Single Molecule Characterization of Amyloid Oligomers
Abstract
1. Introduction
2. Characterization of Amyloid Oligomers by Single Molecule Fluorescence Detection
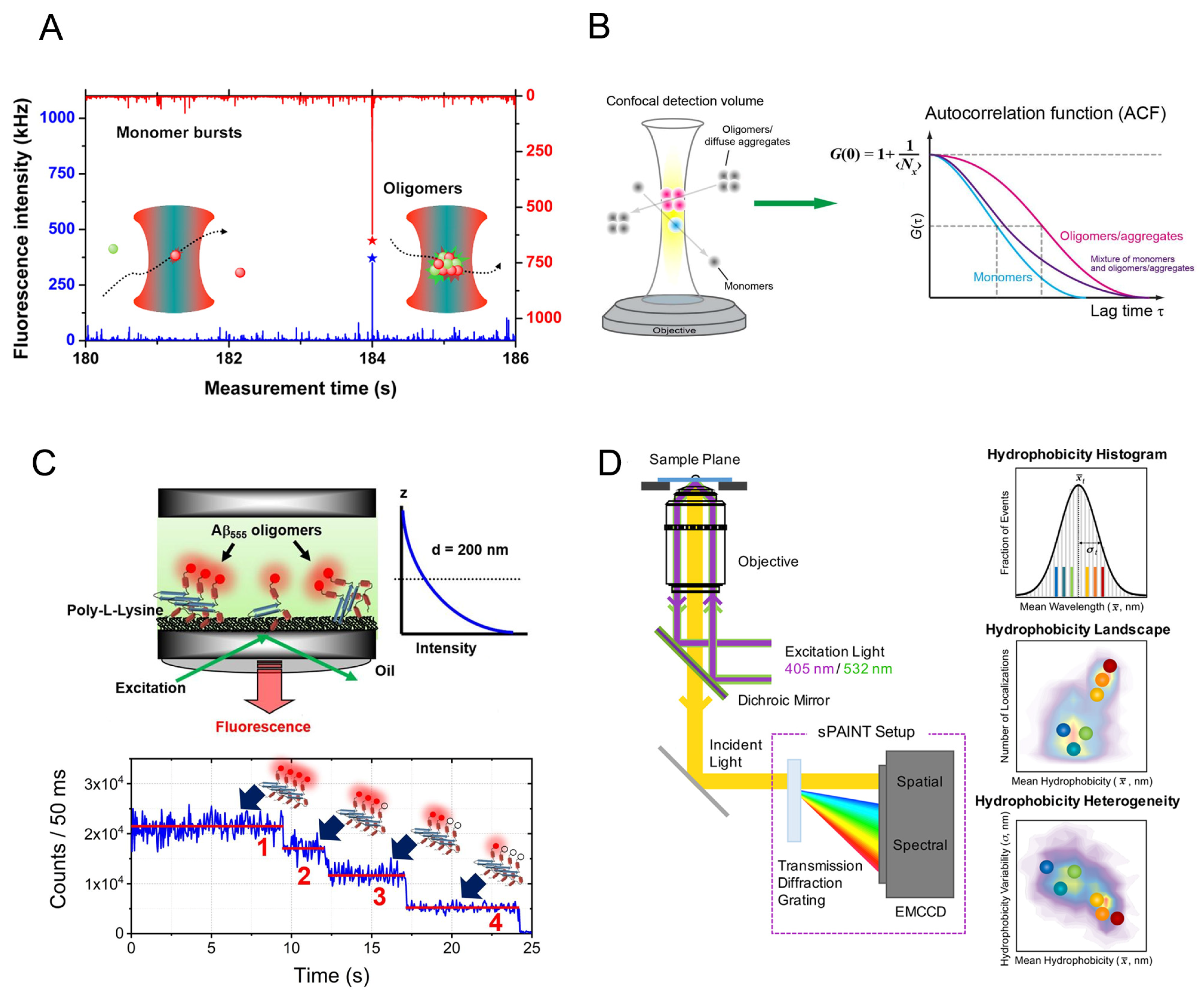
2.1. The Size Distribution of Amyloid Oligomers
2.2. Conformational Diversity of Amyloid Oligomers
2.3. Kinetics and Dynamics of Oligomerization during Amyloid Aggregation
2.4. Thermodynamic Properties of Amyloid Oligomers
2.5. The Effects of Different Factors on the Amyloid Aggregation Pathway
| Factor [with Reference] | Amyloid Protein | Effect on Fibril Formation | Microscopic Mechanism |
|---|---|---|---|
| Heat shock protein 70 (Hsp70) [126] | Tau | Inhibition | Stabilizes monomer and oligomer; sequesters fibril seeds |
| Clusterin [62] | Aβ40 | Inhibition | Interacts with oligomers to form stable complexes |
| αB-crystallin [127] | Aβ40 | Inhibition | Stabilize oligomers to prevent their growth into fibrils |
| GroEL [128] | Aβ42 | Inhibition | Interacts with monomers |
| Clusterin [129] | α-synuclein | Inhibition | Binds to oligomers |
| α2-macroglobulin [129] | α-synuclein | Inhibition | Binds to oligomers |
| Heat shock protein (Hsp27) [130] | α-synuclein | Inhibition | Binds to the fibrillar species to inhibit elongation |
| Polyphenols [138] | α-synuclein | Inhibition | Disaggregates preformed oligomers |
| Arginine [139] | α-synuclein | Inhibition | Changes oligomer conformations |
| Glutamine [139] | α-synuclein | Acceleration | Promotes oligomer formation |
| Nanobodies [107] | α-synuclein | Inhibition | Changes oligomer conformations |
| Short peptide [145] | Ure2 | Acceleration | Promotes oligomer conversion |
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ke, P.C.; Zhou, R.; Serpell, L.C.; Riek, R.; Knowles, T.P.J.; Lashuel, H.A.; Gazit, E.; Hamley, I.W.; Davis, T.P.; Fandrich, M.; et al. Half a century of amyloids: Past, present and future. Chem. Soc. Rev. 2020, 49, 5473–5509. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U. Protein misfolding diseases. Annu Rev. Biochem. 2017, 86, 21–26. [Google Scholar] [CrossRef]
- Soto, C.; Pritzkow, S. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci. 2018, 21, 1332–1340. [Google Scholar] [CrossRef]
- Dobson, C.M.; Knowles, T.P.J.; Vendruscolo, M. The amyloid phenomenon and its significance in biology and medicine. Cold Spring Harb. Perspect. Biol. 2020, 12, a033878. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef]
- Pham, C.L.; Kwan, A.H.; Sunde, M. Functional amyloid: Widespread in Nature, diverse in purpose. Essays Biochem. 2014, 56, 207–219. [Google Scholar]
- Otzen, D.; Riek, R. Functional amyloids. Cold Spring Harb. Perspect. Biol. 2019, 11, a033860. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, A.W.P.; Falcon, B.; He, S.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Crowther, R.A.; Ghetti, B.; Goedert, M.; Scheres, S.H.W. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 2017, 547, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Falcon, B.; Zhang, W.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Vidal, R.; Crowther, R.A.; Ghetti, B.; Scheres, S.H.W.; Goedert, M. Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature 2018, 561, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Gremer, L.; Scholzel, D.; Schenk, C.; Reinartz, E.; Labahn, J.; Ravelli, R.B.G.; Tusche, M.; Lopez-Iglesias, C.; Hoyer, W.; Heise, H.; et al. Fibril structure of amyloid-β(1–42) by cryo-electron microscopy. Science 2017, 358, 116–119. [Google Scholar] [CrossRef] [PubMed]
- Kollmer, M.; Close, W.; Funk, L.; Rasmussen, J.; Bsoul, A.; Schierhorn, A.; Schmidt, M.; Sigurdson, C.J.; Jucker, M.; Fandrich, M. Cryo-EM structure and polymorphism of Aβ amyloid fibrils purified from Alzheimer’s brain tissue. Nat. Commun. 2019, 10, 4760. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Ge, P.; Murray, K.A.; Sheth, P.; Zhang, M.; Nair, G.; Sawaya, M.R.; Shin, W.S.; Boyer, D.R.; Ye, S.; et al. Cryo-EM of full-length α-synuclein reveals fibril polymorphs with a common structural kernel. Nat. Commun. 2018, 9, 3609. [Google Scholar] [CrossRef]
- Cleary, J.P.; Walsh, D.M.; Hofmeister, J.J.; Shankar, G.M.; Kuskowski, M.A.; Selkoe, D.J.; Ashe, K.H. Natural oligomers of the amyloid-β protein specifically disrupt cognitive function. Nat. Neurosci. 2005, 8, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Winner, B.; Jappelli, R.; Maji, S.K.; Desplats, P.A.; Boyer, L.; Aigner, S.; Hetzer, C.; Loher, T.; Vilar, M.; Campioni, S.; et al. In vivo demonstration that α-synuclein oligomers are toxic. Proc. Natl. Acad. Sci. USA 2011, 108, 4194–4199. [Google Scholar] [CrossRef]
- Bemporad, F.; Chiti, F. Protein misfolded oligomers: Experimental approaches, mechanism of formation, and structure-toxicity relationships. Chem. Biol. 2012, 19, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Conway, K.A.; Lee, S.J.; Rochet, J.C.; Ding, T.T.; Williamson, R.E.; Lansbury, P.T., Jr. Acceleration of oligomerization, not fibrillization, is a shared property of both α-synuclein mutations linked to early-onset Parkinson’s disease: Implications for pathogenesis and therapy. Proc. Natl. Acad. Sci. USA 2000, 97, 571–576. [Google Scholar] [CrossRef]
- Guerrero-Munoz, M.J.; Castillo-Carranza, D.L.; Kayed, R. Therapeutic approaches against common structural features of toxic oligomers shared by multiple amyloidogenic proteins. Biochem. Pharmacol. 2014, 88, 468–478. [Google Scholar] [CrossRef]
- LeVine, H., 3rd. Quantification of β-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 1999, 309, 274–284. [Google Scholar]
- Bartolini, M.; Bertucci, C.; Bolognesi, M.L.; Cavalli, A.; Melchiorre, C.; Andrisano, V. Insight into the kinetic of amyloid β (1–42) peptide self-aggregation: Elucidation of inhibitors’ mechanism of action. Chembiochem 2007, 8, 2152–2161. [Google Scholar] [CrossRef]
- Newcomb, C.J.; Moyer, T.J.; Lee, S.S.; Stupp, S.I. Advances in cryogenic transmission electron microscopy for the characterization of dynamic self-assembling nanostructures. Curr. Opin. Colloid Interface Sci. 2012, 17, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.F.; Knowles, T.P.; Dobson, C.M.; Macphee, C.E.; Welland, M.E. Characterization of the nanoscale properties of individual amyloid fibrils. Proc. Natl. Acad. Sci. USA 2006, 103, 15806–15811. [Google Scholar] [CrossRef] [PubMed]
- Carulla, N.; Zhou, M.; Arimon, M.; Gairí, M.; Giralt, E.; Robinson, C.V.; Dobson, C.M. Experimental characterization of disordered and ordered aggregates populated during the process of amyloid fibril formation. Proc. Natl. Acad. Sci. USA 2009, 106, 7828–7833. [Google Scholar] [CrossRef]
- Li, Y.; Lubchenko, V.; Vekilov, P.G. The use of dynamic light scattering and brownian microscopy to characterize protein aggregation. Rev. Sci. Instrum. 2011, 82, 053106. [Google Scholar] [CrossRef]
- Chen, S.W.; Drakulic, S.; Deas, E.; Ouberai, M.; Aprile, F.A.; Arranz, R.; Ness, S.; Roodveldt, C.; Guilliams, T.; De-Genst, E.J.; et al. Structural characterization of toxic oligomers that are kinetically trapped during α-synuclein fibril formation. Proc. Natl. Acad. Sci. USA 2015, 112, E1994–E2003. [Google Scholar] [CrossRef]
- Fusco, G.; Chen, S.W.; Williamson, P.T.F.; Cascella, R.; Perni, M.; Jarvis, J.A.; Cecchi, C.; Vendruscolo, M.; Chiti, F.; Cremades, N.; et al. Structural basis of membrane disruption and cellular toxicity by α-synuclein oligomers. Science 2017, 358, 1440–1443. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Meng, F.; Hao, R.; Wang, C.; Li, F. Study on the structure and membrane disruption of the peptide oligomers constructed by hIAPP18-27 peptide and its d,l-alternating isomer. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183108. [Google Scholar] [CrossRef]
- Ehrnhoefer, D.E.; Bieschke, J.; Boeddrich, A.; Herbst, M.; Masino, L.; Lurz, R.; Engemann, S.; Pastore, A.; Wanker, E.E. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol. 2008, 15, 558–566. [Google Scholar] [CrossRef]
- Ait-Bouziad, N.; Lv, G.; Mahul-Mellier, A.L.; Xiao, S.; Zorludemir, G.; Eliezer, D.; Walz, T.; Lashuel, H.A. Discovery and characterization of stable and toxic Tau/phospholipid oligomeric complexes. Nat. Commun. 2017, 8, 1678. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez Camargo, D.C.; Korshavn, K.J.; Jussupow, A.; Raltchev, K.; Goricanec, D.; Fleisch, M.; Sarkar, R.; Xue, K.; Aichler, M.; Mettenleiter, G.; et al. Stabilization and structural analysis of a membrane-associated hIAPP aggregation intermediate. Elife 2017, 6, e31226. [Google Scholar] [CrossRef]
- Serra-Batiste, M.; Ninot-Pedrosa, M.; Bayoumi, M.; Gairi, M.; Maglia, G.; Carulla, N. Aβ42 assembles into specific β-barrel pore-forming oligomers in membrane-mimicking environments. Proc. Natl. Acad. Sci. USA 2016, 113, 10866–10871. [Google Scholar] [CrossRef]
- Ciudad, S.; Puig, E.; Botzanowski, T.; Meigooni, M.; Arango, A.S.; Do, J.; Mayzel, M.; Bayoumi, M.; Chaignepain, S.; Maglia, G.; et al. Aβ(1–42) tetramer and octamer structures reveal edge conductivity pores as a mechanism for membrane damage. Nat. Commun. 2020, 11, 3014. [Google Scholar] [CrossRef]
- Lerner, E.; Cordes, T.; Ingargiola, A.; Alhadid, Y.; Chung, S.; Michalet, X.; Weiss, S. Toward dynamic structural biology: Two decades of single-molecule Forster resonance energy transfer. Science 2018, 359, eaan1133. [Google Scholar] [CrossRef] [PubMed]
- Voith von Voithenberg, L.; Lamb, D.C. Single pair Forster resonance energy transfer: A versatile tool to investigate protein conformational dynamics. Bioessays 2018, 40, 1700078. [Google Scholar] [CrossRef]
- Bacia, K.; Haustein, E.; Schwille, P. Fluorescence correlation spectroscopy: Principles and applications. Cold Spring Harb. Protoc. 2014, 2014, 709–725. [Google Scholar] [CrossRef]
- Elson, E.L. Introduction to fluorescence correlation spectroscopy-brief and simple. Methods, 2018; 140–141, 3–9. [Google Scholar]
- Kitamura, A.; Kinjo, M. State-of-the-art fluorescence fluctuation-based spectroscopic techniques for the study of protein aggregation. Int. J. Mol. Sci. 2018, 19, 964. [Google Scholar] [CrossRef]
- Meng, F.; Bellaiche, M.M.J.; Kim, J.Y.; Zerze, G.H.; Best, R.B.; Chung, H.S. Highly disordered amyloid-β monomer probed by single-molecule FRET and MD simulation. Biophys. J. 2018, 114, 870–884. [Google Scholar] [CrossRef]
- Elbaum-Garfinkle, S.; Rhoades, E. Identification of an aggregation-prone structure of tau. J. Am. Chem. Soc. 2012, 134, 16607–16613. [Google Scholar] [CrossRef] [PubMed]
- Melo, A.M.; Coraor, J.; Alpha-Cobb, G.; Elbaum-Garfinkle, S.; Nath, A.; Rhoades, E. A functional role for intrinsic disorder in the tau-tubulin complex. Proc. Natl. Acad. Sci. USA 2016, 113, 14336–14341. [Google Scholar] [CrossRef] [PubMed]
- Wickramasinghe, S.P.; Lempart, J.; Merens, H.E.; Murphy, J.; Huettemann, P.; Jakob, U.; Rhoades, E. Polyphosphate initiates Tau aggregation through intra- and intermolecular scaffolding. Biophys. J. 2019, 117, 717–728. [Google Scholar] [CrossRef]
- Ferreon, A.C.; Gambin, Y.; Lemke, E.A.; Deniz, A.A. Interplay of α-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc. Natl. Acad. Sci. USA 2009, 106, 5645–5650. [Google Scholar] [CrossRef] [PubMed]
- Ferreon, A.C.M.; Moran, C.R.; Gambin, Y.; Deniz, A.A. Single-molecule fluorescence studies of intrinsically disordered proteins. Methods Enzymol. 2010, 472, 179–204. [Google Scholar]
- Trexler, A.J.; Rhoades, E. Single molecule characterization of α-synuclein in aggregation-prone states. Biophys. J. 2010, 99, 3048–3055. [Google Scholar] [CrossRef]
- Sevcsik, E.; Trexler, A.J.; Dunn, J.M.; Rhoades, E. Allostery in a disordered protein: Oxidative modifications to α-synuclein act distally to regulate membrane binding. J. Am. Chem. Soc. 2011, 133, 7152–7158. [Google Scholar] [CrossRef]
- Ferreon, A.C.; Moosa, M.M.; Gambin, Y.; Deniz, A.A. Counteracting chemical chaperone effects on the single-molecule α-synuclein structural landscape. Proc. Natl. Acad. Sci. USA 2012, 109, 17826–17831. [Google Scholar] [CrossRef]
- Banerjee, P.R.; Moosa, M.M.; Deniz, A.A. Two-dimensional crowding uncovers a hidden conformation of α-synuclein. Angew Chem. Int. Ed. Engl. 2016, 55, 12789–12792. [Google Scholar] [CrossRef]
- Tsoi, P.S.; Choi, K.J.; Leonard, P.G.; Sizovs, A.; Moosa, M.M.; MacKenzie, K.R.; Ferreon, J.C.; Ferreon, A.C.M. The N-Terminal domain of ALS-linked TDP-43 assembles without misfolding. Angew. Chem. Int. Ed. Engl. 2017, 56, 12590–12593. [Google Scholar] [CrossRef] [PubMed]
- Warner, J.B.T.; Ruff, K.M.; Tan, P.S.; Lemke, E.A.; Pappu, R.V.; Lashuel, H.A. Monomeric Huntingtin exon 1 has similar overall structural features for wild-type and pathological polyglutamine lengths. J. Am. Chem. Soc. 2017, 139, 14456–14469. [Google Scholar] [CrossRef] [PubMed]
- Lou, F.; Yang, J.; Wu, S.; Perrett, S. A co-expression strategy to achieve labeling of individual subunits within a dimeric protein for single molecule analysis. Chem. Commun. (Camb.) 2017, 53, 7986–7989. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, R.; Goodman, J.L.; Mukhopadhyay, S.; Pacheco, C.D.; Lemke, E.A.; Deniz, A.A.; Lindquist, S. Conserved features of intermediates in amyloid assembly determine their benign or toxic states. Proc. Natl. Acad. Sci. USA 2012, 109, 11172–11177. [Google Scholar] [CrossRef]
- Birol, M.; Melo, A.M. Untangling the conformational polymorphism of disordered proteins associated with neurodegeneration at the single-molecule level. Front. Mol. Neurosci. 2019, 12, 309. [Google Scholar] [CrossRef]
- Schuler, B.; Soranno, A.; Hofmann, H.; Nettels, D. Single-Molecule FRET Spectroscopy and the Polymer Physics of Unfolded and Intrinsically Disordered Proteins. Annu Rev. Biophys. 2016, 45, 207–231. [Google Scholar] [CrossRef] [PubMed]
- Metskas, L.A.; Rhoades, E. Single-molecule FRET of intrinsically disordered proteins. Annu Rev. Phys. Chem. 2020, 71, 391–414. [Google Scholar] [CrossRef] [PubMed]
- Cremades, N.; Cohen, S.I.; Deas, E.; Abramov, A.Y.; Chen, A.Y.; Orte, A.; Sandal, M.; Clarke, R.W.; Dunne, P.; Aprile, F.A.; et al. Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell 2012, 149, 1048–1059. [Google Scholar] [CrossRef]
- Shammas, S.L.; Garcia, G.A.; Kumar, S.; Kjaergaard, M.; Horrocks, M.H.; Shivji, N.; Mandelkow, E.; Knowles, T.P.; Mandelkow, E.; Klenerman, D. A mechanistic model of tau amyloid aggregation based on direct observation of oligomers. Nat. Commun. 2015, 6, 7025. [Google Scholar] [CrossRef]
- Iljina, M.; Garcia, G.A.; Horrocks, M.H.; Tosatto, L.; Choi, M.L.; Ganzinger, K.A.; Abramov, A.Y.; Gandhi, S.; Wood, N.W.; Cremades, N.; et al. Kinetic model of the aggregation of α-synuclein provides insights into prion-like spreading. Proc. Natl. Acad. Sci. USA 2016, 113, E1206–E1215. [Google Scholar] [CrossRef]
- Yang, J.; Dear, A.J.; Michaels, T.C.T.; Dobson, C.M.; Knowles, T.P.J.; Wu, S.; Perrett, S. Direct observation of oligomerization by single molecule fluorescence reveals a multistep aggregation mechanism for the yeast prion protein Ure2. J. Am. Chem. Soc. 2018, 140, 2493–2503. [Google Scholar] [CrossRef]
- Orte, A.; Clarke, R.; Klenerman, D. Single-molecule two-colour coincidence detection to probe biomolecular associations. Biochem. Soc. Trans. 2010, 38, 914–918. [Google Scholar] [CrossRef]
- Orte, A.; Birkett, N.R.; Clarke, R.W.; Devlin, G.L.; Dobson, C.M.; Klenerman, D. Direct characterization of amyloidogenic oligomers by single-molecule fluorescence. Proc. Natl. Acad. Sci. USA 2008, 105, 14424–14429. [Google Scholar] [CrossRef] [PubMed]
- Horrocks, M.H.; Li, H.; Shim, J.-U.U.; Ranasinghe, R.T.; Clarke, R.W.; Huck, W.T.; Abell, C.; Klenerman, D. Single molecule fluorescence under conditions of fast flow. Anal. Chem. 2012, 84, 179–185. [Google Scholar] [CrossRef]
- Horrocks, M.H.; Tosatto, L.; Dear, A.J.; Garcia, G.A.; Iljina, M.; Cremades, N.; Dalla Serra, M.; Knowles, T.P.; Dobson, C.M.; Klenerman, D. Fast flow microfluidics and single-molecule fluorescence for the rapid characterization of α-synuclein oligomers. Anal. Chem. 2015, 87, 8818–8826. [Google Scholar] [CrossRef]
- Narayan, P.; Orte, A.; Clarke, R.W.; Bolognesi, B.; Hook, S.; Ganzinger, K.A.; Meehan, S.; Wilson, M.R.; Dobson, C.M.; Klenerman, D. The extracellular chaperone clusterin sequesters oligomeric forms of the amyloid-β(1-40) peptide. Nat. Struct. Mol. Biol. 2012, 19, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Varela, J.A.; Rodrigues, M.; De, S.; Flagmeier, P.; Gandhi, S.; Dobson, C.M.; Klenerman, D.; Lee, S.F. Optical structural analysis of individual α-synuclein oligomers. Angew. Chem. Int. Ed. Engl. 2018, 57, 4886–4890. [Google Scholar] [CrossRef]
- Horrocks, M.H.; Lee, S.F.; Gandhi, S.; Magdalinou, N.K.; Chen, S.W.; Devine, M.J.; Tosatto, L.; Kjaergaard, M.; Beckwith, J.S.; Zetterberg, H.; et al. Single-Molecule Imaging of Individual Amyloid Protein Aggregates in Human Biofluids. ACS Chem. Neurosci. 2016, 7, 399–406. [Google Scholar] [CrossRef]
- Sang, J.C.; Meisl, G.; Thackray, A.M.; Hong, L.; Ponjavic, A.; Knowles, T.P.J.; Bujdoso, R.; Klenerman, D. Direct observation of murine prion protein replication in vitro. J. Am. Chem. Soc. 2018, 140, 14789–14798. [Google Scholar] [CrossRef]
- Ries, J.; Udayar, V.; Soragni, A.; Hornemann, S.; Nilsson, K.P.; Riek, R.; Hock, C.; Ewers, H.; Aguzzi, A.A.; Rajendran, L. Superresolution imaging of amyloid fibrils with binding-activated probes. ACS Chem. Neurosci. 2013, 4, 1057–1061. [Google Scholar] [CrossRef]
- Bongiovanni, M.N.; Godet, J.; Horrocks, M.H.; Tosatto, L.; Carr, A.R.; Wirthensohn, D.C.; Ranasinghe, R.T.; Lee, J.E.; Ponjavic, A.; Fritz, J.V.; et al. Multi-dimensional super-resolution imaging enables surface hydrophobicity mapping. Nat. Commun. 2016, 7, 13544. [Google Scholar] [CrossRef]
- Lee, J.E.; Sang, J.C.; Rodrigues, M.; Carr, A.R.; Horrocks, M.H.; De, S.; Bongiovanni, M.N.; Flagmeier, P.; Dobson, C.M.; Wales, D.J.; et al. Mapping surface hydrophobicity of α-synuclein oligomers at the nanoscale. Nano Lett. 2018, 18, 7494–7501. [Google Scholar] [CrossRef] [PubMed]
- Sang, J.C.; Lee, J.E.; Dear, A.J.; De, S.; Meisl, G.; Thackray, A.M.; Bujdoso, R.; Knowles, T.P.J.; Klenerman, D. Direct observation of prion protein oligomer formation reveals an aggregation mechanism with multiple conformationally distinct species. Chem. Sci. 2019, 10, 4588–4597. [Google Scholar] [CrossRef] [PubMed]
- Spehar, K.; Ding, T.; Sun, Y.; Kedia, N.; Lu, J.; Nahass, G.R.; Lew, M.D.; Bieschke, J. Super-resolution imaging of amyloid structures over extended times by using transient binding of single thioflavin T molecules. Chembiochem 2018, 19, 1944–1948. [Google Scholar] [CrossRef]
- Torra, J.; Bondia, P.; Gutierrez-Erlandsson, S.; Sot, B.; Flors, C. Long-term STED imaging of amyloid fibers with exchangeable thioflavin T. Nanoscale 2020, 12, 15050–15053. [Google Scholar] [CrossRef]
- Castello, F.; Paredes, J.M.; Ruedas-Rama, M.J.; Martin, M.; Roldan, M.; Casares, S.; Orte, A. Two-step amyloid aggregation: Sequential lag phase intermediates. Sci. Rep. 2017, 7, 40065. [Google Scholar] [CrossRef] [PubMed]
- Dresser, L.; Hunter, P.; Yendybayeva, F.; Hargreaves, A.L.; Howard, J.A.L.; Evans, G.J.O.; Leake, M.C.; Quinn, S.D. Amyloid-β oligomerization monitored by single-molecule stepwise photobleaching. Methods 2020. [CrossRef]
- Nath, S.; Meuvis, J.; Hendrix, J.; Carl, S.A.; Engelborghs, Y. Early aggregation steps in α-synuclein as measured by FCS and FRET: Evidence for a contagious conformational change. Biophys. J. 2010, 98, 1302–1311. [Google Scholar] [CrossRef]
- Basak, S.; Prasad, G.V.; Varkey, J.; Chattopadhyay, K. Early sodium dodecyl sulfate induced collapse of alpha-synuclein correlates with its amyloid formation. ACS Chem. Neurosci. 2015, 6, 239–246. [Google Scholar] [CrossRef]
- Joshi, N.; Basak, S.; Kundu, S.; De, G.; Mukhopadhyay, A.; Chattopadhyay, K. Attenuation of the early events of α-synuclein aggregation: A fluorescence correlation spectroscopy and laser scanning microscopy study in the presence of surface-coated Fe3O4 nanoparticles. Langmuir 2015, 31, 1469–1478. [Google Scholar] [CrossRef]
- Wennmalm, S.; Chmyrov, V.; Widengren, J.; Tjernberg, L. Highly sensitive FRET-FCS detects amyloid β-peptide oligomers in solution at physiological concentrations. Anal. Chem. 2015, 87, 11700–11705. [Google Scholar] [CrossRef]
- Guan, Y.; Cao, K.J.; Cantlon, A.; Elbel, K.; Theodorakis, E.A.; Walsh, D.M.; Yang, J.; Shah, J.V. Real-time monitoring of Alzheimer’s-related amyloid aggregation via probe enhancement-fluorescence correlation spectroscopy. ACS Chem. Neurosci. 2015, 6, 1503–1508. [Google Scholar] [CrossRef]
- Tiiman, A.; Jelic, V.; Jarvet, J.; Jaremo, P.; Bogdanovic, N.; Rigler, R.; Terenius, L.; Graslund, A.; Vukojevic, V. Amyloidogenic nanoplaques in blood serum of patients with Alzheimer’s disease revealed by time-resolved thioflavin T fluorescence intensity fluctuation analysis. J. Alzheimers Dis. 2019, 68, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Wong, P.T.; Lee, E.L.; Gafni, A.; Steel, D.G. Determination of the Oligomer Size of Amyloidogenic Protein β-Amyloid(1–40) by Single-Molecule Spectroscopy. Biophys. J. 2009, 97, 912–921. [Google Scholar] [CrossRef]
- Johnson, R.D.; Schauerte, J.A.; Wisser, K.C.; Gafni, A.; Steel, D.G. Direct observation of single amyloid-β(1-40) oligomers on live cells: Binding and growth at physiological concentrations. PLoS ONE 2011, 6, e23970. [Google Scholar] [CrossRef]
- Ding, H.; Schauerte, J.A.; Steel, D.G.; Gafni, A. β-Amyloid (1-40) peptide interactions with supported phospholipid membranes: A single-molecule study. Biophys. J. 2012, 103, 1500–1509. [Google Scholar] [CrossRef]
- Lukiw, W.; Chang, C.-C.; Althaus, J.C.; Carruthers, C.J.L.; Sutton, M.A.; Steel, D.G.; Gafni, A. Synergistic interactions between Alzheimer’s Aβ40 and Aβ42 on the surface of primary neurons revealed by single molecule microscopy. PLoS ONE 2013, 8, e82139. [Google Scholar]
- Johnson, R.D.; Steel, D.G.; Gafni, A. Structural evolution and membrane interactions of Alzheimer’s amyloid-β peptide oligomers: New knowledge from single-molecule fluorescence studies. Protein Sci. 2014, 23, 869–883. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.; Das, A.; Dey, A.; Maiti, S. Membrane affinity of individual toxic protein oligomers determined at the single-molecule level. Phys. Chem. Chem. Phys. 2020, 22, 14613–14620. [Google Scholar] [CrossRef]
- Zijlstra, N.; Blum, C.; Segers-Nolten, I.M.; Claessens, M.M.; Subramaniam, V. Molecular composition of sub-stoichiometrically labeled α-synuclein oligomers determined by single-molecule photobleaching. Angew. Chem. Int. Ed. Engl. 2012, 51, 8821–8824. [Google Scholar] [CrossRef]
- Zijlstra, N.; Claessens, M.M.; Blum, C.; Subramaniam, V. Elucidating the aggregation number of dopamine-induced α-synuclein oligomeric assemblies. Biophys. J. 2014, 106, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Mannini, B.; Mulvihill, E.; Sgromo, C.; Cascella, R.; Khodarahmi, R.; Ramazzotti, M.; Dobson, C.M.; Cecchi, C.; Chiti, F. Toxicity of protein oligomers is rationalized by a function combining size and surface hydrophobicity. ACS Chem. Biol. 2014, 9, 2309–2317. [Google Scholar] [CrossRef]
- Campioni, S.; Mannini, B.; Zampagni, M.; Pensalfini, A.; Parrini, C.; Evangelisti, E.; Relini, A.; Stefani, M.; Dobson, C.M.; Cecchi, C.; et al. A causative link between the structure of aberrant protein oligomers and their toxicity. Nat. Chem. Biol. 2010, 6, 140–147. [Google Scholar] [CrossRef]
- Tosatto, L.; Horrocks, M.H.; Dear, A.J.; Knowles, T.P.; Dalla Serra, M.; Cremades, N.; Dobson, C.M.; Klenerman, D. Single-molecule FRET studies on α-synuclein oligomerization of Parkinson’s disease genetically related mutants. Sci. Rep. 2015, 5, 16696. [Google Scholar] [CrossRef]
- Castello, F.; Casares, S.; Ruedas-Rama, M.J.; Orte, A. The first step of amyloidogenic aggregation. J. Phys. Chem. B 2015, 119, 8260–8267. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Culyba, E.K.; Powers, E.T.; Kelly, J.W. Amyloid-β forms fibrils by nucleated conformational conversion of oligomers. Nat. Chem. Biol. 2011, 7, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Saric, A.; Chebaro, Y.C.; Knowles, T.P.; Frenkel, D. Crucial role of nonspecific interactions in amyloid nucleation. Proc. Natl. Acad. Sci. USA 2014, 111, 17869–17874. [Google Scholar] [CrossRef]
- Saric, A.; Michaels, T.C.T.; Zaccone, A.; Knowles, T.P.J.; Frenkel, D. Kinetics of spontaneous filament nucleation via oligomers: Insights from theory and simulation. J. Chem. Phys. 2016, 145, 211926. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.; Waudby, C.A.; Devlin, G.L.; Cohen, S.I.; Aguzzi, A.; Vendruscolo, M.; Terentjev, E.M.; Welland, M.E.; Dobson, C.M. An analytical solution to the kinetics of breakable filament assembly. Science 2009, 326, 1533–1537. [Google Scholar] [CrossRef]
- Cohen, S.I.; Vendruscolo, M.; Welland, M.E.; Dobson, C.M.; Terentjev, E.M.; Knowles, T.P. Nucleated polymerization with secondary pathways. I. Time evolution of the principal moments. J. Chem. Phys. 2011, 135, 065105. [Google Scholar] [CrossRef]
- Cohen, S.I.A.; Vendruscolo, M.; Dobson, C.M.; Knowles, T.P.J. From macroscopic measurements to microscopic mechanisms of protein aggregation. J. Mol. Biol. 2012, 421, 160–171. [Google Scholar] [CrossRef]
- Cohen, S.I.; Linse, S.; Luheshi, L.M.; Hellstrand, E.; White, D.A.; Rajah, L.; Otzen, D.E.; Vendruscolo, M.; Dobson, C.M.; Knowles, T.P. Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl. Acad. Sci. USA 2013, 110, 9758–9763. [Google Scholar] [CrossRef]
- Xu, L.Q.; Wu, S.; Buell, A.K.; Cohen, S.I.A.; Chen, L.J.; Hu, W.H.; Cusack, S.A.; Itzhaki, L.S.; Zhang, H.; Knowles, T.P.J.; et al. Influence of specific HSP70 domains on fibril formation of the yeast prion protein Ure2. Phil. Trans. R. Soc. B 2013, 368, 20110410. [Google Scholar] [CrossRef]
- Meisl, G.; Kirkegaard, J.B.; Arosio, P.; Michaels, T.C.; Vendruscolo, M.; Dobson, C.M.; Linse, S.; Knowles, T.P. Molecular mechanisms of protein aggregation from global fitting of kinetic models. Nat. Protoc. 2016, 11, 252–272. [Google Scholar] [CrossRef] [PubMed]
- Kundel, F.; Hong, L.; Falcon, B.; McEwan, W.A.; Michaels, T.C.T.; Meisl, G.; Esteras, N.; Abramov, A.Y.; Knowles, T.J.P.; Goedert, M.; et al. Measurement of Tau filament fragmentation provides insights into prion-like spreading. ACS Chem. Neurosci. 2018, 9, 1276–1282. [Google Scholar] [CrossRef]
- Kjaergaard, M.; Dear, A.J.; Kundel, F.; Qamar, S.; Meisl, G.; Knowles, T.P.J.; Klenerman, D. Oligomer diversity during the aggregation of the repeat region of Tau. ACS Chem. Neurosci. 2018, 9, 3060–3071. [Google Scholar] [CrossRef] [PubMed]
- Buell, A.K.; Galvagnion, C.; Gaspar, R.; Sparr, E.; Vendruscolo, M.; Knowles, T.P.; Linse, S.; Dobson, C.M. Solution conditions determine the relative importance of nucleation and growth processes in α-synuclein aggregation. Proc. Natl. Acad. Sci. USA 2014, 111, 7671–7676. [Google Scholar] [CrossRef] [PubMed]
- Ruschak, A.M.; Miranker, A.D. Fiber-dependent amyloid formation as catalysis of an existing reaction pathway. Proc. Natl. Acad. Sci. USA 2007, 104, 12341–12346. [Google Scholar] [CrossRef]
- Dear, A.J.; Michaels, T.C.T.; Meisl, G.; Klenerman, D.; Wu, S.; Perrett, S.; Linse, S.; Dobson, C.M.; Knowles, T.P.J. Kinetic diversity of amyloid oligomers. Proc. Natl. Acad. Sci. USA 2020, 117, 12087–12094. [Google Scholar] [CrossRef] [PubMed]
- Arosio, P.; Michaels, T.C.; Linse, S.; Mansson, C.; Emanuelsson, C.; Presto, J.; Johansson, J.; Vendruscolo, M.; Dobson, C.M.; Knowles, T.P. Kinetic analysis reveals the diversity of microscopic mechanisms through which molecular chaperones suppress amyloid formation. Nat. Commun. 2016, 7, 10948. [Google Scholar] [CrossRef] [PubMed]
- Iljina, M.; Hong, L.; Horrocks, M.H.; Ludtmann, M.H.; Choi, M.L.; Hughes, C.D.; Ruggeri, F.S.; Guilliams, T.; Buell, A.K.; Lee, J.-E.E.; et al. Nanobodies raised against monomeric ɑ-synuclein inhibit fibril formation and destabilize toxic oligomeric species. BMC Biol. 2017, 15, 57. [Google Scholar] [CrossRef]
- Michaels, T.C.T.; Saric, A.; Curk, S.; Bernfur, K.; Arosio, P.; Meisl, G.; Dear, A.J.; Cohen, S.I.A.; Dobson, C.M.; Vendruscolo, M.; et al. Dynamics of oligomer populations formed during the aggregation of Alzheimer’s Aβ42 peptide. Nat. Chem. 2020, 12, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Spires-Jones, T.L.; Attems, J.; Thal, D.R. Interactions of pathological proteins in neurodegenerative diseases. Acta Neuropathol. 2017, 134, 187–205. [Google Scholar] [CrossRef] [PubMed]
- Frost, D.; Gorman, P.M.; Yip, C.M.; Chakrabartty, A. Co-incorporation of Aβ40 and Aβ42 to form mixed pre-fibrillar aggregates. Eur. J. Biochem. 2003, 270, 654–663. [Google Scholar] [CrossRef]
- Hasegawa, K.; Yamaguchi, I.; Omata, S.; Gejyo, F.; Naiki, H. Interaction between Aβ(1–42) and Aβ(1–40) in Alzheimer’s β-amyloid fibril formation in vitro. Biochemistry 1999, 38, 15514–15521. [Google Scholar] [CrossRef] [PubMed]
- Cukalevski, R.; Yang, X.; Meisl, G.; Weininger, U.; Bernfur, K.; Frohm, B.; Knowles, T.P.J.; Linse, S. The Aβ40 and Aβ42 peptides self-assemble into separate homomolecular fibrils in binary mixtures but cross-react during primary nucleation. Chem. Sci. 2015, 6, 4215–4233. [Google Scholar] [CrossRef]
- Iljina, M.; Garcia, G.A.; Dear, A.J.; Flint, J.; Narayan, P.; Michaels, T.C.; Dobson, C.M.; Frenkel, D.; Knowles, T.P.; Klenerman, D. Quantitative analysis of co-oligomer formation by amyloid-β peptide isoforms. Sci. Rep. 2016, 6, 28658. [Google Scholar] [CrossRef]
- Sierecki, E.; Giles, N.; Bowden, Q.; Polinkovsky, M.E.; Steinbeck, J.; Arrioti, N.; Rahman, D.; Bhumkar, A.; Nicovich, P.R.; Ross, I.; et al. Nanomolar oligomerization and selective co-aggregation of α-synuclein pathogenic mutants revealed by single-molecule fluorescence. Sci. Rep. 2016, 6, 37630. [Google Scholar] [CrossRef]
- Nubling, G.; Bader, B.; Levin, J.; Hildebrandt, J.; Kretzschmar, H.; Giese, A. Synergistic influence of phosphorylation and metal ions on tau oligomer formation and coaggregation with α-synuclein at the single molecule level. Mol. Neurodegener. 2012, 7, 35. [Google Scholar] [CrossRef]
- Iljina, M.; Dear, A.J.; Garcia, G.A.; De, S.; Tosatto, L.; Flagmeier, P.; Whiten, D.R.; Michaels, T.C.T.; Frenkel, D.; Dobson, C.M.; et al. Quantifying co-oligomer formation by α-synuclein. ACS Nano 2018, 12, 10855–10866. [Google Scholar] [CrossRef] [PubMed]
- Flagmeier, P.; De, S.; Wirthensohn, D.C.; Lee, S.F.; Vincke, C.; Muyldermans, S.; Knowles, T.P.J.; Gandhi, S.; Dobson, C.M.; Klenerman, D. Ultrasensitive measurement of Ca(2+) influx into lipid vesicles induced by protein aggregates. Angew. Chem. Int. Ed. Engl. 2017, 56, 7750–7754. [Google Scholar] [CrossRef] [PubMed]
- Mannini, B.; Chiti, F. Chaperones as Suppressors of Protein Misfolded Oligomer Toxicity. Front. Mol. Neurosci. 2017, 10, 98. [Google Scholar] [CrossRef]
- Siddiqi, M.K.; Alam, P.; Chaturvedi, S.K.; Shahein, Y.E.; Khan, R.H. Mechanisms of protein aggregation and inhibition. Front. Biosci. 2017, 9, 1–20. [Google Scholar]
- Doig, A.J.; Derreumaux, P. Inhibition of protein aggregation and amyloid formation by small molecules. Curr. Opin. Struct. Biol. 2015, 30, 50–56. [Google Scholar] [CrossRef]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. In vivo aspects of protein folding and quality control. Science 2016, 353, aac4354. [Google Scholar] [CrossRef]
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. [Google Scholar] [CrossRef]
- Wentink, A.; Nussbaum-Krammer, C.; Bukau, B. Modulation of amyloid states by molecular chaperones. Cold Spring Harb. Perspect. Biol. 2019, 11, a033969. [Google Scholar] [CrossRef]
- Voss, K.; Combs, B.; Patterson, K.R.; Binder, L.I.; Gamblin, T.C. Hsp70 alters tau function and aggregation in an isoform specific manner. Biochemistry 2012, 51, 888–898. [Google Scholar] [CrossRef] [PubMed]
- Patterson, K.R.; Ward, S.M.; Combs, B.; Voss, K.; Kanaan, N.M.; Morfini, G.; Brady, S.T.; Gamblin, T.C.; Binder, L.I. Heat shock protein 70 prevents both tau aggregation and the inhibitory effects of preexisting tau aggregates on fast axonal transport. Biochemistry 2011, 50, 10300–10310. [Google Scholar] [CrossRef]
- Kundel, F.; De, S.; Flagmeier, P.; Horrocks, M.H.; Kjaergaard, M.; Shammas, S.L.; Jackson, S.E.; Dobson, C.M.; Klenerman, D. Hsp70 inhibits the nucleation and elongation of Tau and sequesters Tau aggregates with high affinity. ACS Chem. Biol. 2018, 13, 636–646. [Google Scholar] [CrossRef]
- Narayan, P.; Meehan, S.; Carver, J.A.; Wilson, M.R.; Dobson, C.M.; Klenerman, D. Amyloid-β oligomers are sequestered by both intracellular and extracellular chaperones. Biochemistry 2012, 51, 9270–9276. [Google Scholar] [CrossRef] [PubMed]
- Walti, M.A.; Steiner, J.; Meng, F.; Chung, H.S.; Louis, J.M.; Ghirlando, R.; Tugarinov, V.; Nath, A.; Clore, G.M. Probing the mechanism of inhibition of amyloid-β(1–42)-induced neurotoxicity by the chaperonin GroEL. Proc. Natl. Acad. Sci. USA 2018, 115, E11924–E11932. [Google Scholar] [CrossRef] [PubMed]
- Whiten, D.R.; Cox, D.; Horrocks, M.H.; Taylor, C.G.; De, S.; Flagmeier, P.; Tosatto, L.; Kumita, J.R.; Ecroyd, H.; Dobson, C.M.; et al. Single-molecule characterization of the interactions between extracellular chaperones and toxic α-synuclein oligomers. Cell Rep. 2018, 23, 3492–3500. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.; Whiten, D.R.; Brown, J.W.P.; Horrocks, M.H.; San Gil, R.; Dobson, C.M.; Klenerman, D.; van Oijen, A.M.; Ecroyd, H. The small heat shock protein Hsp27 binds α-synuclein fibrils, preventing elongation and cytotoxicity. J. Biol. Chem. 2018, 293, 4486–4497. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Gong, H.; Xiao, H.; Petersen, R.B.; Zheng, L.; Huang, K. Inhibiting toxic aggregation of amyloidogenic proteins: A therapeutic strategy for protein misfolding diseases. Biochim. Biophys. Acta 2013, 1830, 4860–4871. [Google Scholar] [CrossRef]
- Michaels, T.C.T.; Saric, A.; Meisl, G.; Heller, G.T.; Curk, S.; Arosio, P.; Linse, S.; Dobson, C.M.; Vendruscolo, M.; Knowles, T.P.J. Thermodynamic and kinetic design principles for amyloid-aggregation inhibitors. Proc. Natl. Acad. Sci. USA 2020, 24251–24257. [Google Scholar] [CrossRef] [PubMed]
- Linse, S. Mechanism of amyloid protein aggregation and the role of inhibitors. Pure Appl. Chem. 2019, 91, 211–229. [Google Scholar] [CrossRef]
- Giorgetti, S.; Greco, C.; Tortora, P.; Aprile, F.A. Targeting amyloid aggregation: An overview of strategies and mechanisms. Int. J. Mol. Sci. 2018, 19, 2677. [Google Scholar] [CrossRef]
- Aguzzi, A.; O’Connor, T. Protein aggregation diseases: Pathogenicity and therapeutic perspectives. Nat. Rev. Drug Discov. 2010, 9, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Kwon, Y.T. Degradation of misfolded proteins in neurodegenerative diseases: Therapeutic targets and strategies. Exp. Mol. Med. 2015, 47, e147. [Google Scholar] [CrossRef]
- Zaman, M.; Khan, A.N.; Wahiduzzaman; Zakariya, S.M.; Khan, R.H. Protein misfolding, aggregation and mechanism of amyloid cytotoxicity: An overview and therapeutic strategies to inhibit aggregation. Int. J. Biol. Macromol. 2019, 134, 1022–1037. [Google Scholar] [CrossRef] [PubMed]
- Caruana, M.; Hogen, T.; Levin, J.; Hillmer, A.; Giese, A.; Vassallo, N. Inhibition and disaggregation of α-synuclein oligomers by natural polyphenolic compounds. FEBS Lett. 2011, 585, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Kundu, A.; Chattopadhyay, K. Small molecules attenuate the interplay between conformational fluctuations, early oligomerization and amyloidosis of α-synuclein. Sci. Rep. 2018, 8, 5481. [Google Scholar] [CrossRef] [PubMed]
- De Genst, E.; Messer, A.; Dobson, C.M. Antibodies and protein misfolding: From structural research tools to therapeutic strategies. Biochim Biophys. Acta 2014, 1844, 1907–1919. [Google Scholar] [CrossRef]
- De Genst, E.; Chan, P.-H.; Pardon, E.; Hsu, S.-T.D.; Kumita, J.R.; Christodoulou, J.; Menzer, L.; Chirgadze, D.Y.; Robinson, C.V.; Muyldermans, S.; et al. A nanobody binding to non-amyloidogenic regions of the protein human lysozyme enhances partial unfolding but inhibits amyloid fibril formation. J. Phys. Chem. B 2013, 117, 13245–13258. [Google Scholar] [CrossRef]
- Emadi, S.; Liu, R.; Yuan, B.; Schulz, P.; McAllister, C.; Lyubchenko, Y.; Messer, A.; Sierks, M.R. Inhibiting aggregation of α-synuclein with human single chain antibody fragments. Biochemistry 2004, 43, 2871–2878. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Kotia, V.; Ghosh, D.; Mohite, G.M.; Kumar, A.; Maji, S.K. Curcumin modulates α-synuclein aggregation and toxicity. ACS Chem. Neurosci. 2013, 4, 393–407. [Google Scholar] [CrossRef]
- Cremers, C.M.; Knoefler, D.; Gates, S.; Martin, N.; Dahl, J.U.; Lempart, J.; Xie, L.; Chapman, M.R.; Galvan, V.; Southworth, D.R.; et al. Polyphosphate: A conserved modifier of amyloidogenic processes. Mol. Cell 2016, 63, 768–780. [Google Scholar] [CrossRef]
- Yang, J.; Dear, A.J.; Yao, Q.Q.; Liu, Z.; Dobson, C.M.; Knowles, T.P.J.; Wu, S.; Perrett, S. Amelioration of aggregate cytotoxicity by catalytic conversion of protein oligomers into amyloid fibrils. Nanoscale 2020, 12, 18663–18672. [Google Scholar] [CrossRef]
- König, I.; Zarrine-Afsar, A.; Aznauryan, M.; Soranno, A.; Wunderlich, B.; Dingfelder, F.; Stüber, J.C.; Plückthun, A.; Nettels, D.; Schuler, B. Single-molecule spectroscopy of protein conformational dynamics in live eukaryotic cells. Nat. Methods 2015, 12, 773–779. [Google Scholar] [CrossRef]
- Crawford, R.; Torella, J.P.; Aigrain, L.; Plochowietz, A.; Gryte, K.; Uphoff, S.; Kapanidis, A.N. Long-lived intracellular single-molecule fluorescence using electroporated molecules. Biophys. J. 2013, 105, 2439–2450. [Google Scholar] [CrossRef] [PubMed]
- Aigrain, L.; Sustarsic, M.; Crawford, R.; Plochowietz, A.; Kapanidis, A.N. Internalization and observation of fluorescent biomolecules in living microorganisms via electroporation. J. Vis. Exp. 2015, 52208. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Perrett, S.; Wu, S. Single Molecule Characterization of Amyloid Oligomers. Molecules 2021, 26, 948. https://doi.org/10.3390/molecules26040948
Yang J, Perrett S, Wu S. Single Molecule Characterization of Amyloid Oligomers. Molecules. 2021; 26(4):948. https://doi.org/10.3390/molecules26040948
Chicago/Turabian StyleYang, Jie, Sarah Perrett, and Si Wu. 2021. "Single Molecule Characterization of Amyloid Oligomers" Molecules 26, no. 4: 948. https://doi.org/10.3390/molecules26040948
APA StyleYang, J., Perrett, S., & Wu, S. (2021). Single Molecule Characterization of Amyloid Oligomers. Molecules, 26(4), 948. https://doi.org/10.3390/molecules26040948








