An Estimation Algorithm for General Linear Single Particle Tracking Models with Time-Varying Parameters
Abstract
1. Introduction
2. Methods
2.1. Background on EM
2.1.1. EM for Fixed Parameter Estimation
2.1.2. EM Using Local Likelihood
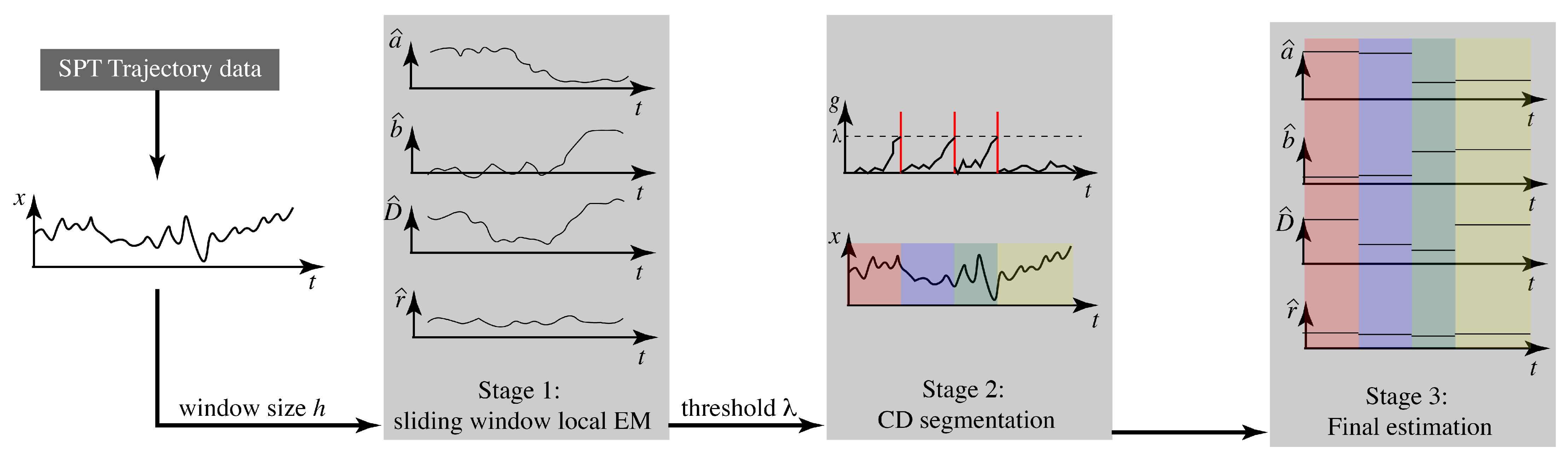
2.2. Algorithm Stages
2.2.1. Stage 1: Sliding Window Estimation with Local Likelihood
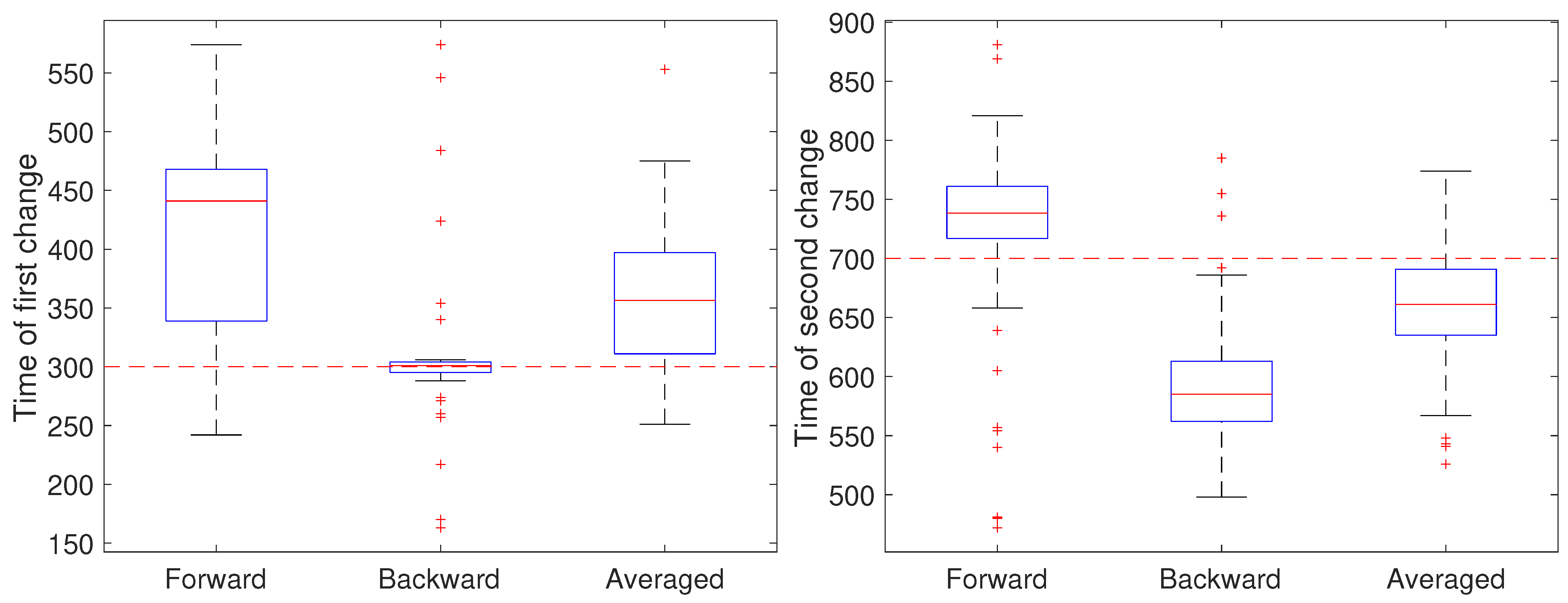
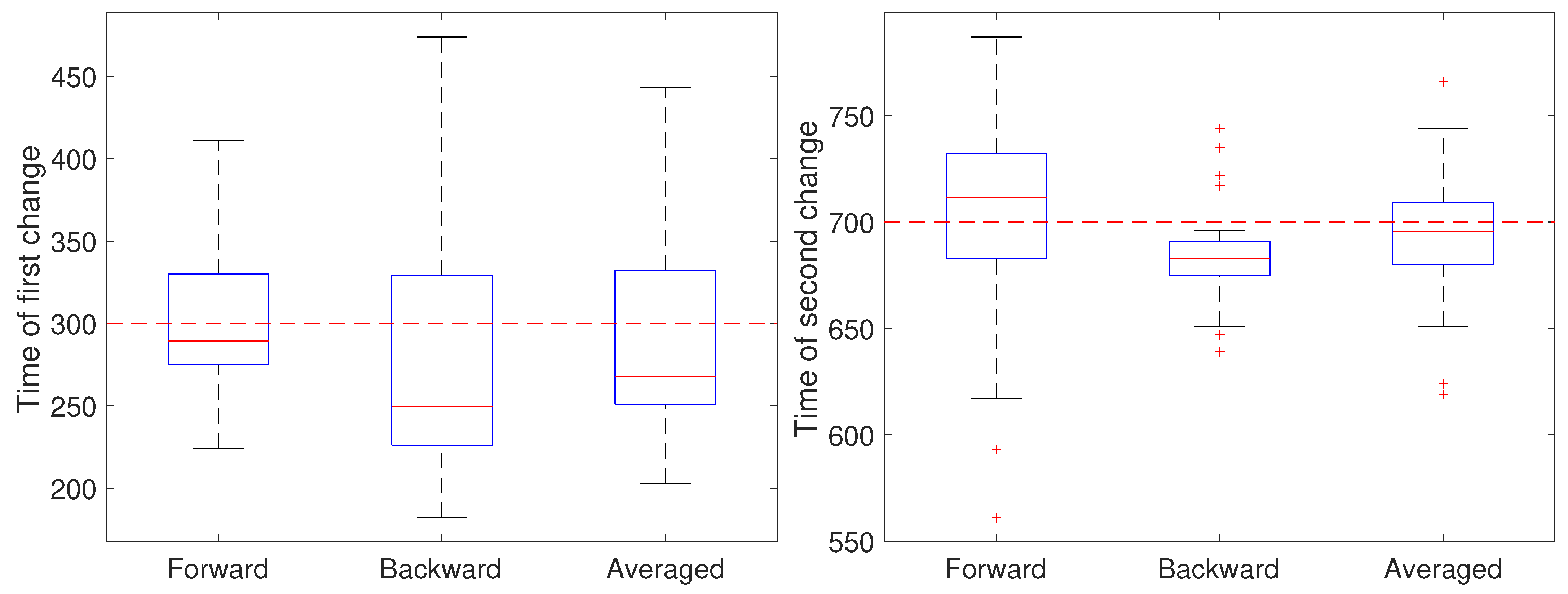
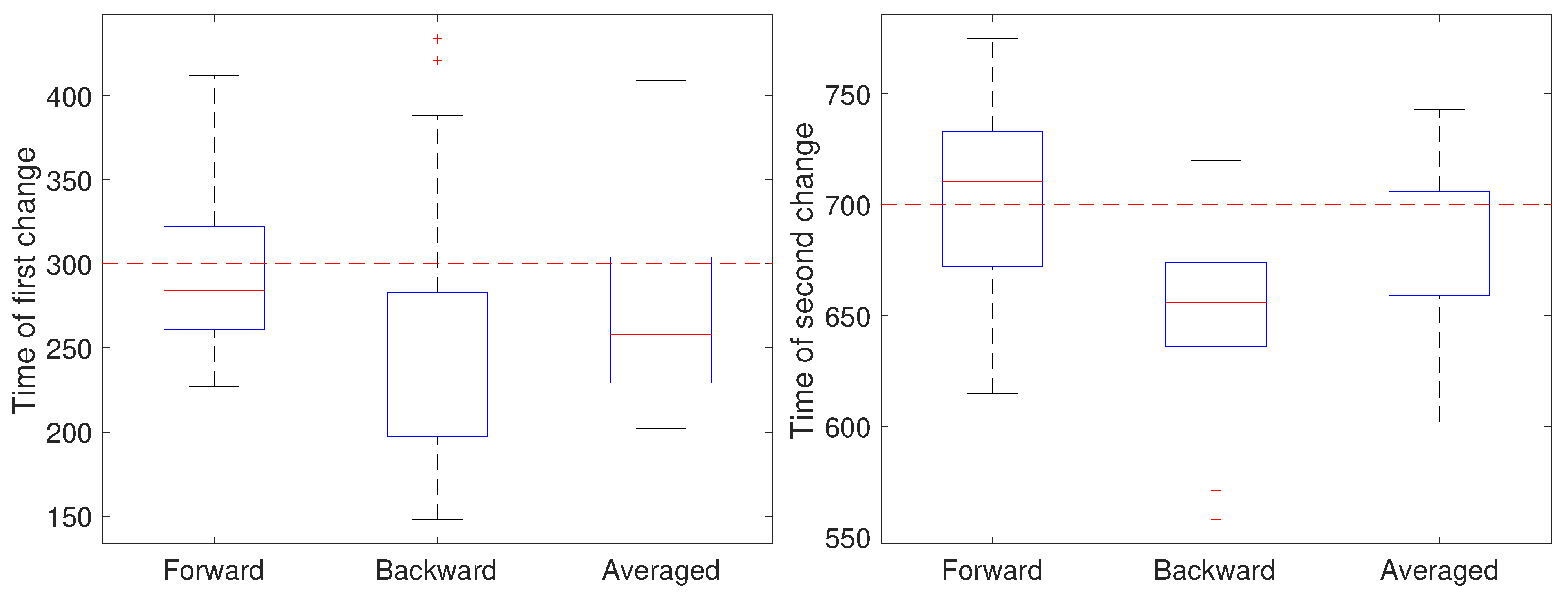
2.2.2. Stage 2: Change Detection
2.2.3. Stage 3: Final Estimation and the Complete Algorithm
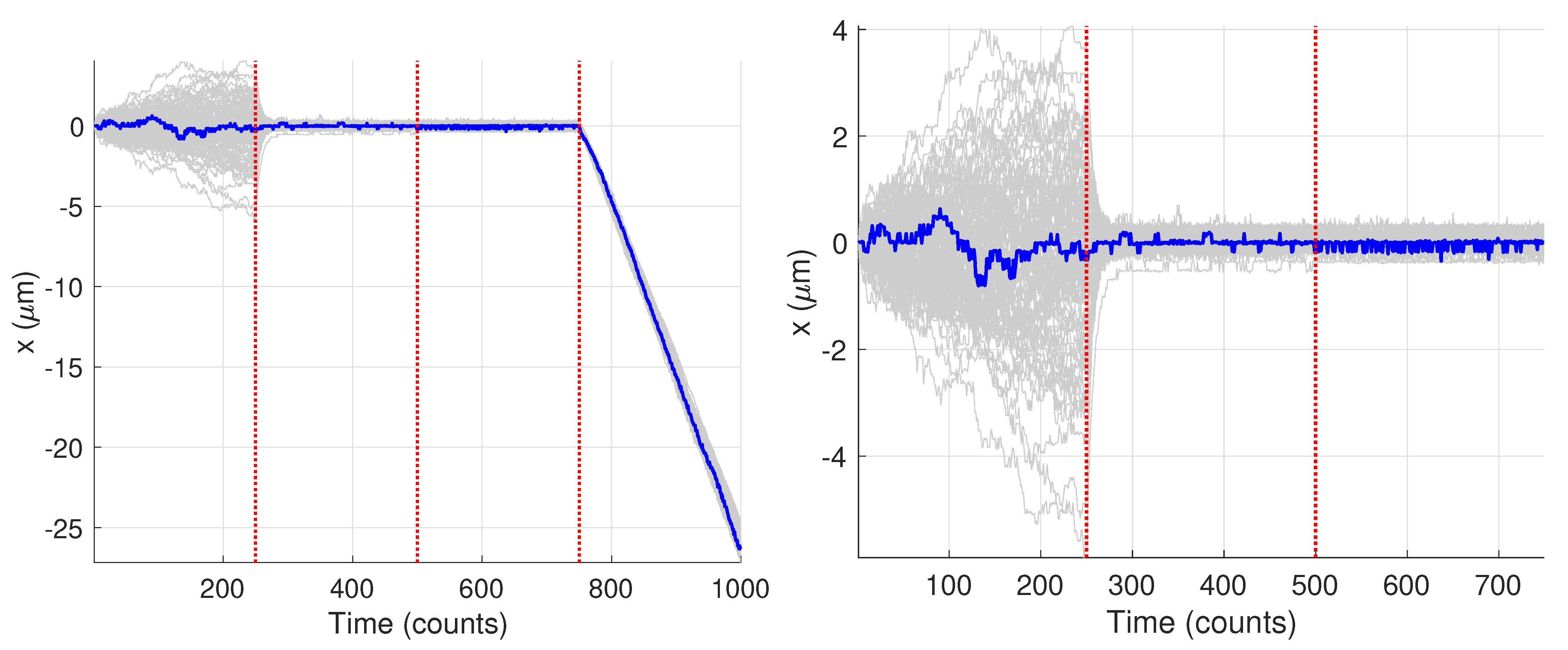
2.3. Generation of Synthetic Data
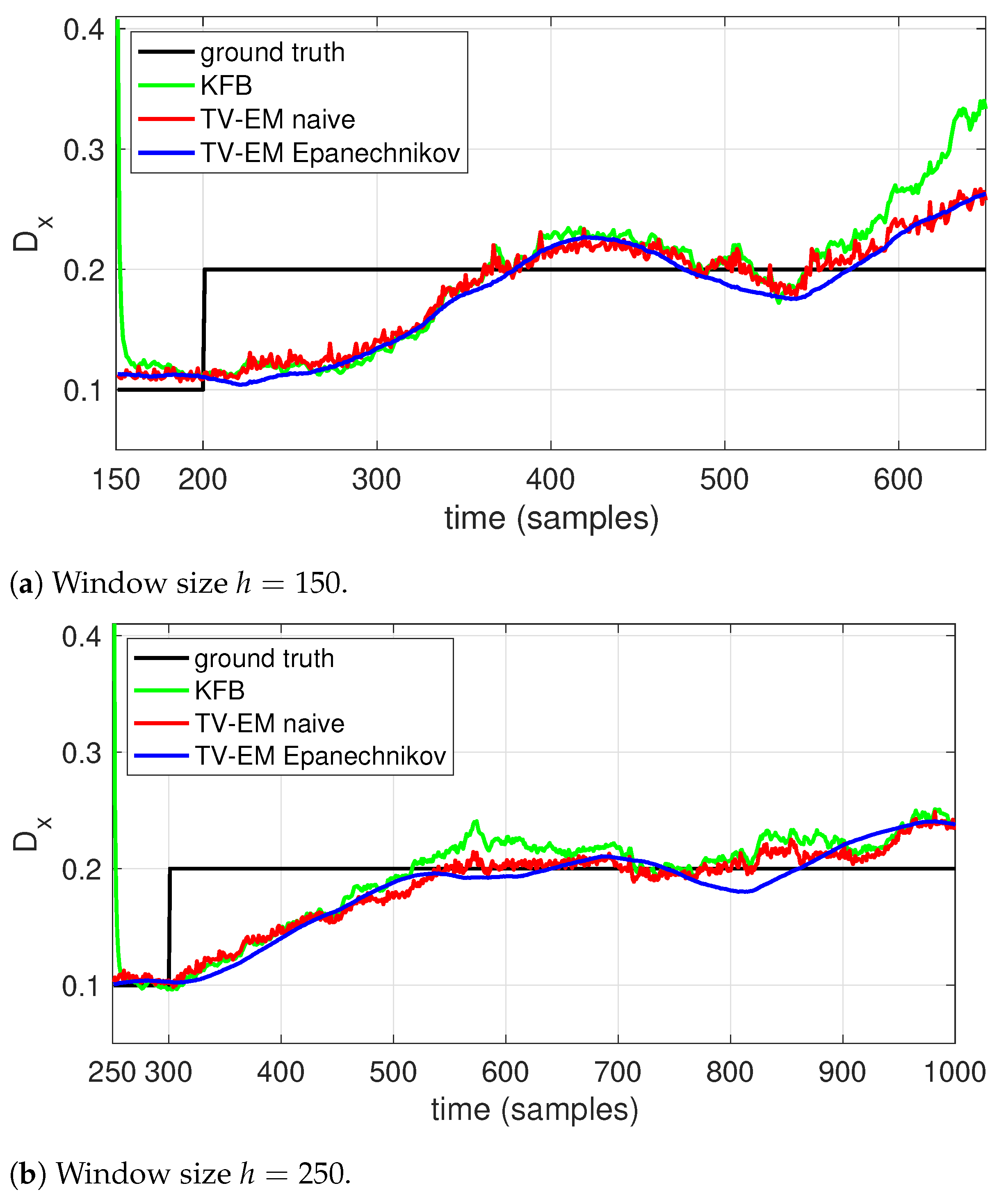
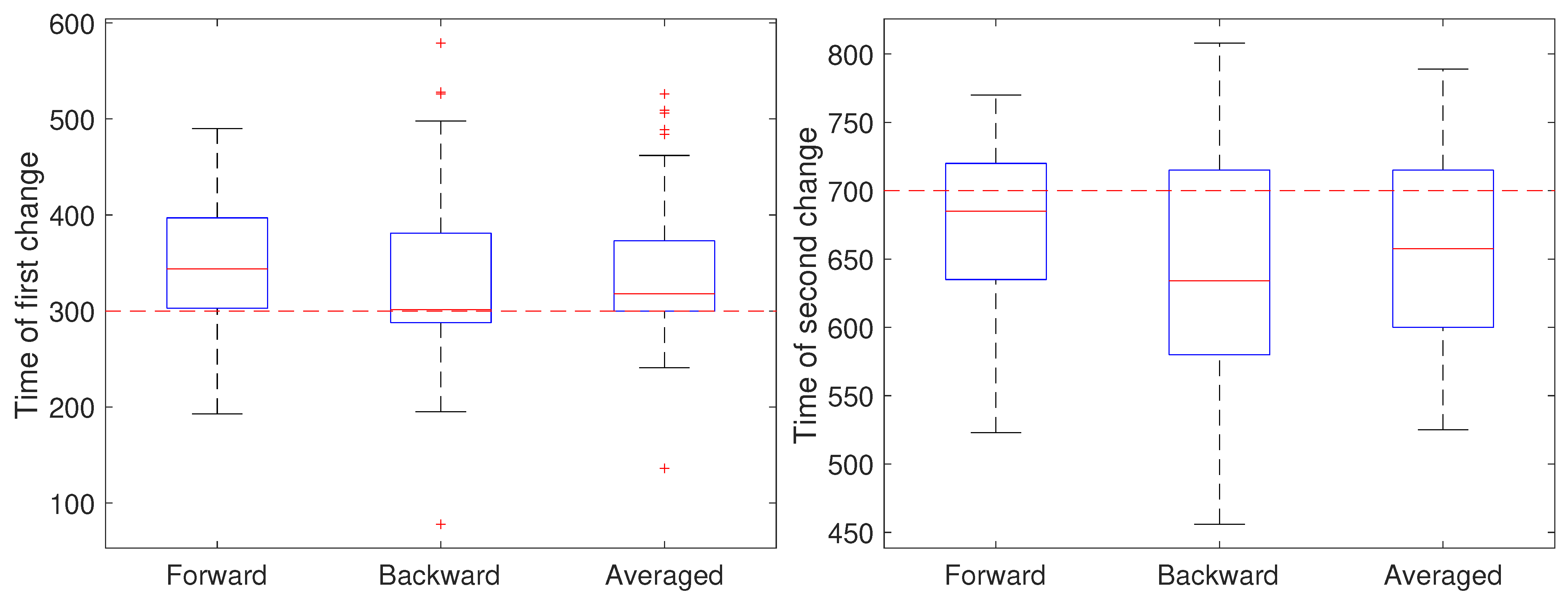
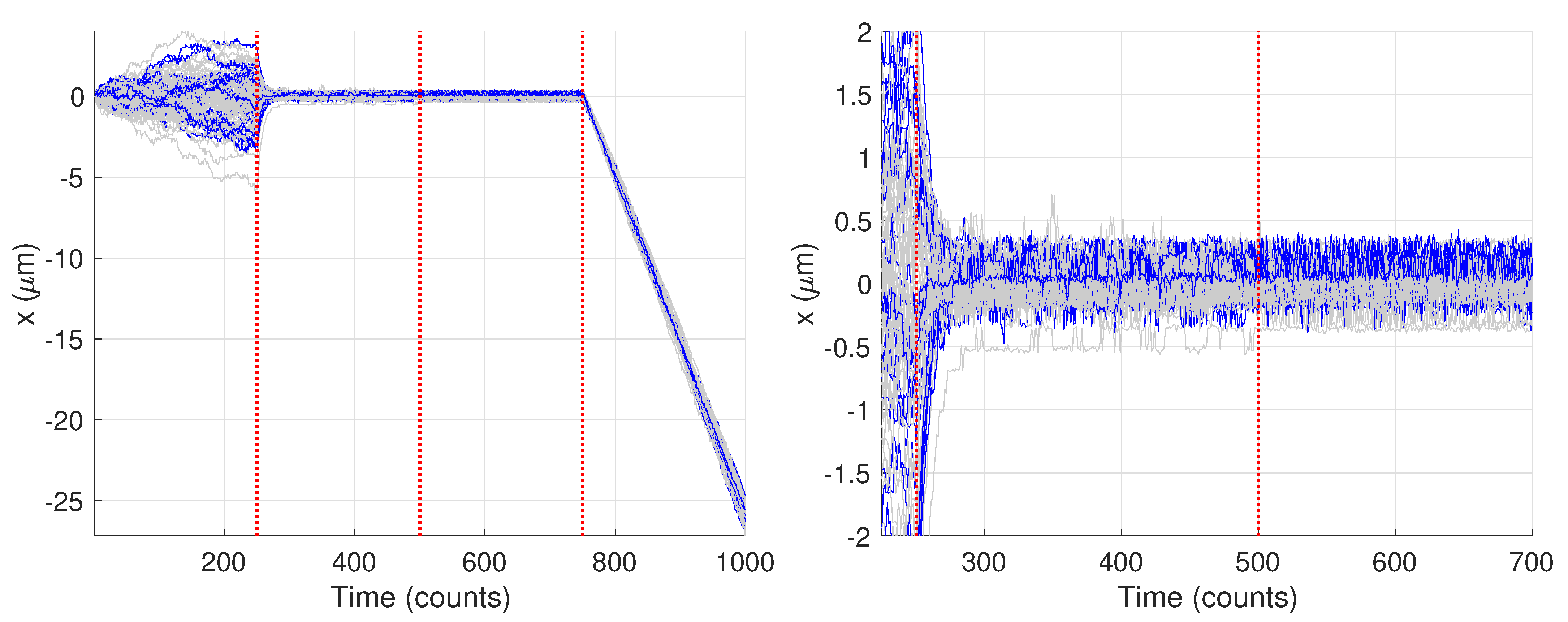
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shen, H.; Tauzin, L.J.; Baiyasi, R.; Wang, W.; Moringo, N.; Shuang, B.; Landes, C.F. Single particle tracking: From theory to biophysical applications. Chem. Rev. 2017, 117, 7331–7376. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, X.; Liu, H.; Wei, L.; Xiao, L. Recent advances in optical microscopic methods for single-particle tracking in biological samples. Anal. Bioanal. Chem. 2019, 143, 1–19. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, G. Three-dimensional single particle tracking and its applications in confined environments. Annu. Rev. Anal. Chem. 2020, 13, 1–23. [Google Scholar] [CrossRef]
- Saxton, M.J.; Jacobson, K. Single-particle tracking: Applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 1997, 26, 373–399. [Google Scholar] [CrossRef]
- Monnier, N.; Guo, S.M.; Mori, M.; He, J.; Lénárt, P.; Bathe, M. Bayesian approach to MSD-based analysis of particle motion in live cells. Biophys. J. 2012, 103, 616–626. [Google Scholar] [CrossRef]
- Manzo, C.; Garcia-Parajo, M.F. A review of progress in single particle tracking: From methods to biophysical insights. Rep. Prog. Phys. 2015, 78, 124601. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.L.; Wang, Z.G.; Xie, H.Y.; Liu, A.A.; Lamb, D.C.; Pang, D.W. Single-virus tracking: From imaging methodologies to virological applications. Chem. Rev. 2020, 120, 1936–1979. [Google Scholar] [CrossRef] [PubMed]
- Berglund, A.J. Statistics of camera-based single-particle tracking. Phys. Rev. E 2010, 82, 11917. [Google Scholar] [CrossRef] [PubMed]
- Michalet, X. Mean square displacement analysis of single-particle trajectories with localization error: Brownian motion in an isotropic medium. Phys. Rev. E 2010, 82, 41914. [Google Scholar] [CrossRef]
- Michalet, X.; Berglund, A.J. Optimal diffusion coefficient estimation in single-particle tracking. Phys. Rev. E 2012, 85, 61916. [Google Scholar] [CrossRef] [PubMed]
- Boyer, D.; Dean, D.S.; Mejía-Monasterio, C.; Oshanin, G. Optimal least-squares estimators of the diffusion constant from a single Brownian trajectory. Eur. Phys. J. Spec. Top. 2013, 216, 57–71. [Google Scholar] [CrossRef]
- Calderon, C.P. Motion blur filtering: A statistical approach for extracting confinement forces and diffusivity from a single blurred trajectory. Phys. Rev. E 2016, 93, 53303. [Google Scholar] [CrossRef]
- Ashley, T.T.; Andersson, S.B. Method for simultaneous localization and parameter estimation in particle tracking experiments. Phys. Rev. E 2015, 92, 52707. [Google Scholar] [CrossRef]
- Lin, Y.; Andersson, S.B. Simultaneous localization and parameter estimation for single particle tracking via sigma points based EM. In Proceedings of the IEEE Conference on Decision and Control, Nice, France, 11–13 December 2019; pp. 6467–6472. [Google Scholar]
- Vega, A.R.; Freeman, S.A.; Grinstein, S.; Jaqaman, K. Multistep track segmentation and motion classification for transient mobility analysis. Biophys. J. 2018, 114, 1018–1025. [Google Scholar] [CrossRef]
- Ashley, T.T.; Andersson, S.B. A sequential Monte Carlo framework for the system identification of jump Markov state space models. In Proceedings of the American Control Conference, Portland, OR, USA, 4–6 June 2014; pp. 1144–1149. [Google Scholar]
- Calderon, C.P. Data-driven techniques for detecting dynamical state changes in noisily measured 3D single-molecule trajectories. Molecules 2014, 19, 18381–18398. [Google Scholar] [CrossRef]
- Calderon, C.P.; Bloom, K. Inferring latent states and refining force estimates via hierarchical Dirichlet process modeling in single particle tracking experiments. PLoS ONE 2015, 10, e0137633. [Google Scholar] [CrossRef]
- Helmuth, J.A.; Burckhardt, C.J.; Koumoutsakos, P.; Greber, U.F.; Sbalzarini, I.F. A novel supervised trajectory segmentation algorithm identifies distinct types of human adenovirus motion in host cells. J. Struct. Biol. 2007, 159, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Huet, S.; Karatekin, E.; Tran, V.S.; Fanget, I.; Cribier, S.; Henry, J.P. Analysis of transient behavior in complex trajectories: Application to secretory vesicle dynamics. Biophys. J. 2006, 91, 3542–3559. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.L.; Perillo, E.P.; Liu, C.; Yu, P.; Chou, C.K.; Hung, M.C.; Dunn, A.K.; Yeh, H.C. Segmentation of 3D trajectories acquired by TSUNAMI microscope: An application to EGFR trafficking. Biophys. J. 2016, 111, 2214–2227. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.; Kroll, A.; Haramagatti, C.R.; Lipinski, H.G.; Wiemann, M. Classification and segmentation of nanoparticle diffusion trajectories in cellular micro environments. PLoS ONE 2017, 12, e0170165. [Google Scholar] [CrossRef]
- Janczura, J.; Kowalek, P.; Loch-Olszewska, H.; Szwabiński, J.; Weron, A. Classification of particle trajectories in living cells: Machine learning versus statistical testing hypothesis for fractional anomalous diffusion. Phys. Rev. E 2020, 102, 032402. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Korabel, N.; Chen, R.; Johnston, M.; Gavrilova, A.; Allan, V.J.; Fedotov, S.; Waigh, T.A. Deciphering anomalous heterogeneous intracellular transport with neural networks. eLife 2020, 9, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Godoy, B.I.; Lin, Y.; Agüero, J.C.; Andersson, S.B. A 2-step algorithm for the estimation of time-varying single particle tracking models using Maximum Likelihood. In Proceedings of the Asian Control Conference, Kitakyushu, Japan, 9–12 June 2019. [Google Scholar]
- Godoy, B.I.; Vickers, N.A.; Lin, Y.; Andersson, S.B. Estimation of general time-varying single particle tracking linear models using local likelihood. In Proceedings of the European Control Conference, Saint Petersburg, Russia, 12–15 May 2020; pp. 527–533. [Google Scholar]
- Soderström, T. Discrete-Time Stochastic Systems; Springer: London, UK, 2002. [Google Scholar]
- Ljung, L. System Identification: Theory for the User; Prentice-Hall: Upper Saddle River, NJ, USA, 1999. [Google Scholar]
- Goodwin, G.; Agüero, J. Approximate EM algorithms for parameter and state estimation in nonlinear stochastic models. In Proceedings of the IEEE Conference on Decision and Control and the European Control Conference, Seville, Spain, 12–15 December 2005; pp. 368–373. [Google Scholar]
- Lange, K. A gradient algorithm locally equivalent to the EM algorithm. J. R. Stat. Soc. Ser. B 1995, 57, 425–437. [Google Scholar] [CrossRef]
- Cappé, O.; Moulines, E.; Rydeén, T. Inference in Hidden Markov Models, 2nd ed.; Springer: New York, NY, USA, 2005. [Google Scholar]
- Dempster, A.P.; Laird, N.M.; Rubin, D.B. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. 1977, 39, 1–38. [Google Scholar]
- Gray, A.; Markel, J. Distance measures for speech processing. IEEE Trans. Acoust. Speech Signal Process. 1976, 24, 380–437. [Google Scholar] [CrossRef]
- Gray, R.; Buzo, A.; Gray, A.; Matsuyama, Y. Distortion measures for speeach processing. IEEE Trans. Acoust. Speech Signal Process. 1980, 28, 367–376. [Google Scholar] [CrossRef]
- Baseville, M. Edge detection using sequential methods for change in level. Part II: Sequential detection of change in a mean. IEEE Trans. Acoust. Speech Signal Process. 1981, 29, 32–50. [Google Scholar] [CrossRef]
- Bohlin, T. Analysis of EEG signals with changing spectra using a short word Kalman estimator. Math. Biosci. 1977, 35, 221–259. [Google Scholar] [CrossRef]
- Isaksson, A.; Wennberg, A.; Zetterberg, L. Computer analysis of EEG signals with parametric models. Proc. IEEE 1981, 69, 451–461. [Google Scholar] [CrossRef]
- Gustafson, D.; Willsky, A.; Wang, J.; Lancaster, M.; Triebwasser, J. ECG/VCG rhythm diagnosis using statistical signal analysis, Part II: Identification of transient rhythms. IEEE Trans. Biomed. Eng. 1978, 25, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Türkcan, S.; Masson, J.B. Bayesian decision tree for the classification of the mode of motion in single-molecule trajectories. PLoS ONE 2013, 8, e82799. [Google Scholar] [CrossRef]
- Lanoiselée, Y.; Grebenkov, D.S. Unraveling intermittent features in single-particle trajectories by a local convex hull method. Phys. Rev. E 2017, 96, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hubicka, K.; Janczura, J. Time-dependent classification of protein diffusion types: A statistical detection of mean-squared-displacement exponent transitions. Phys. Rev. E 2020, 101, 1–13. [Google Scholar] [CrossRef]
- Vickers, N.A.; Andersson, S.B. Monte Carlo simulation of Brownian motion using a piezo-actuated microscope stage. In Proceedings of the American Control Conference, Philadelphia, PA, USA, 10–12 July 2019; pp. 567–572. [Google Scholar]
- Chenouard, N.; Smal, I.; de Chaumont, F.; Maška, M.; Sbalzarini, I.F.; Gong, Y.; Cardinale, J.; Carthel, C.; Coraluppi, S.; Winter, M.; et al. Objective comparison of particle tracking methods. Nat. Methods 2014, 11, 281–289. [Google Scholar] [CrossRef]
- Sage, D.; Pham, T.A.; Babcock, H.; Lukes, T.; Pengo, T.; Chao, J.; Velmurugan, R.; Herbert, A.; Agrawal, A.; Colabrese, S.; et al. Super-resolution fight club: Assessment of 2D and 3D single-molecule localization software. Nat. Methods 2019, 16, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Bronshtein, I.; Israel, Y.; Kepten, E.; Mai, S.; Shav-Tal, Y.; Barkai, E.; Garini, Y. Transient Anomalous Diffus. Telomeres Nucl. Mamm. Cells 2009, 103, 18102. [Google Scholar]
- Metzler, R.; Jeon, J.H.; Cherstvy, A.G.; Barkai, E. Anomalous diffusion models and their properties: Non-stationarity, non-ergodicity, and ageing at the centenary of single particle tracking. Phys. Chem. Chem. Phys. 2014, 16, 24128–24164. [Google Scholar] [CrossRef] [PubMed]
- Barkai, E.; Burov, S. Packets of Diffusing Particles Exhibit Universal Exponential Tails. Phys. Rev. Lett. 2020, 124, 60603. [Google Scholar] [CrossRef] [PubMed]
- van Kampen, N.G. Stochastic Processes in Physics and Chemistry; Elsevier: Amsterdam, The Netherlands, 1992. [Google Scholar]
- Gibson, S.; Ninness, B. Robust maximum-likelihood estimation of multivariable dynamic systems. Automatica 2005, 41, 1667–1682. [Google Scholar] [CrossRef]
- Hewitt, E.; Hewitt, R. The Gibbs-Wilbraham phenomenon: An episode in Fourier analysis. Arch. Hist. Exact Sci. 1979, 21, 129–160. [Google Scholar] [CrossRef]
- Anderson, B.D.O.; Moore, J.B. Optimal Filtering; Prentice-Hall Inc.: Upper Saddle River, NJ, USA, 1979. [Google Scholar]
- Mehra, R.K. Identification of stochastic linear dynamic systems. In Proceedings of the IEEE Symposium on Adaptive Processes, University Park, PA, USA, 17–19 November 1969. [Google Scholar]
- Mehra, R. Approaches to adaptive filtering. IEEE Trans. Autom. Control 1972, 17, 693–698. [Google Scholar] [CrossRef]
- Sobolic, F.; Bernstein, D.S. Kalman-filter-based time-varying parameter estimation via retrospective optimization of the process noise covariance. In Proceedings of the American Control Conference, Boston, MA, USA, 6–8 July 2016. [Google Scholar]
- Basseville, M.; Benveniste, A. (Eds.) Detection of Abrupt Changes in Signals and Dynamical Systems; Springer: Berlin/Heidelberg, Germany, 1986. [Google Scholar]
- Basseville, M.; Nikiforov, I. Detection of Abrupt Changes: Theory and Application; Prentice-Hall: Upper Saddle River, NJ, USA, 1993. [Google Scholar]
- Markel, J.; Gray, A. Linear Prediction of Speech, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 1976. [Google Scholar]
- Burnecki, K.; Weron, A. Algorithms for testing of fractional dynamics: A practical guide to ARFIMA modelling. J. Stat. Mech. Theory Exp. 2014, 2014, P10036. [Google Scholar] [CrossRef]
- Burnecki, K.; Sikora, G.; Weron, A.; Tamkun, M.M.; Krapf, D. Identifying diffusive motions in single-particle trajectories on the plasma membrane via fractional time-series models. Phys. Rev. E 2019, 99, 1–10. [Google Scholar] [CrossRef]
- Basseville, M.; Benveniste, A. Sequential detection of abrupt changes in spectral characteristics of digital signals. IEEE Trans. Inf. Theory 1983, 29, 709–724. [Google Scholar] [CrossRef]
- Long, C.J.; Brown, E.N.; Triantafyllou, C.; Wald, L.L.; Solo, V. Nonstationary noise estimation in functional MRI. Neuroimage 2005, 28, 890–903. [Google Scholar] [CrossRef]
- Thompson, R.E.; Larson, D.R.; Webb, W.W. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002, 82, 2775–2783. [Google Scholar] [CrossRef]
- Lakadamyali, M.; Rust, M.J.; Babcock, H.P.; Zhuang, X. Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. USA 2003, 100, 9280–9285. [Google Scholar] [CrossRef] [PubMed]
- Godoy, B.I.; Lin, Y.; Andersson, S.B. A time-varying approach to single particle tracking with a nonlinear observation model. In Proceedings of the American Control Conference, Denver, CO, USA, 1–3 July 2020; pp. 5151–5156. [Google Scholar]
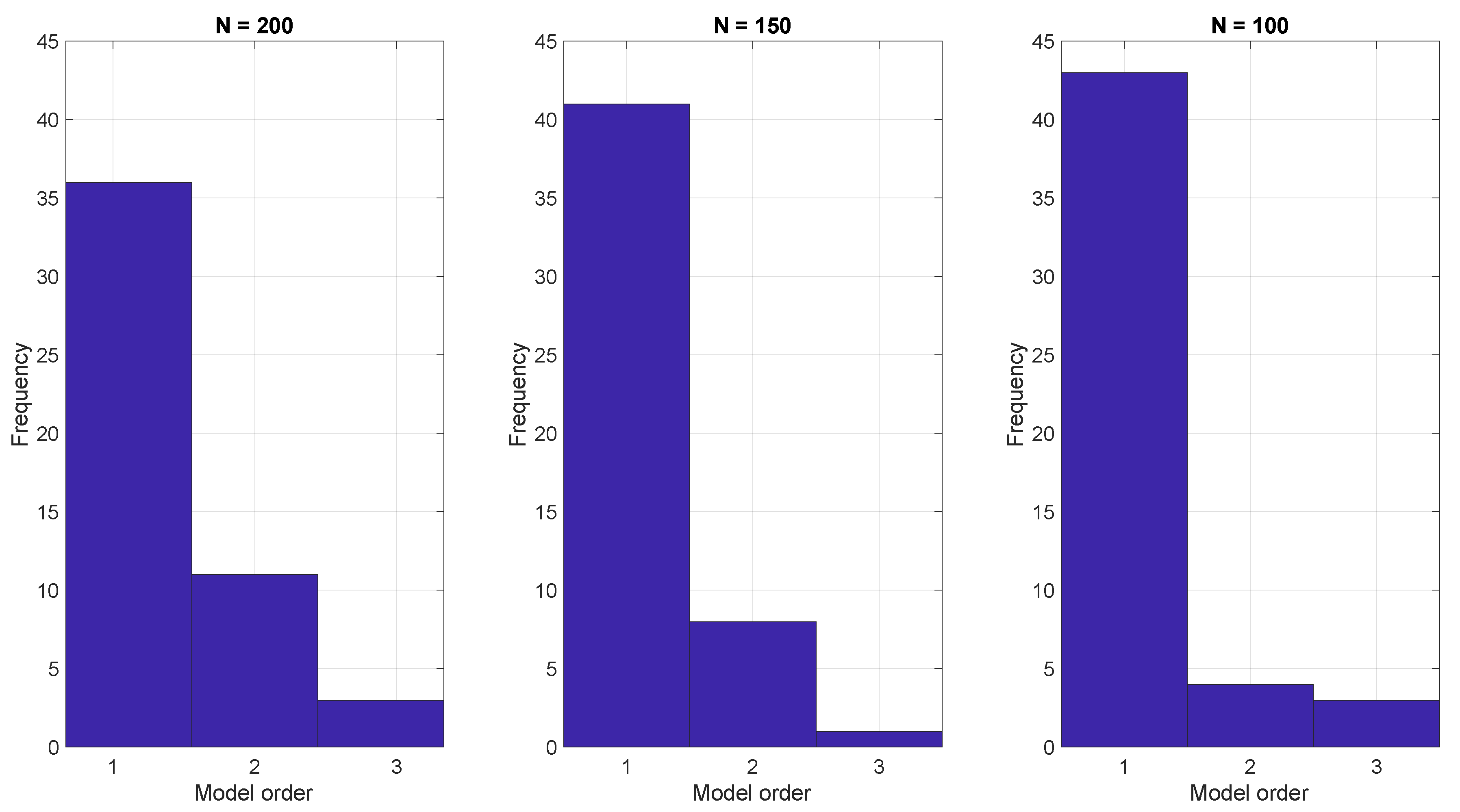

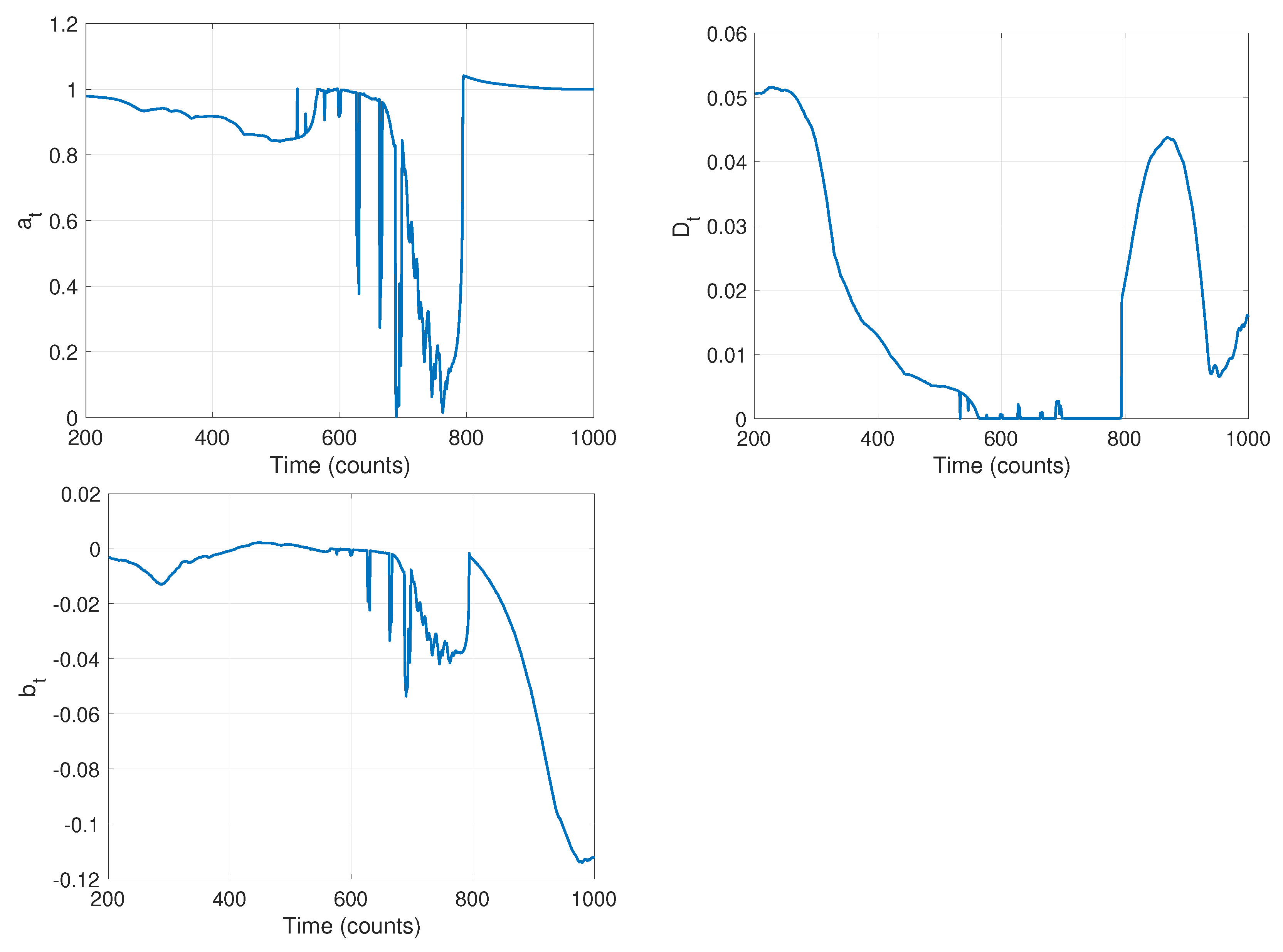
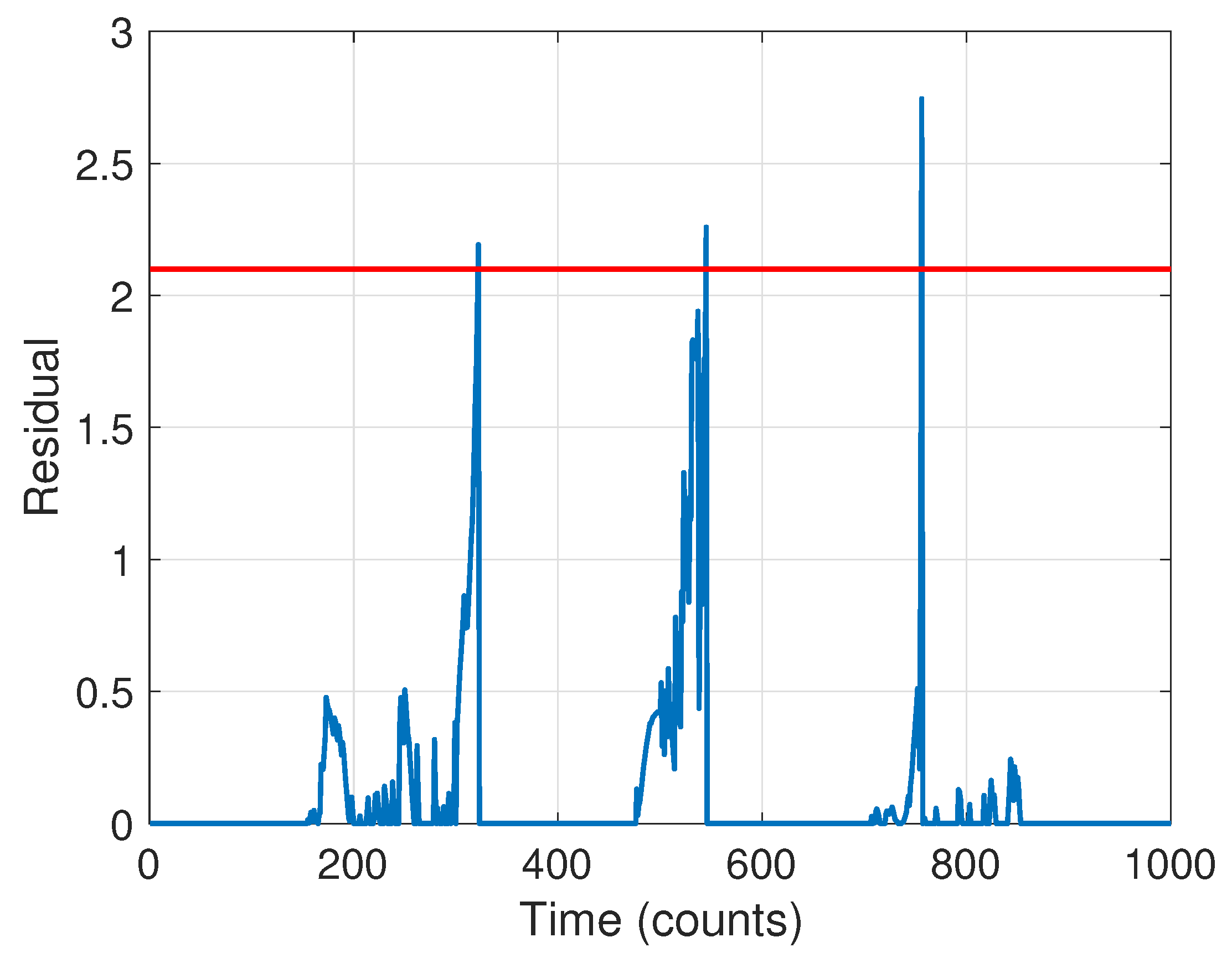
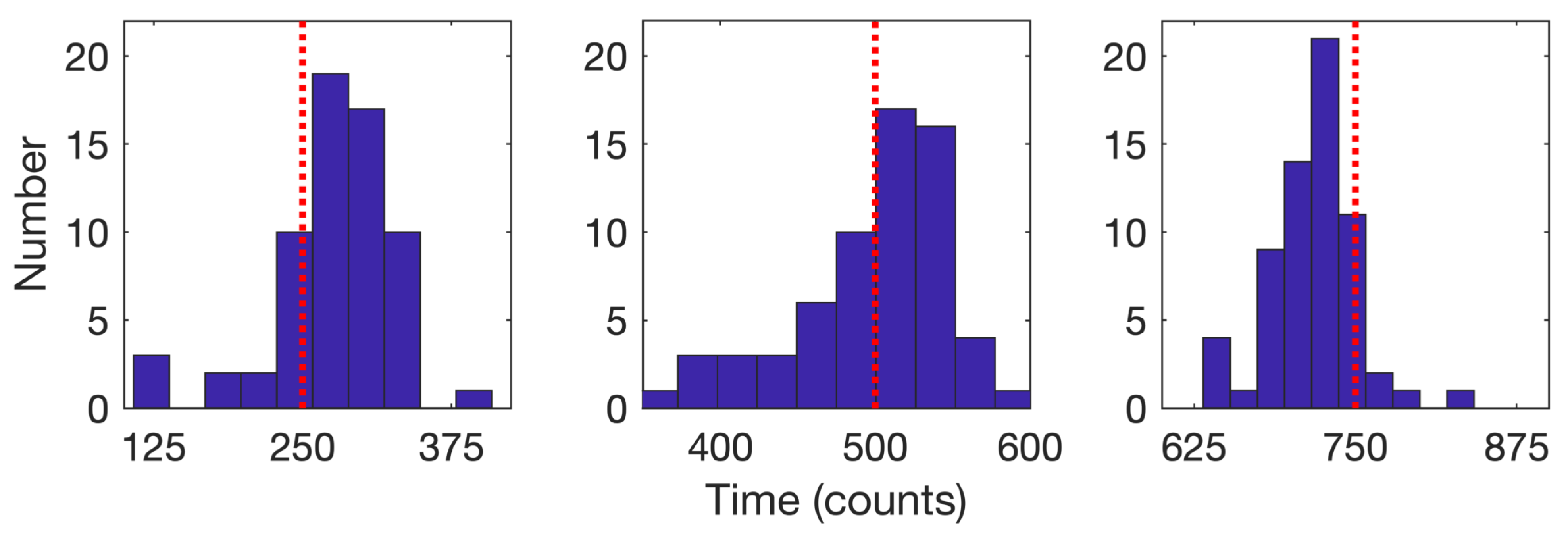

| Time Step | ||||
|---|---|---|---|---|
| Parameter | 1–250 | 251–500 | 501–750 | 751–1000 |
| [unitless] | 1 | 0.86 | 1 | 1 |
| [m] | 0 | 0 | 0 | −0.1 |
| [m/s] | 0.1 | 0.01 | 0 | 0.01 |
| Time Step | ||||
|---|---|---|---|---|
| Parameter | 1–284 | 285–483 | 483–745 | 746–1000 |
| [unitless] | 0.9691 (3.1% error) | 0.8661 (0.7% error) | 0.2482 (75.2% error) | 1.0006 (0.06% error) |
| True | 1.0 | 0.86 | 1.0 | 1.0 |
| [m] | −0.0015 (1.5% error) | 0.0007 (0.7% error) | −0.0463 (4.63% error) | −0.098 (2.0% error) |
| True | 0 | 0 | 0 | −0.1 |
| [m/s] | 0.0408 (52% error) | 0.0064 (36% error) | 0.0094 (N/A % error) | 0.0112 (12% error) |
| True | 0.1 | 0.01 | 0 | 0.01 |
| [m] | 0.0041 | 0.002 | 0.0054 | 0.0052 |
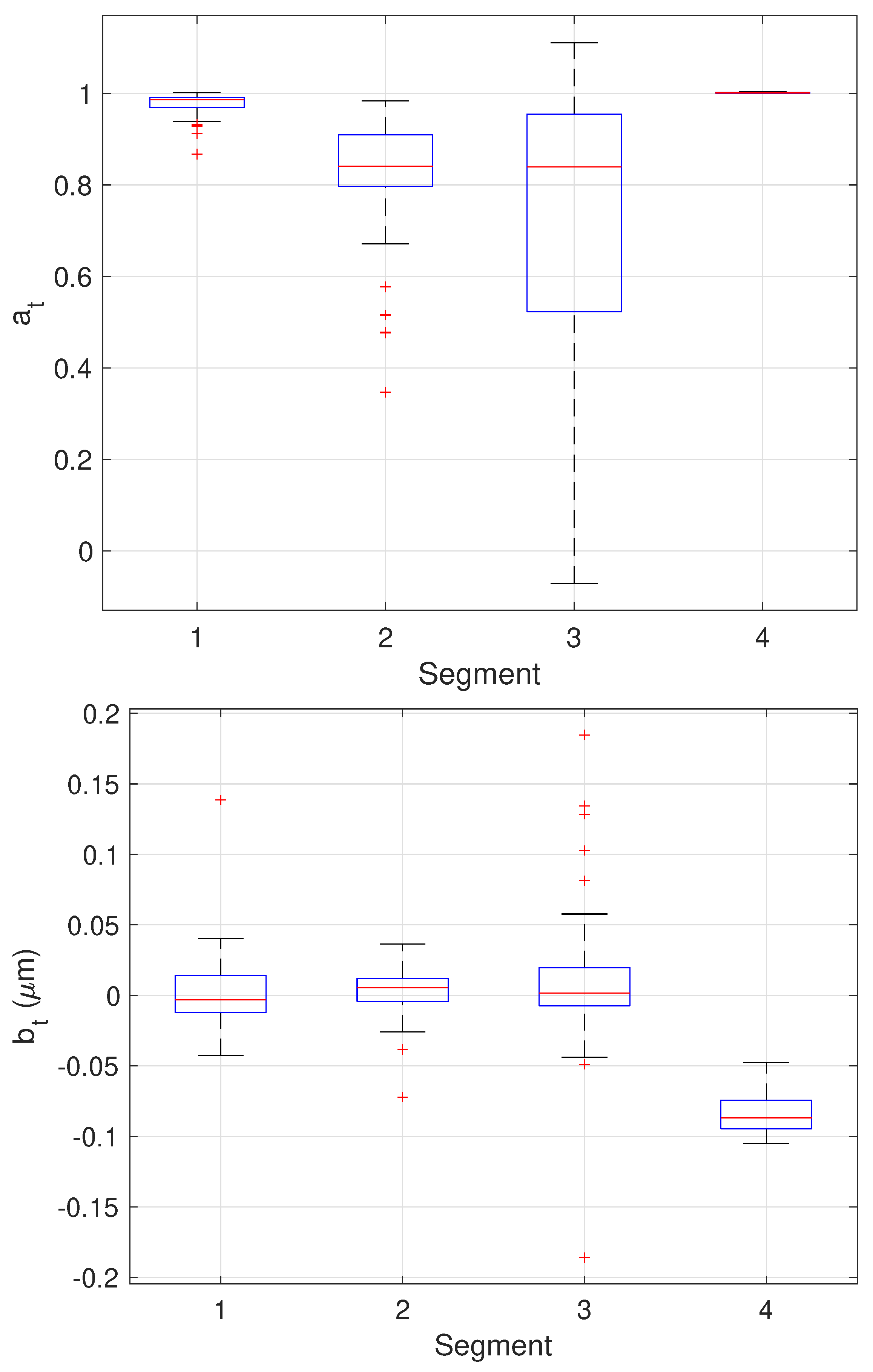
| Parameter | Segment 1 | Segment 2 | Segment 3 | Segment 4 |
|---|---|---|---|---|
| [unitless] | 0.9768 ± 0.0237 | 0.8282 ± 0.127 | 0.7085 ± 0.3185 | 1.002 ± 0.0011 |
| True | 1.0 | 0.86 | 1.0 | 1.0 |
| [m] | 0.0011 ± 0.025 | 0.0029 ± 0.0165 | 0.0109 ± 0.047 | −0.0836 ± 0.015 |
| True | 0 | 0 | 0 | −0.1 |
| [m/s] | 0.0874 ± 0.023 | 0.01934 ± 0.025 | 0.0030 ± 0.0083 | 0.0264 ± 0.0141 |
| True | 0.1 | 0.01 | 0 | 0.01 |
| [m] | 0.0045 ± 0.0022 | 0.0045 ± 0.0031 | 0.0046 ± 0.0034 | 0.0049 ± 0.0016 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Godoy, B.I.; Vickers, N.A.; Andersson, S.B. An Estimation Algorithm for General Linear Single Particle Tracking Models with Time-Varying Parameters. Molecules 2021, 26, 886. https://doi.org/10.3390/molecules26040886
Godoy BI, Vickers NA, Andersson SB. An Estimation Algorithm for General Linear Single Particle Tracking Models with Time-Varying Parameters. Molecules. 2021; 26(4):886. https://doi.org/10.3390/molecules26040886
Chicago/Turabian StyleGodoy, Boris I., Nicholas A. Vickers, and Sean B. Andersson. 2021. "An Estimation Algorithm for General Linear Single Particle Tracking Models with Time-Varying Parameters" Molecules 26, no. 4: 886. https://doi.org/10.3390/molecules26040886
APA StyleGodoy, B. I., Vickers, N. A., & Andersson, S. B. (2021). An Estimation Algorithm for General Linear Single Particle Tracking Models with Time-Varying Parameters. Molecules, 26(4), 886. https://doi.org/10.3390/molecules26040886






