Oligonucleotide-Based Approaches to Inhibit Dengue Virus Replication
Abstract
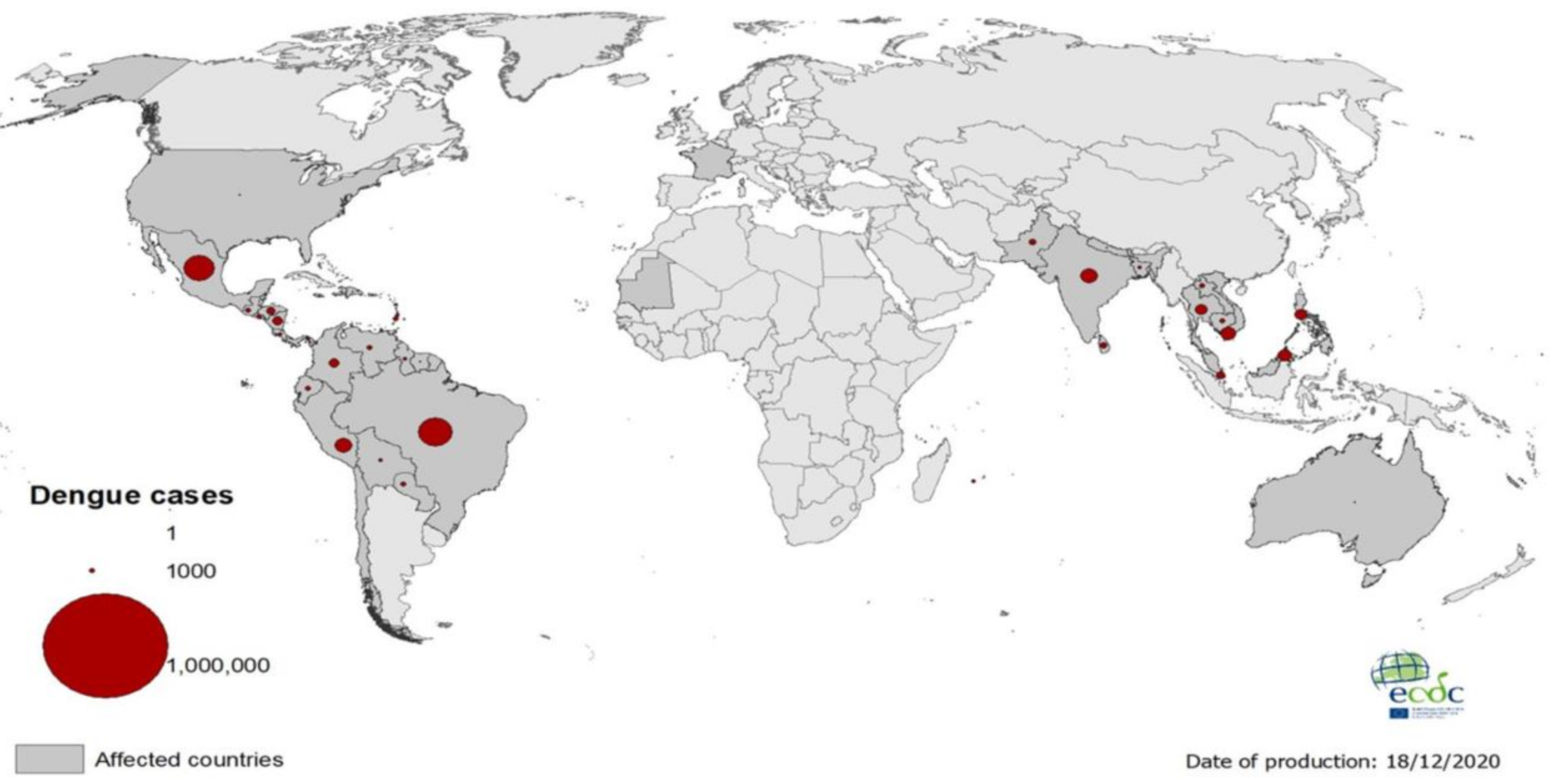
:1. Introduction
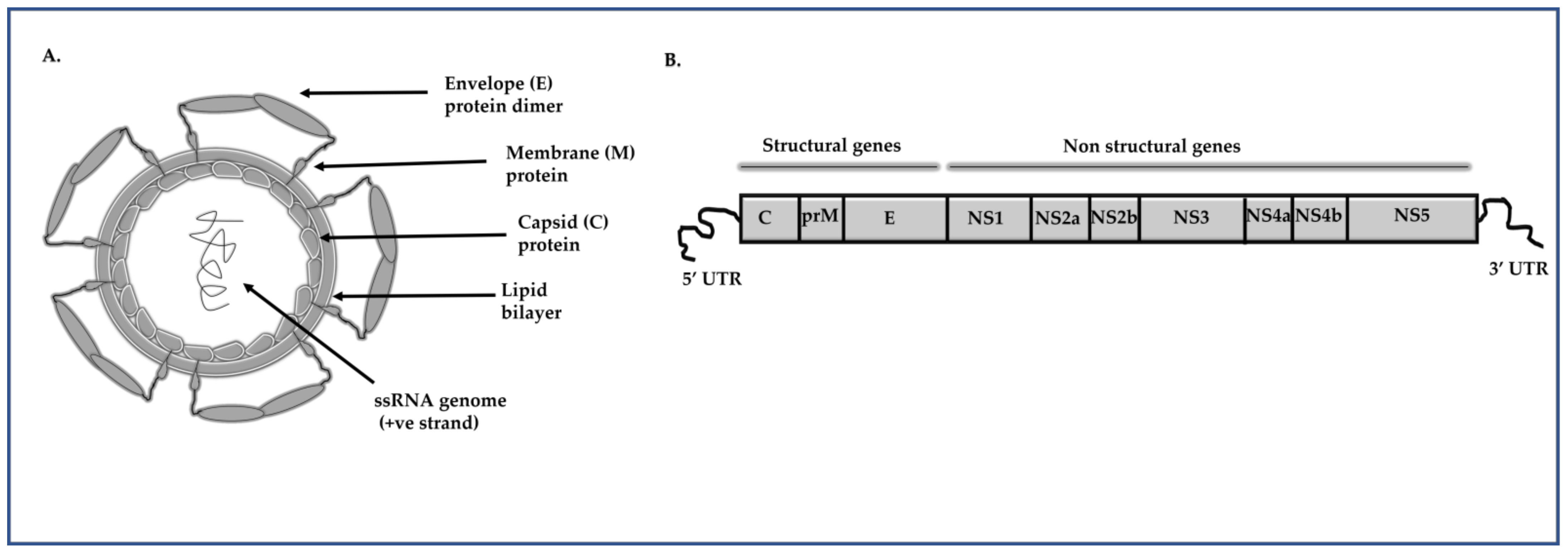
2. Oligonucleotide-Based Approaches to Inhibit DENV Replication
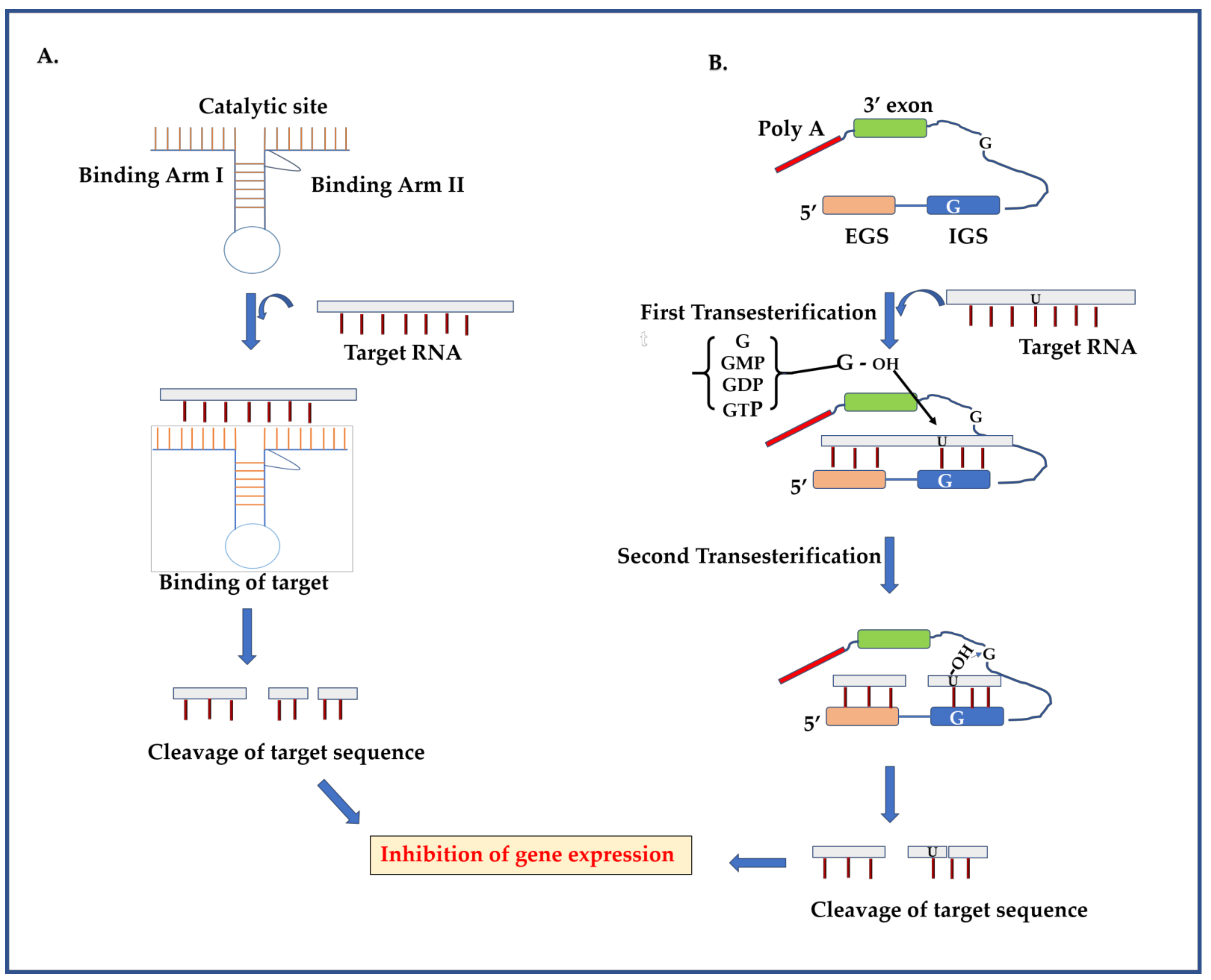
2.1. Ribozyme
2.1.1. Ribozyme Mechanism
2.1.2. Targeting DENV Genome Using Ribozymes
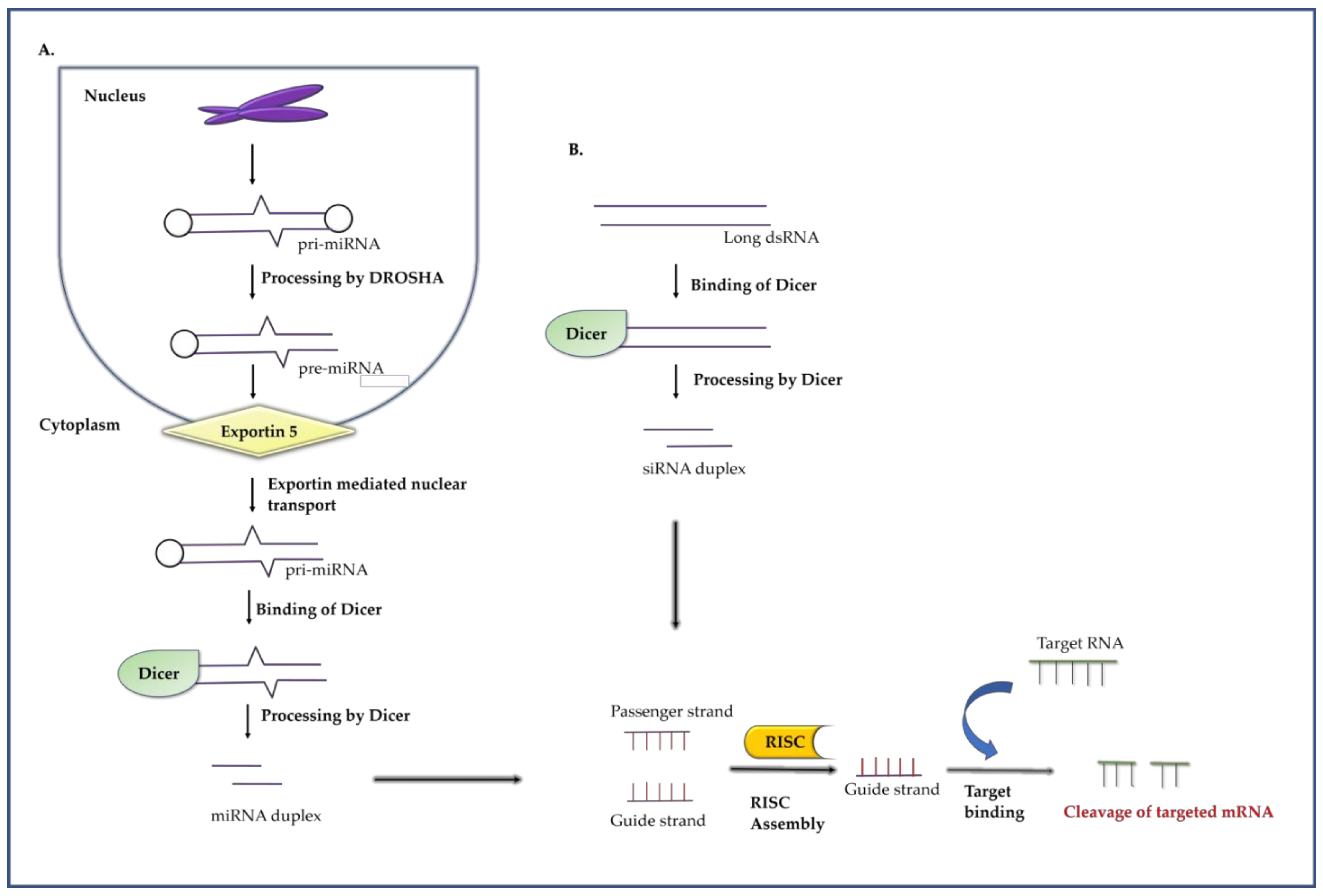
2.2. RNA Interference
2.2.1. RNA Interference Mechanism
2.2.2. RNAi as an Antiviral against DENV
2.3. Aptamer
2.3.1. Mechanism of Aptamer and Identification of Target Specific Aptamers
2.3.2. Inhibition of DENV Replication by Aptamer
2.4. Morpholinos
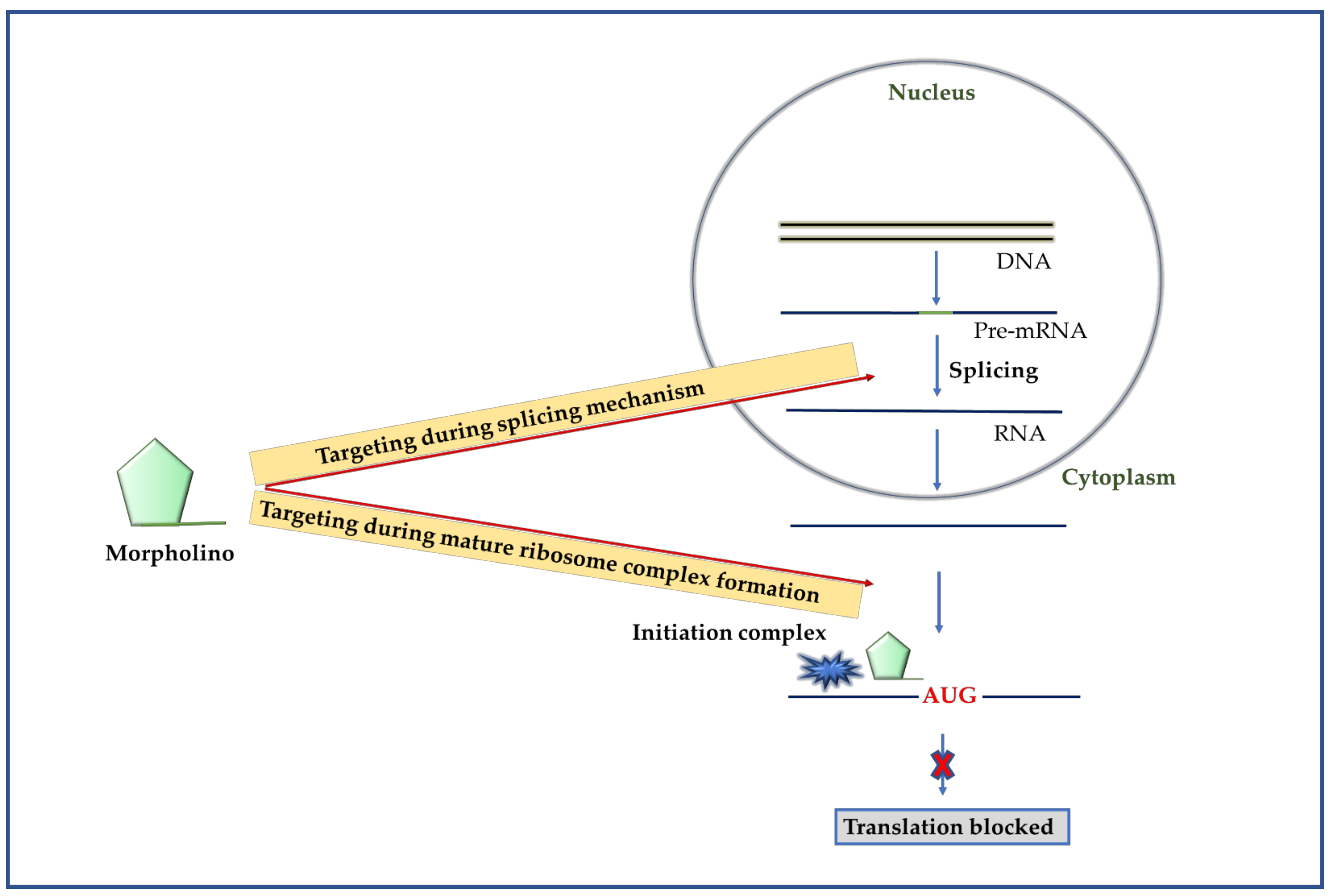
2.4.1. Mechanism of Morpholino
2.4.2. Targeting DENV Using Morpholino
3. Other Oligonucleotide-Based Methods Reported for DENV Inhibition
4. Challenges Associated with Oligonucleotides-Based Antiviral Methods and Possible Ways to Overcome the Hurdles
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cuypers:, L.; Libin, P.J.; Simmonds, P.; Nowé, A.; Muñoz-Jordán, J.; Alcantara, L.C.J.; Vandamme, A.-M.; Santiago, G.A.; Theys, K. Time to harmonize dengue Nomenclature and classification. Viruses 2018, 10, 569. [Google Scholar]
- Hung, T.M.; Shepard, D.S.; Bettis, A.A.; Nguyen, H.A.; McBride, A.; Clapham, H.E.; Turner, H.C. Productivity costs from a dengue episode in Asia: A systematic literature review. BMC Infect. Dis. 2020, 20, 1–18. [Google Scholar]
- Hwang, E.-H.; Kim, G.; Oh, H.; An, Y.J.; Kim, J.; Kim, J.H.; Hwang, E.-S.; Park, J.-H.; Hong, J.; Koo, B.-S. Molecular and evolutionary analysis of dengue virus serotype 2 isolates from Korean travelers in 2015. Arch. Virol. 2020, 165, 1739–1748. [Google Scholar]
- Murugesan, A.; Manoharan, M. Dengue Virus. In Emerging and Reemerging Viral Pathogens; Elsevier: Amsterdam, The Netherlands, 2020; pp. 281–359. [Google Scholar]
- Lozach, P.-Y.; Burleigh, L.; Staropoli, I.; Navarro-Sanchez, E.; Harriague, J.; Virelizier, J.-L.; Rey, F.A.; Desprès, P.; Arenzana-Seisdedos, F.; Amara, A. Dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin (DC-SIGN)-mediated enhancement of dengue virus infection is independent of DC-SIGN internalization signals. J. Biol. Chem. 2005, 280, 23698–23708. [Google Scholar]
- Carro, A.C.; Piccini, L.E.; Damonte, E.B. Blockade of dengue virus entry into myeloid cells by endocytic inhibitors in the presence or absence of antibodies. PLoS Negl. Tropical. Dis. 2018, 12, e0006685. [Google Scholar]
- Marchiori, E.; Hochhegger, B.; Zanetti, G. Pulmonary manifestations of dengue. J. Bras. Pneumol. 2020, 46. [Google Scholar] [CrossRef]
- World Health Organization. Dengue and Severe Dengue; World Health Organization, Regional Office for the Eastern Mediterranean: Cairo, Egypt, 2014. [Google Scholar]
- Zonetti, L.F.; Coutinho, M.C.; de Araujo, A.S.; Letters, P. Molecular aspects of the dengue virus infection process: A review. Protein Pept. Lett. 2018, 25, 712–719. [Google Scholar]
- Singh, A.; Bisht, P.; Bhattacharya, S.; Guchhait, P. Role of platelet cytokines in Dengue virus infection. Front. Cell Infect. Microbiol. 2020, 10, 561366. [Google Scholar]
- Masri, M.F.B.; Rathore, A.P.; John, A.L.S. Therapeutics for Dengue. Curr. Treat. Options Infect. Dis. 2019, 11, 199–214. [Google Scholar]
- Low, J.G.; Ooi, E.E.; Vasudevan, S.G. Current status of dengue therapeutics research and development. J. Infect. Dis. 2017, 215, S96–S102. [Google Scholar]
- Jasamai, M.; Yap, W.B.; Sakulpanich, A.; Jaleel, A. Current prevention and potential treatment options for dengue infection. J. Pharm. Pharm. Sci. 2019, 22, 440–456. [Google Scholar]
- Wilder-Smith, A. Dengue vaccine development by the year 2020: Challenges and prospects. Curr. Opin. Virol. 2020, 43, 71–78. [Google Scholar]
- Thomas, S.J.; Yoon, I.-K. A review of Dengvaxia®: Development to deployment. Hum. Vaccin Immunother. 2019, 15, 2295–2314. [Google Scholar]
- Powers, C.N.; Setzer, W.N. An in-silico investigation of phytochemicals as antiviral agents against dengue fever. Comb. Chem High. Throughput Screen. 2016, 19, 516–536. [Google Scholar]
- Sivaraman, D.; Pradeep, P.S. Exploration of bioflavonoids targeting dengue virus NS5 RNA-dependent RNA polymerase: In silico molecular docking approach. J. App. Pharm. Sci. 2020, 10, 016–022. [Google Scholar]
- Tricou, V.; Minh, N.N.; Van, T.P.; Lee, S.J.; Farrar, J.; Wills, B.; Tran, H.T.; Simmons, C.P. A randomized controlled trial of chloroquine for the treatment of dengue in Vietnamese adults. PLos Negl. Trop. Dis. 2010, 4, e785. [Google Scholar]
- Nguyen, N.M.; Tran, C.N.B.; Phung, L.K.; Duong, K.T.H.; Huynh, H.l.A.; Farrar, J.; Nguyen, Q.T.H.; Tran, H.T.; Nguyen, C.V.V.; Merson, L. A randomized, double-blind placebo controlled trial of balapiravir, a polymerase inhibitor, in adult dengue patients. J. Infect. Dis. 2013, 207, 1442–1450. [Google Scholar]
- Dhama, K.; Karthik, K.; Khandia, R.; Munjal, A.; Tiwari, R.; Rana, R.; Khurana, S.K.; Ullah, S.; Khan, R.U.; Alagawany, M. Medicinal and therapeutic potential of herbs and plant metabolites/extracts countering viral pathogens-current knowledge and future prospects. Curr. Drug Metab. 2018, 19, 236–263. [Google Scholar]
- Weinberg, C.E.; Weinberg, Z.; Hammann, C. Novel ribozymes: Discovery, catalytic mechanisms, and the quest to understand biological function. Nucleic Acids Res. 2019, 47, 9480–9494. [Google Scholar]
- Tanner, N.K. Ribozymes: The characteristics and properties of catalytic RNAs. FEMS Microbiol. Rev. 1999, 23, 257–275. [Google Scholar]
- Sczakiel, G.; Nedbal, W. The potential of ribozymes as antiviral agents. Trends Microbiol. 1995, 3, 213–217. [Google Scholar]
- Menke, A.; Hobom, G. Antiviral ribozymes. Mol. Biotechnol. 1997, 8, 17–33. [Google Scholar]
- Rossi, J.J. Ribozyme therapy for HIV infection. Adv Drug Deliv. Rev. 2000, 44, 71–78. [Google Scholar]
- Lee, C.H.; Han, S.R.; Lee, S.W. Therapeutic applications of group I intron-based trans-splicing ribozymes. Wiley Interdiscip Rev. RNA 2018, 9, e1466. [Google Scholar]
- Nawtaisong, P.; Keith, J.; Fraser, T.; Balaraman, V.; Kolokoltsov, A.; Davey, R.A.; Higgs, S.; Mohammed, A.; Rongsriyam, Y.; Komalamisra, N. Effective suppression of Dengue fever virus in mosquito cell cultures using retroviral transduction of hammerhead ribozymes targeting the viral genome. Virol. J. 2009, 6, 73. [Google Scholar]
- Carter, J.R.; Keith, J.H.; Barde, P.V.; Fraser, T.S.; Fraser, M.J. Targeting of highly conserved Dengue virus sequences with anti-Dengue virus trans-splicing group I introns. BMC Mol. Biol. 2010, 11, 84. [Google Scholar]
- Carter, J.R.; Keith, J.H.; Fraser, T.S.; Dawson, J.L.; Kucharski, C.A.; Horne, K.M.; Higgs, S.; Fraser, M.J. Effective suppression of dengue virus using a novel group-I intron that induces apoptotic cell death upon infection through conditional expression of the Bax C-terminal domain. Virol. J. 2014, 11, 1–18. [Google Scholar]
- Carter, J.R.; Taylor, S.; Fraser, T.S.; Kucharski, C.A.; Dawson, J.L.; Fraser Jr, M.J. Suppression of the arboviruses dengue and chikungunya using a dual-acting group-I intron coupled with conditional expression of the Bax C-terminal domain. PLoS ONE. 2015, 10, e0139899. [Google Scholar]
- Napoli, C.; Lemieux, C.; Jorgensen, R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant. Cell. 1990, 2, 279–289. [Google Scholar]
- Sen, G.L.; Blau, H.M. A brief history of RNAi: The silence of the genes. FASEB J. 2006, 20, 1293–1299. [Google Scholar]
- Qureshi, A.; Tantray, V.G.; Kirmani, A.R.; Ahangar, A.G. A review on current status of antiviral siRNA. Rev. Med. Virol. 2018, 28, e1976. [Google Scholar]
- Lam, J.K.; Chow, M.Y.; Zhang, Y.; Leung, S.W. siRNA versus miRNA as therapeutics for gene silencing. Mol. Ther. Nucleic Acids. 2015, 4, e252. [Google Scholar]
- Idrees, S.; Ashfaq, U.A. RNAi: Antiviral therapy against dengue virus. Asian Pac. J. Trop Biomed. 2013, 3, 232–236. [Google Scholar]
- Agrawal, N.; Dasaradhi, P.; Mohmmed, A.; Malhotra, P.; Bhatnagar, R.K.; Mukherjee, S.K. RNA interference: Biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 2003, 67, 657–685. [Google Scholar]
- Parashar, D.; Rajendran, V.; Shukla, R.; Sistla, R. Lipid-based nanocarriers for delivery of small interfering RNA for therapeutic use. Eur. J. Pharm. Sci. 2020, 142, 105159. [Google Scholar]
- Zhang, W.; Singam, R.; Hellermann, G.; Kong, X.; San Juan, H.; Lockey, R.F.; Wu, S.-J.; Porter, K.; Mohapatra, S.S. Attenuation of dengue virus infection by adeno-associated virus-mediated siRNA delivery. Genet. Vaccines Ther. 2004, 2, 8. [Google Scholar]
- Subramanya, S.; Kim, S.-S.; Abraham, S.; Yao, J.; Kumar, M.; Kumar, P.; Haridas, V.; Lee, S.-K.; Shultz, L.D.; Greiner, D. Targeted delivery of small interfering RNA to human dendritic cells to suppress dengue virus infection and associated proinflammatory cytokine production. J Virol. 2010, 84, 2490–2501. [Google Scholar]
- Yue, J.; Wu, X.; Wu, Y.; Li, X.; Jiang, L.; Li, Q.; Li, L.; Yang, X. Study on the inhibitory effect of RNA interference on replication of dengue virus. Bing Du Xue Bao. 2010, 26, 373–378. [Google Scholar]
- Stein, D.A.; Perry, S.T.; Buck, M.D.; Oehmen, C.S.; Fischer, M.A.; Poore, E.; Smith, J.L.; Lancaster, A.M.; Hirsch, A.J.; Slifka, M.K. Inhibition of dengue virus infections in cell cultures and in AG129 mice by a small interfering RNA targeting a highly conserved sequence. J. Virol. 2011, 85, 10154–10166. [Google Scholar]
- Ang, F.; Wong, A.P.Y.; Ng, M.M.-L.; Chu, J.J.H. Small interference RNA profiling reveals the essential role of human membrane trafficking genes in mediating the infectious entry of dengue virus. Virol. J. 2010, 7, 24. [Google Scholar]
- Alhoot, M.A.; Wang, S.M.; Sekaran, S.D. RNA interference mediated inhibition of dengue virus multiplication and entry in HepG2 cells. PLoS ONE. 2012, 7, e34060. [Google Scholar]
- Villegas-Rosales, P.M.; Méndez-Tenorio, A.; Ortega-Soto, E.; Barrón, B.L. Bioinformatics prediction of siRNAs as potential antiviral agents against dengue viruses. Bioinformation. 2012, 8, 519. [Google Scholar]
- Villegas, P.M.; Ortega, E.; Villa-Tanaca, L.; Barrón, B.L.; Torres-Flores, J. Inhibition of dengue virus infection by small interfering RNAs that target highly conserved sequences in the NS4B or NS5 coding regions. Arch Virol. 2018, 163, 1331–1335. [Google Scholar]
- Yan, H.; Zhou, Y.; Liu, Y.; Deng, Y.; Puthiyakunnon, S.; Chen, X. MiR-252 of the Asian tiger mosquito Aedes albopictus regulates dengue virus replication by suppressing the expression of the dengue virus envelope protein. J Med. Virol. 2014, 86, 1428–1436. [Google Scholar]
- Wen, W.; He, Z.; Jing, Q.; Hu, Y.; Lin, C.; Zhou, R.; Wang, X.; Su, Y.; Yuan, J.; Chen, Z. Cellular microRNA-miR-548g-3p modulates the replication of dengue virus. J. Infect. 2015, 70, 631–640. [Google Scholar]
- Castillo, J.A.; Castrillón, J.C.; Diosa-Toro, M.; Betancur, J.G.; St Laurent, G.; Smit, J.M.; Urcuqui-Inchima, S.J.B.i.d. Complex interaction between dengue virus replication and expression of miRNA-133a. BMC Inf Dis. 2015, 16, 29. [Google Scholar]
- Castrillón-Betancur, J.C.; Urcuqui-Inchima, S. Overexpression of miR-484 and miR-744 in Vero cells alters Dengue virus replication. Mem Inst Oswaldo Cruz. 2017, 112, 281–291. [Google Scholar]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990, 249, 505–510. [Google Scholar]
- Berzal-Herranz, A.; Romero-López, C. Two Examples of RNA Aptamers with Antiviral Activity. Are Aptamers the Wished Antiviral Drugs? Pharmaceuticals 2020, 13, 157. [Google Scholar]
- González, V.M.; Martín, M.E.; Fernández, G.; García-Sacristán, A. Use of aptamers as diagnostics tools and antiviral agents for human viruses. Pharmaceuticals 2016, 9, 78. [Google Scholar]
- Kedzierski, S.; Khoshnejad, M.; Caltagirone, G.T. Synthetic antibodies: The emerging field of aptamers. BioProcess J. 2012, 11, 46–49. [Google Scholar]
- Bouchard, P.; Hutabarat, R.; Thompson, K. Discovery and development of therapeutic aptamers. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 237–257. [Google Scholar]
- Marton, S.; Reyes-Darias, J.A.; Sánchez-Luque, F.J.; Romero-López, C.; Berzal-Herranz, A. In vitro and ex vivo selection procedures for identifying potentially therapeutic DNA and RNA molecules. Molecules 2010, 15, 4610–4638. [Google Scholar]
- Kulkarni, R. Antibody-Dependent Enhancement of Viral Infections. In Dynamics of Immune Activation in Viral Diseases; Springer: Singapore, 2020; pp. 9–41. [Google Scholar]
- Balinsky, C.A.; Schmeisser, H.; Ganesan, S.; Singh, K.; Pierson, T.C.; Zoon, K.C. Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J. Virol. 2013, 87, 13094–13106. [Google Scholar]
- Chen, H.-L.; Hsiao, W.-H.; Lee, H.-C.; Wu, S.-C.; Cheng, J.-W. Selection and characterization of DNA aptamers targeting all four serotypes of dengue viruses. PLoS ONE. 2015, 10, e0131240. [Google Scholar]
- Jung, J.I.; Han, S.R.; Lee, S.-W. Development of RNA aptamer that inhibits methyltransferase activity of dengue virus. Biotechnol Lett. 2018, 40, 315–324. [Google Scholar]
- Summerton, J.E. Invention and early history of morpholinos: From pipe dream to practical products. In Morpholino Oligomers. Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; Volume 1565, pp. 1–15. [Google Scholar]
- Mendell, J.R.; Rodino-Klapac, L.R.; Sahenk, Z.; Roush, K.; Bird, L.; Lowes, L.P.; Alfano, L.; Gomez, A.M.; Lewis, S.; Kota, J. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann. Neurol. 2013, 74, 637–647. [Google Scholar]
- Summerton, J.; Weller, D. Morpholino antisense oligomers: Design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997, 7, 187–195. [Google Scholar]
- Morcos, P.A.; Li, Y.; Jiang, S. Vivo-Morpholinos: A non-peptide transporter delivers Morpholinos into a wide array of mouse tissues. Biotechniques 2008, 45, 613–623. [Google Scholar]
- Nan, Y.; Zhang, Y.-J. Antisense phosphorodiamidate morpholino oligomers as novel antiviral compounds. Front. Microbiol. 2018, 9, 750. [Google Scholar]
- Kinney, R.M.; Huang, C.Y.-H.; Rose, B.C.; Kroeker, A.D.; Dreher, T.W.; Iversen, P.L.; Stein, D.A. Inhibition of dengue virus serotypes 1 to 4 in vero cell cultures with morpholino oligomers. J. Virol. 2005, 79, 5116–5128. [Google Scholar]
- Holden, K.L.; Stein, D.A.; Pierson, T.C.; Ahmed, A.A.; Clyde, K.; Iversen, P.L.; Harris, E. Inhibition of dengue virus translation and RNA synthesis by a morpholino oligomer targeted to the top of the terminal 3′ stem–loop structure. Virology 2006, 344, 439–452. [Google Scholar]
- Stein, D.A.; Huang, C.Y.-H.; Silengo, S.; Amantana, A.; Crumley, S.; Blouch, R.E.; Iversen, P.L.; Kinney, R.M. Treatment of AG129 mice with antisense morpholino oligomers increases survival time following challenge with dengue 2 virus. J. Antimicrob Chemother. 2008, 62, 555–565. [Google Scholar]
- Phumesin, P.; Junking, M.; Panya, A.; Yongpitakwattana, P.; Noisakran, S.; Limjindaporn, T.; Yenchitsomanus, P.-T. Vivo-morpholino oligomers strongly inhibit dengue virus replication and production. Arch. Virol. 2018, 163, 867–876. [Google Scholar]
- Phumesin, P.; Junking, M.; Panya, A.; Yongpitakwattana, P.; Noisakran, S.; Limjindaporn, T.; Yenchitsomanus, P.-T. Inhibition of dengue virus replication in monocyte-derived dendritic cells by vivo-morpholino oligomers. Virus Res. 2019, 260, 123–128. [Google Scholar]
- Bayat, H.; Naderi, F.; Khan, A.H.; Memarnejadian, A.; Rahimpour, A. The impact of crispr-cas system on antiviral therapy. Adv. Pharm Bull. 2018, 8, 591. [Google Scholar]
- Zhang, R.; Miner, J.J.; Gorman, M.J.; Rausch, K.; Ramage, H.; White, J.P.; Zuiani, A.; Zhang, P.; Fernandez, E.; Zhang, Q. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature 2016, 535, 164–168. [Google Scholar]
- Savidis, G.; McDougall, W.M.; Meraner, P.; Perreira, J.M.; Portmann, J.M.; Trincucci, G.; John, S.P.; Aker, A.M.; Renzette, N.; Robbins, D.R. Identification of Zika virus and dengue virus dependency factors using functional genomics. Cell Rep. 2016, 16, 232–246. [Google Scholar]
- Marceau, C.D.; Puschnik, A.S.; Majzoub, K.; Ooi, Y.S.; Brewer, S.M.; Fuchs, G.; Swaminathan, K.; Mata, M.A.; Elias, J.E.; Sarnow, P. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 2016, 535, 159–163. [Google Scholar]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.; Kellner, M.J.; Regev, A. RNA targeting with CRISPR–Cas13. Nature 2017, 550, 280–284. [Google Scholar]
- Kumar, N.; Marx, D. Deciphering the Self-Cleavage Reaction Mechanism of Hairpin Ribozyme. J. Phys. Chem B. 2020. [Google Scholar]
- Xu, W.; Jiang, X.; Huang, L. RNA Interference Technology. Compr. Biotechnol. 2019, 560–575. [Google Scholar]
- Buglak, A.A.; Samokhvalov, A.V.; Zherdev, A.V.; Dzantiev, B.B. Methods and Applications of In Silico Aptamer Design and Modeling. Int. J. Mol. Sci. 2020, 21, 8420. [Google Scholar]
- Roberts, T.C.; Langer, R.; Wood, M.J. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar]
- Perreault, J.; Weinberg, Z.; Roth, A.; Popescu, O.; Chartrand, P.; Ferbeyre, G.; Breaker, R.R. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput Biol. 2011, 7, e1002031. [Google Scholar]
- Hedberg, A.; Johansen, S.D. Nuclear group I introns in self-splicing and beyond. Mob. DNA. 2013, 4, 17. [Google Scholar]
- Svoboda, P. Key Mechanistic Principles and Considerations Concerning RNA Interference. Front. Plant. Sci. 2020, 11. [Google Scholar]
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent advances in aptamer discovery and applications. Molecules 2019, 24, 941. [Google Scholar]
- Quijano, E.; Bahal, R.; Ricciardi, A.; Saltzman, W.M.; Glazer, P.M. Therapeutic Peptide Nucleic Acids: Principles, Limitations, and Opportunities. Yale J. Biol. Med. 2017, 90, 583. [Google Scholar]
- Hertel, K.J.; Herschlag, D.; Uhlenbeck, O.C. Specificity of hammerhead ribozyme cleavage. EMBO J. 1996, 15, 3751–3757. [Google Scholar]
- Kalra, P.; Dhiman, A.; Cho, W.C.; Bruno, J.G.; Sharma, T.K. Simple methods and rational design for enhancing aptamer sensitivity and specificity. Front. Mol. Biosci. 2018, 5, 41. [Google Scholar]
- Blum, M.; De Robertis, E.M.; Wallingford, J.B.; Niehrs, C. Morpholinos: Antisense and sensibility. Dev. Cell. 2015, 35, 145–149. [Google Scholar]
- Kharma, N.; Varin, L.; Abu-Baker, A.; Ouellet, J.; Najeh, S.; Ehdaeivand, M.-R.; Belmonte, G.; Ambri, A.; Rouleau, G.; Perreault, J. Automated design of hammerhead ribozymes and validation by targeting the PABPN1 gene transcript. Nucleic Acids Res. 2016, 44, e39. [Google Scholar]
- Bartoszewski, R.; Sikorski, A.F. Editorial focus: Understanding off-target effects as the key to successful RNAi therapy. Cell Mol. Biol. Lett. 2019, 24, 1–23. [Google Scholar]
- Zhu, G.; Chen, X. Aptamer-based targeted therapy. Adv. Drug Deliv. Rev. 2018, 134, 65–78. [Google Scholar]
- Eisen, J.S.; Smith, J.C. Controlling morpholino experiments: Don’t stop making antisense. Development 2008, 135, 1735–1743. [Google Scholar]
- Chu, C.-Y.; Rana, T.M. Potent RNAi by short RNA triggers. RNA 2008, 14, 1714–1719. [Google Scholar]
- Zou, X.; Wu, J.; Gu, J.; Shen, L.; Mao, L. Application of aptamers in virus detection and antiviral therapy. Front. Microbiol. 2019, 10, 1462. [Google Scholar]
- Moulton, J.D.; Yan, Y.L. Using Morpholinos to control gene expression. Curr. Protoc. Mol. Biol. 2008, 83, 26–28. [Google Scholar]
- Westhof, E.; Fritsch, V. RNA folding: Beyond Watson–Crick pairs. Structure 2000, 8, R55–R65. [Google Scholar]
- Kim, D.H.; Rossi, J.J. RNAi mechanisms and applications. Biotechniques 2008, 44, 613–616. [Google Scholar]
- Hidding, J. A therapeutic battle: Antibodies vs. Aptamers. Nanosci. Master Program. 2016, 1–20. [Google Scholar]
- Xiao, G.; Wesolowski, D.; Izadjoo, M.; Altman, S. Morpholino oligonucleotides do not participate perfectly in standard Watson-Crick complexes with RNA. RNA 2010, 16, 2218–2225. [Google Scholar]
- Delley, C.L.; Liu, L.; Sarhan, M.F.; Abate, A.R. Combined aptamer and transcriptome sequencing of single cells. Sci. Rep. 2018, 8, 1–8. [Google Scholar]
- Bedell, V.M.; Westcot, S.E.; Ekker, S.C. Lessons from morpholino-based screening in zebrafish. Brief. Funct Genomics. 2011, 10, 181–188. [Google Scholar]
- Lewin, A.S.; Hauswirth, W.W. Ribozyme gene therapy: Applications for molecular medicine. Trends Mol. Med. 2001, 7, 221–228. [Google Scholar]
- Heigwer, F.; Port, F.; Boutros, M. RNA interference (RNAi) screening in Drosophila. Genetics 2018, 208, 853–874. [Google Scholar]
- Esposito, C.L.; Catuogno, S.; Condorelli, G.; Ungaro, P.; De Franciscis, V. Aptamer chimeras for therapeutic delivery: The challenging perspectives. Genes (Basel). 2018, 9, 529. [Google Scholar]
- Lakhin, A.; Tarantul, V.; Gening, L. Aptamers: Problems, solutions and prospects. Acta Naturae. 2013, 5, 34–43. [Google Scholar]
- Burnett, J.C.; Rossi, J.J. RNA-based therapeutics: Current progress and future prospects. Chem. Biol. 2012, 19, 60–71. [Google Scholar]
- Ovcharenko, D.; Jarvis, R.; Hunicke-Smith, S.; Kelnar, K.; Brown, D. High-throughput RNAi screening in vitro: From cell lines to primary cells. RNA 2005, 11, 985–993. [Google Scholar]
- Scott, W.G. Ribozymes. Curr. Opin. Struct Biol. 2007, 17, 280–286. [Google Scholar]
- Joga, M.R.; Zotti, M.J.; Smagghe, G.; Christiaens, O. RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: What we know so far. Front. Physiol. 2016, 7, 553. [Google Scholar]
- Phylactou, L.A.; Kilpatrick, M.W.; Wood, M.J. Ribozymes as therapeutic tools for genetic disease. Hum. Mol. Genet. 1998, 7, 1649–1653. [Google Scholar]
- Berkhout, B. RNAi-mediated antiviral immunity in mammals. Curr. Opin. Virol. 2018, 32, 9–14. [Google Scholar]
- Song, K.-M.; Lee, S.; Ban, C. Aptamers and their biological applications. Sensors 2012, 12, 612–631. [Google Scholar]
- Pandey, S.N.; Lee, Y.-C.; Yokota, T.; Chen, Y.-W. Morpholino treatment improves muscle function and pathology of Pitx1 transgenic mice. Mol. Ther. 2014, 22, 390–396. [Google Scholar]
- Gurav, B.; Srinivasan, G. Antisense oligonucleotides as therapeutics and their delivery. Curr. Sci. 2017, 112, 490–498. [Google Scholar]
- Da Pieve, C.; Blackshaw, E.; Missailidis, S.; Perkins, A.C. PEGylation and biodistribution of an anti-MUC1 aptamer in MCF-7 tumor-bearing mice. Bioconjug Chem. 2012, 23, 1377–1381. [Google Scholar]
- Deas, T.S.; Bennett, C.J.; Jones, S.A.; Tilgner, M.; Ren, P.; Behr, M.J.; Stein, D.A.; Iversen, P.L.; Kramer, L.D.; Bernard, K.A.; et al. In vitro resistance selection and in vivo efficacy of morpholino oligomers against West Nile virus. Antimicrob. Agents Chemother. 2007, 51, 2470–2482. [Google Scholar]
- Kugelman, J.R.; Sanchez-Lockhart, M.; Andersen, K.G.; Gire, S.; Park, D.J.; Sealfon, R.; Lin, A.E.; Wohl, S.; Sabeti, P.C.; Kuhn, J.H. Evaluation of the potential impact of Ebola virus genomic drift on the efficacy of sequence-based candidate therapeutics. mBio 2015, 6, e02227-14. [Google Scholar]
- Seelam Prabhakar, P.; Manderville, R.A.; Wetmore, S.D. Impact of the Position of the Chemically Modified 5-Furyl-2′-Deoxyuridine Nucleoside on the Thrombin DNA Aptamer–Protein Complex: Structural Insights into Aptamer Response from MD Simulations. Molecules 2019, 24, 2908. [Google Scholar]
- Seley-Radtke, K. Discovery, Design, Synthesis, and Application of Nucleoside/Nucleotides. Molecules 2020, 25, 1526. [Google Scholar]
- Alves Ferreira-Bravo, I.; Cozens, C.; Holliger, P.; DeStefano, J.J. Selection of 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res. 2015, 43, 9587–9599. [Google Scholar]
- Eremeeva, E.; Fikatas, A.; Margamuljana, L.; Abramov, M.; Schols, D.; Groaz, E.; Herdewijn, P. Highly stable hexitol based XNA aptamers targeting the vascular endothelial growth factor. Nucleic Acids Res. 2019, 47, 4927–4939. [Google Scholar]
- Taylor, A.I.; Houlihan, G.; Holliger, P. Beyond DNA and RNA: The expanding toolbox of synthetic genetics. Cold Spring Harb Perspect Biol. 2019, 11, a032490. [Google Scholar]

| Ribozyme | RNA Interference | Aptamer | Morpholino | |
|---|---|---|---|---|
| Mode of Action | Chemical catalysis reaction [76] | RISC mediated breakage [77] | Intermolecular hybridization-based inhibition [78] | Steric blocking of translation [79] |
| Structural Components | Functional domain and structural domain [80,81] | Guide strand and Passenger strand [82] | Small ss DNA, RNA or synthetic XNA [83] | Nucleotide bases attached with methylenemorpholine rings/oligonucleotide analogs [84] |
| Specificity | High [85] | High [82] | High [86] | High [87] |
| Off Target Effects | Limited [88] | Extensive [89] | Limited [90] | Extensive [91] |
| Target site | 3–10 nt [88] | 18–20 nt [92] | 30–100 nt [93] | 5′UTR-first 25 nt [94] |
| Recognition Mechanism | Watson-crick base pairing [95] | Watson-crick base pairing [96] | Similar like antigen-antibody interaction [97] | Watson-crick base pairing [98] |
| Required Sequence Information | Transcriptome [21] | Transcriptome [77] | Transcriptome/Transcription factor [99] | Transcription start site (TSS) [100] |
| Delivery | Direct delivery, viral vector mediated [101] | Bathing, feeding, injection, transfection, transduction, transgenic [102] | Viral vector, carrier mediated [103] | Lipid-based delivery system [79] |
| Time to Effect | May require a week [88] | 2–3 days [88] | Several days [104] | 4–6 days [94] |
| Cytotoxicity | High [105] | Variable to high [106] | Non-Toxic [104] | Variable to high [105] |
| Cost | Low [107] | Low [108] | Low [83] | High [91] |
| Immunogenicity | Moderate [109] | High [110] | Less [111] | Less [112] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panda, K.; Alagarasu, K.; Parashar, D. Oligonucleotide-Based Approaches to Inhibit Dengue Virus Replication. Molecules 2021, 26, 956. https://doi.org/10.3390/molecules26040956
Panda K, Alagarasu K, Parashar D. Oligonucleotide-Based Approaches to Inhibit Dengue Virus Replication. Molecules. 2021; 26(4):956. https://doi.org/10.3390/molecules26040956
Chicago/Turabian StylePanda, Kingshuk, Kalichamy Alagarasu, and Deepti Parashar. 2021. "Oligonucleotide-Based Approaches to Inhibit Dengue Virus Replication" Molecules 26, no. 4: 956. https://doi.org/10.3390/molecules26040956
APA StylePanda, K., Alagarasu, K., & Parashar, D. (2021). Oligonucleotide-Based Approaches to Inhibit Dengue Virus Replication. Molecules, 26(4), 956. https://doi.org/10.3390/molecules26040956







