness and their Disease or Organ Category. Front. Chem. 2018, 6, 162. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Luna, J.; Grisoni, F.; Schneider, G. Drug discovery with explainable artificial intelligence. Nat. Mach. Intell. 2020, 2, 573–584. [Google Scholar] [CrossRef]
- DeepChem v. 2.3.0. Available online: https://github.com/deepchem/deepchem (accessed on 1 January 2020).
- García-Sosa, A.T. Benford’s Law in Medicinal Chemistry: Implications for Drug Design. Future Med. Chem. 2019, 11, 2247–2253. [Google Scholar] [CrossRef]
- Kleinstreuer, N.C.; Ceger, P.; Watt, E.D.; Martin, M.; Houck, K.; Browne, P.; Thomas, R.S.; Casey, W.M.; Dix, D.J.; Allen, D.; et al. Development and Validation of a Computational Model for Androgen Receptor Activity. Chem. Res. Toxicol. 2017, 30, 946–964. [Google Scholar] [CrossRef] [PubMed]
- Zorn, K.M.; Foil, D.H.; Lane, T.R.; Hillwalker, W.; Feifarek, D.J.; Jones, F.; Klaren, W.D.; Brinkman, A.M.; Ekins, S. Comparison of Machine Learning Models for the Androgen Receptor. Environ. Sci. Technol. 2020, 54, 13690–13700. [Google Scholar] [CrossRef]
- Martinez-Mayorga, K.; Madariaga-Mazon, A.; Medina-Franco, J.L.; Maggiora, G. The impact of chemoinformatics on drug discovery in the pharmaceutical industry. Expert Opin. Drug Discov. 2020, 15, 293–306. [Google Scholar] [CrossRef]
- Forssén, P.; Multia, E.; Samuelsson, J.; Andersson, M.; Aastrup, T.; Altun, S.; Wallinder, D.; Wallbing, L.; Liangsupree, T.; Riekkola, M.-L.; et al. Reliable strategy for analysis of complex biosensor data. Anal. Chem. 2018, 90, 5366–5374. [Google Scholar] [CrossRef]
- Forssén, P.; Samuelsson, J.; Lacki, K.; Fornstedt, T. Advanced analysis of biosensor data for SARS-CoV-2 RBD and ACE2 interactions. Anal. Chem. 2020, 92, 11520–11524. [Google Scholar] [CrossRef]
- Dix, D.J.; Houck, K.A.; Martin, M.T.; Richard, A.M.; Setzer, R.W.; Kavlock, R.J. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol. Sci. 2007, 95, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Tice, R.R.; Austin, C.P.; Kavlock, R.J.; Bucher, J.R. Improving the human hazard characterization of chemicals: A Tox21 update. Environ. Health Perspect. 2013, 21, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef]
- Pandas Tools v. 0.25.3. Python Data Analysis Library. Available online: https://pandas.pydata.org/ (accessed on 1 January 2020).
- PubChem. National Institutes of Health (NIH). Available online: https://pubchem.ncbi.nlm.nih.gov (accessed on 1 January 2020).
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Schrödinger, LLC. Protein Preparation Wizard; Schrödinger, LLC: New York, NY, USA, 2019. [Google Scholar]
- Schrödinger, LLC. Virtual Screening Workflow; Schrödinger, LLC: New York, NY, USA, 2019. [Google Scholar]
- García-Sosa, A.T.; Sild, S.; Maran, U. Docking and Virtual Screening Using Distributed Grid Technology. SQER 2009, 28, 815–821. [Google Scholar] [CrossRef]
- Viira, B.; Selyutina, A.; García-Sosa, A.T.; Karonen, M.; Sinkkonen, J.; Merits, A.; Maran, U. Design, Discovery, Modelling, Synthesis, and Biological Evaluation of Novel and Small, Low Toxicity s-Triazine Derivatives as HIV 1 Nonnucleoside Reverse Transcriptase Inhibitors. Bioorg. Med. Chem. 2016, 24, 2519–2529. [Google Scholar] [CrossRef]
- Glisic, S.; Sencanski, M.; Perovic, V.; Stevanovic, S.; García-Sosa, A.T. Arginase Flavonoid Anti-Leishmanial in Silico Inhibitors Flagged against Anti-Targets. Molecules 2016, 21, 589. [Google Scholar] [CrossRef]
- Beans, M. Version 5.3.8; ChemAxon: Budapest, Hungary, 2010; Available online: http://www.chemaxon.com (accessed on 1 January 2020).
- Open Babel. An Open Chemical Toolbox. Available online: http://openbabel.org (accessed on 1 November 2020).
- Winter, R.; Montanari, F.; Noé, F.; Clevert, D.-A. Learning continuous and data-driven molecular descriptors by translating equivalent chemical representations. Chem. Sci. 2019, 10, 1692–1701. [Google Scholar] [CrossRef] [PubMed]
- Scikit-Learn v. 0.21.3. Machine Learning in Python. Available online: https://scikit-learn.org/stable/ (accessed on 1 January 2020).
- Gonzalez-Medina, M.; Medina-Franco, J.L. Platform for Unified Molecular Analysis: PUMA. J. Chem. Inf. Model. 2017, 57, 1735–1740. [Google Scholar] [CrossRef] [PubMed]
- t-Map. Reymond Group. Available online: https://tmap.gdb.tools/index.html (accessed on 18 September 2020).
- MHFP6 Fingerprints. Available online: https://github.com/reymond-group/mhfp/tree/master/mhfp (accessed on 18 September 2020).
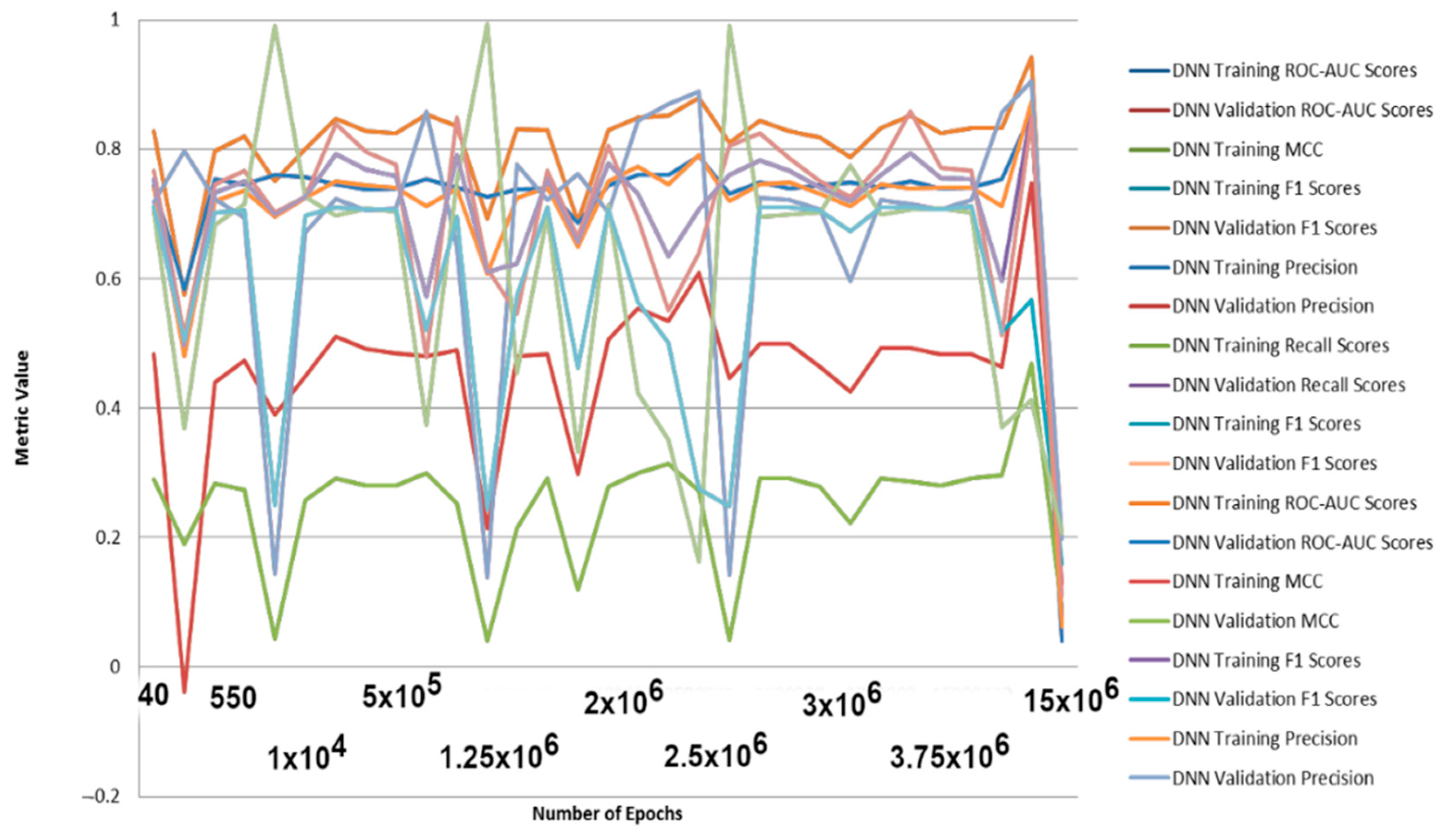
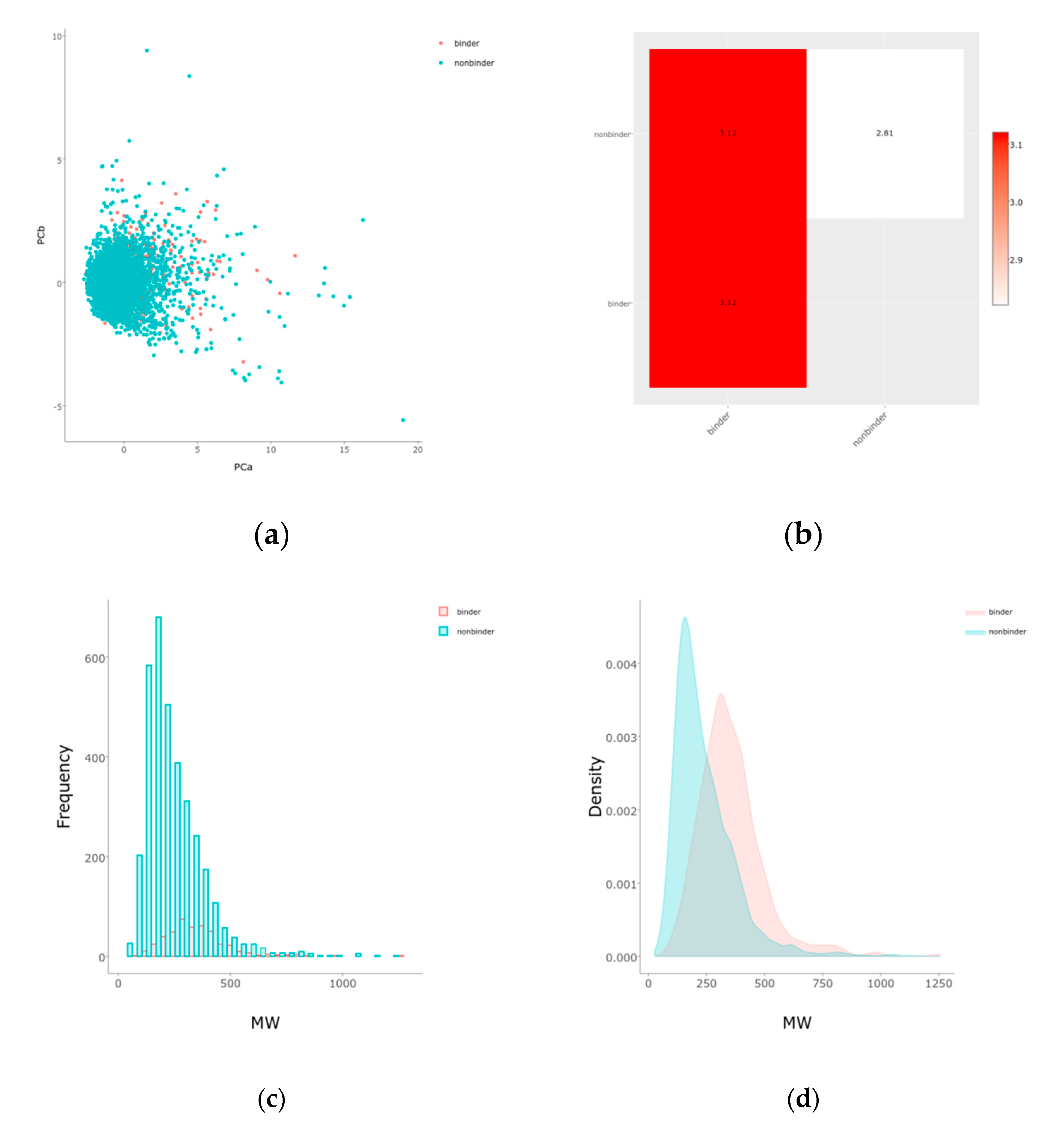

| Method | Train ± s.d. | Valid ± s.d. | Best Hyperparameters |
|---|---|---|---|
| RF classifier myfeats (I) | AUC 0.9999 ± 0.0009; MCC 0.9951 ± 0.0153; F1 0.9963 ± 0.0153; Prec. 0.9976 ± 0.011; Recall 0.9951 ± 0.0198 | AUC 0.7564 ± 0.0105; MCC 0.297435 ± 0.0478; F1 0.5805 ± 0.1041 (3 × 106 epochs); Prec. 0.8856 ± 0.0148(1.5 × 105 epochs); Recall 0.4481 ± 0.0866 (3 × 106 epochs) | eightfold cross-validation, (19 runs, ‘sqrt’), 2.25 × 106 epochs |
| DNN classifier myfeats (II) | AUC 0.9424 ± 0.0655; MCC 0.7472 ± 0.1283; F1 0.8608 ± 0.0754; Prec. 0.8732 ± 0.063; Recall 0.8585 ± 0.1092(4.5 × 106 epochs) | AUC 0.8686 ± 0.0398; MCC 0.4685 ± 0.0892; F1 0.7943 ± 0.1617 (4.5 × 106 epochs); Prec. 0.9052 ± 0.1988; Recall 0.8585 ± 0.2054 (4.5 × 106 epochs) | Learning rate: 0.00047, weight decay penalty: 2.637 × 106, 2.5 × 106 epochs |
| GraphConv CNN (VII) | AUC 1.0 | AUC 0.7264 | −(50 runs, ‘sqrt’) |
| RF classifier CDDD features (V) | AUC 0.9997 | AUC 0.7308 | (18, ‘sqrt’) |
| DNN classifier CDDD features (VI) | AUC 0.8498 | AUC 0.7563 | Learning rate: 0.00067, weight decay penalty: 4.073 × 106, 2.5 × 106 epochs |
| RF regression myfeats (III) | R2 = 0.8817 | R2 = −0.0520 | (10 runs, ‘log2’) |
| DNN regression myfeats (IV) | R2 = 0.2721 | R2 = −0.1926 | fourfold cross-validation, learning rate: 0.000359 weight decay penalty: 8.831 × 106, nb. epochs: 20 |
| CAS | Name | Structure | Agonist | Antagonist | Predicted | Correct |
|---|---|---|---|---|---|---|
| 52806-53-8 | hydroxyflutamide | 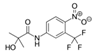 | NA | Strong | 1 | Yes |
| 90357-06-5 | bicalutamide | 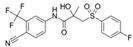 | NA | Strong | 1 | Yes |
| 122-14-5 | fenitrothion | 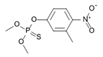 | NA | Strong | 0 | X |
| 63612-50-0 | nilutamide | 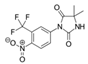 | Negative | Moderate | 0 | X |
| 427-51-0 | cyproterone acetate | 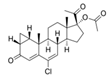 | Weak | Moderate | 1 | Yes |
| 80-05-7 | bisphenol a | 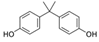 | NA | Moderate/weak | 1 | Yes |
| 330-55-2 | linuron | 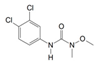 | NA | Moderate/weak | 0 | X |
| 13311-84-7 | flutamide | 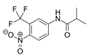 | Negative | Moderate/weak | 0 | X |
| 67747-09-5 | prochloraz | 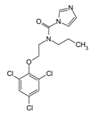 | Negative | Moderate/weak | 1 | Yes |
| 789-02-6 | o,p′-ddt | 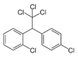 | Negative | Weak | 1 | Yes |
| 60168-88-9 | fenarimol | 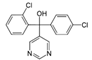 | Negative | Very weak | 0 | Yes |
| 58-18-4 | methyl testosterone | 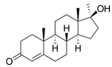 | Strong | Negative | 1 | Yes |
| 58-22-0 | testosterone | 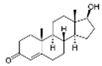 | Strong | Negative | 1 | Yes |
| 63-05-8 | 4-androstenedione | 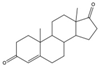 | Moderate | Negative | 1 | Yes |
| 1912-24-9 | atrazine | 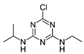 | Negative | Negative | 0 | Yes |
| 52918-63-5 | deltamethrin | 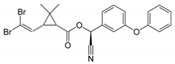 | Negative | Negative | 0 | Yes |
| 10161-33-8 | 17b-trenbolone |  | Strong | NA | 1 | Yes |
| 797-63-7 | levonorgestrel | 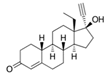 | Strong | NA | 1 | Yes |
| 68-22-4 | norethindrone |  | Strong | NA | 1 | Yes |
| 521-18-6 | 5a-dihydrotestosterone | 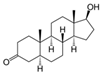 | Strong | NA | 1 | Yes |
| CAS | Name | Structure | Agonist | Antagonist | Predicted by II | Correct |
|---|---|---|---|---|---|---|
| 58-18-4 | methyl testosterone |  | Strong | Negative | 1 | Yes |
| 57-91-0 | 17α-estradiol | 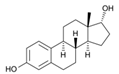 | Inactive | 1 | X | |
| 63-05-8 | 4-androstenedione |  | Moderate | Negative | 1 | Yes |
| 486-66-8 | daidzein | 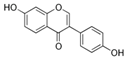 | Inactive | 1 | X | |
| 98319-26-7 | finasteride | 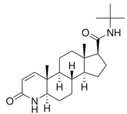 | Inactive | 0 (RF) | Yes | |
| 57-85-2 | testosterone propionate |  | Strong | Inactive | 1 | X |
| 51-28-5 | 2,4-dinitrophenol | 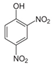 | Negative | Negative | 0 | Yes |
| 129453-61-8 | fulvestrant | 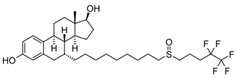 | Inactive | 1 | X | |
| 84-74-2 | dibutyl phtalate | 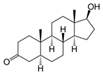 | Active | 0 | X | |
| 117-81-7 | diethylhexyl phtalate | 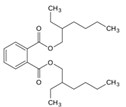 | Active | 0 | X | |
| 13194-48-4 | ethoprop | 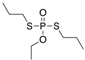 | Active | 0 | X |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Sosa, A.T. Androgen Receptor Binding Category Prediction with Deep Neural Networks and Structure-, Ligand-, and Statistically Based Features. Molecules 2021, 26, 1285. https://doi.org/10.3390/molecules26051285
García-Sosa AT. Androgen Receptor Binding Category Prediction with Deep Neural Networks and Structure-, Ligand-, and Statistically Based Features. Molecules. 2021; 26(5):1285. https://doi.org/10.3390/molecules26051285
Chicago/Turabian StyleGarcía-Sosa, Alfonso T. 2021. "Androgen Receptor Binding Category Prediction with Deep Neural Networks and Structure-, Ligand-, and Statistically Based Features" Molecules 26, no. 5: 1285. https://doi.org/10.3390/molecules26051285
APA StyleGarcía-Sosa, A. T. (2021). Androgen Receptor Binding Category Prediction with Deep Neural Networks and Structure-, Ligand-, and Statistically Based Features. Molecules, 26(5), 1285. https://doi.org/10.3390/molecules26051285






