DNA Manipulation and Single-Molecule Imaging
Abstract
1. Introduction
1.1. Statistical Average in Biological Fields
1.2. Flexibility of DNA Molecule as a Polymer
1.3. Flexibility of Genomic DNA Molecules
2. DNA Manipulation
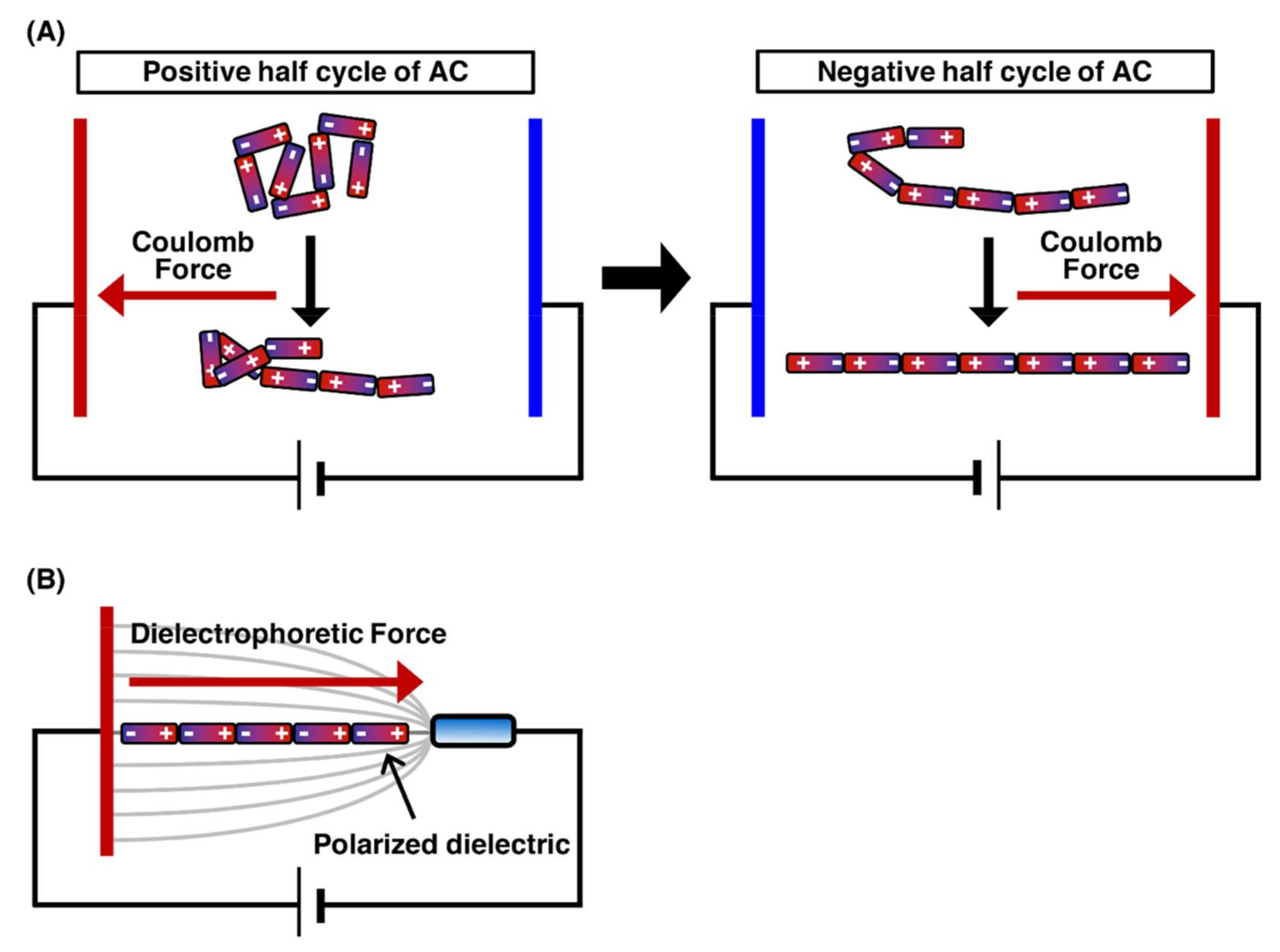
2.1. DNA Manipulation by Orientation Forces
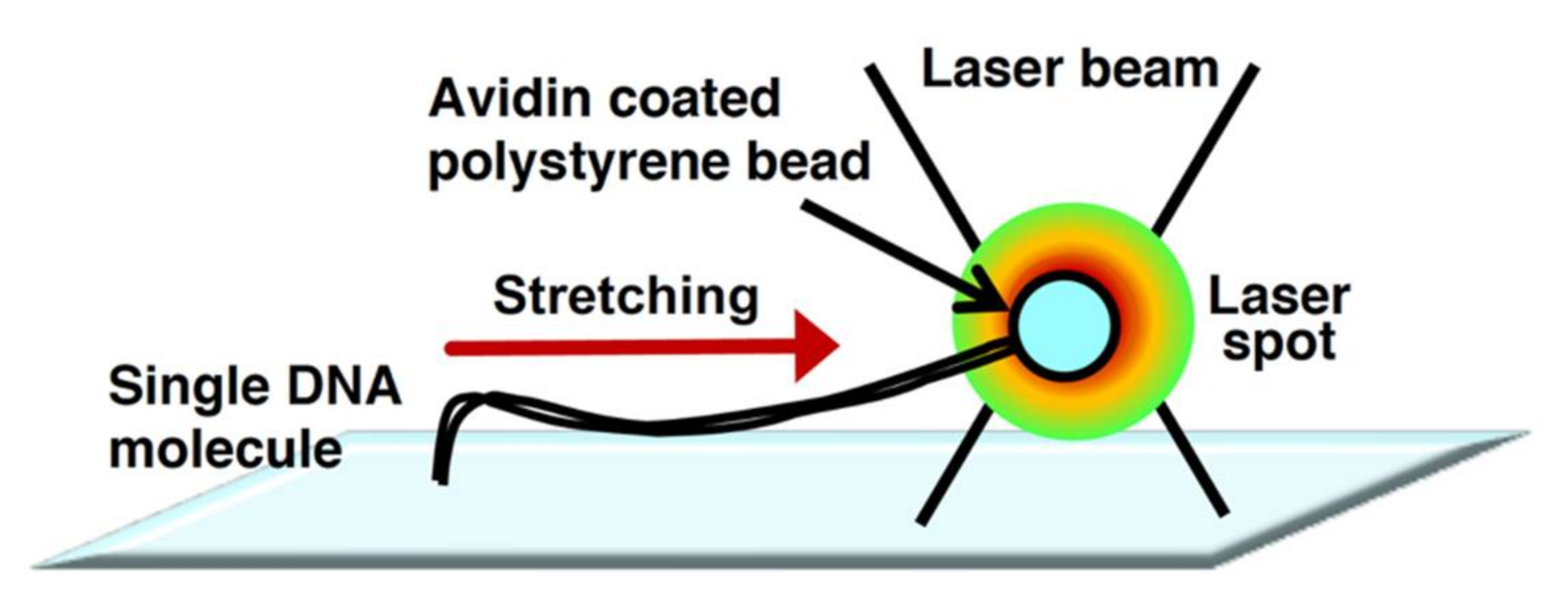
2.2. DNA Manipulation by Optical Tweezers
2.3. Manipulation of Genomic DNA by Laser Trapping
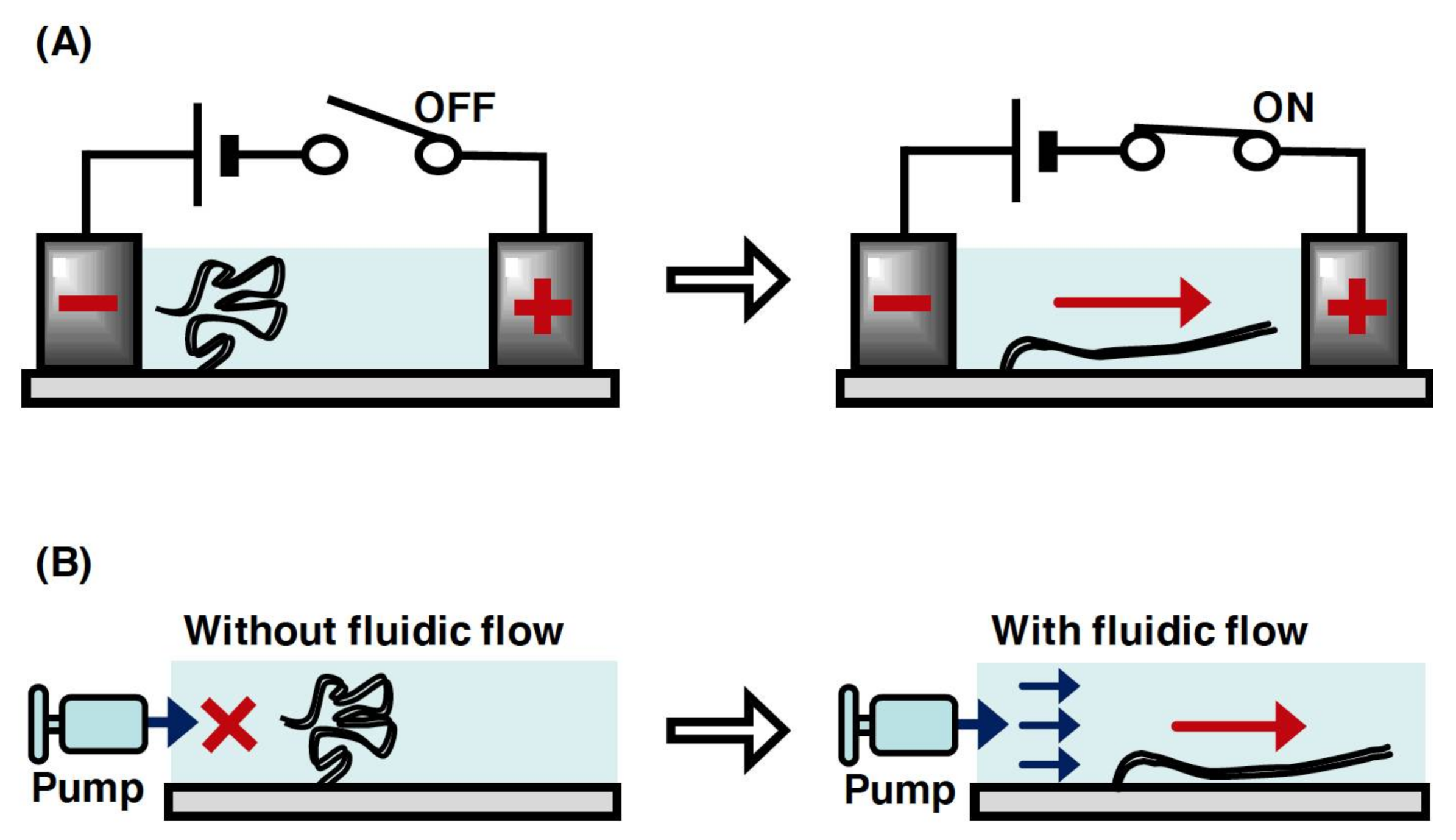
2.4. DNA Manipulation by Electric Field and Fluid Flow
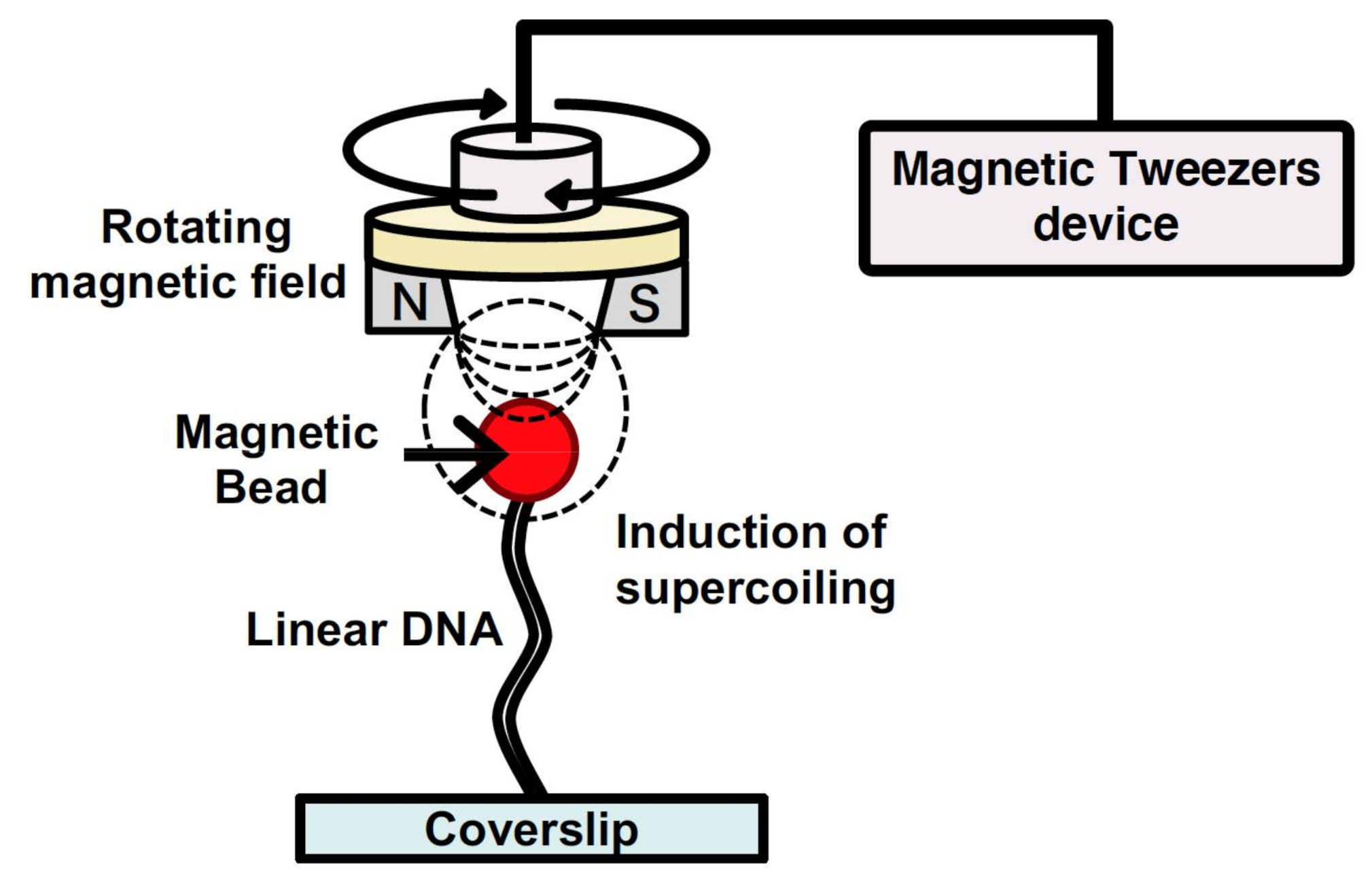
2.5. DNA Manipulation by Magnetic Tweezers
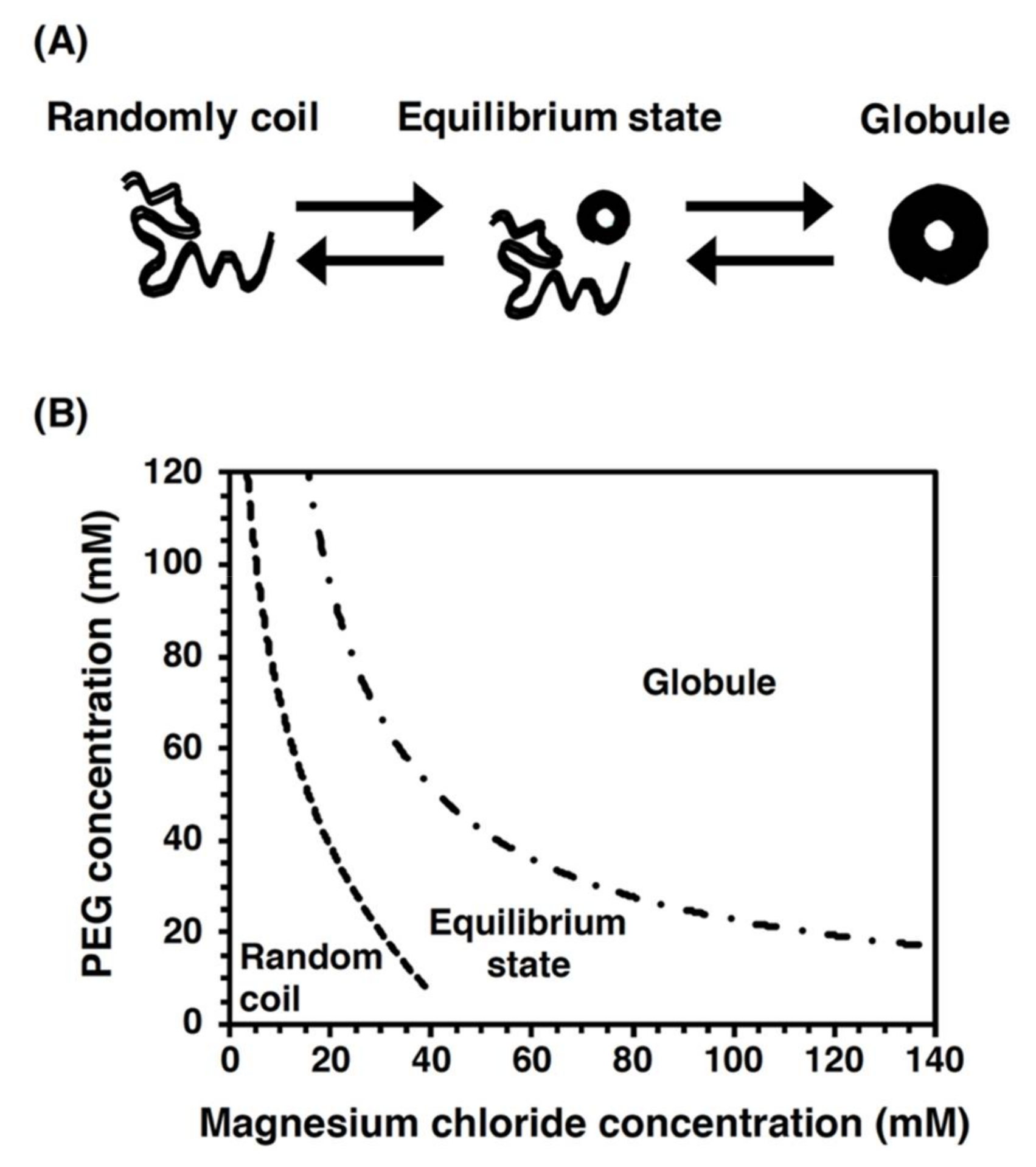
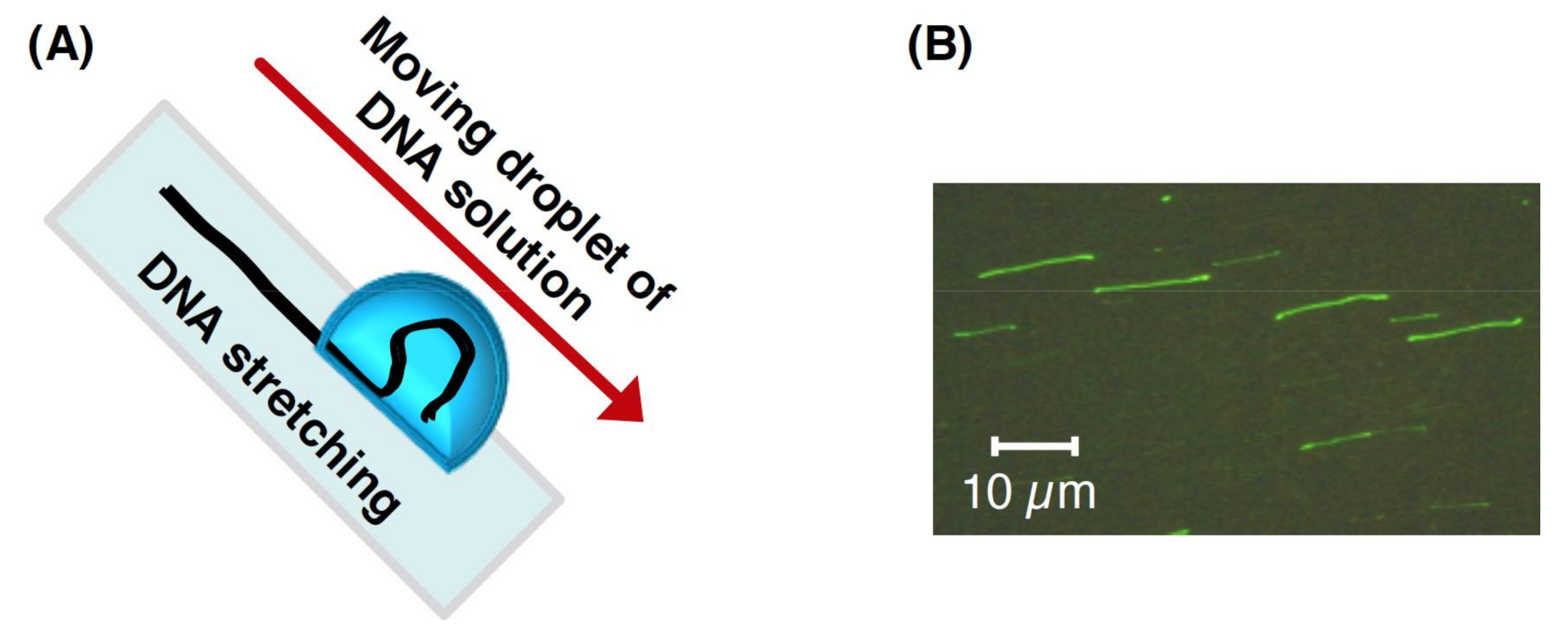
2.6. DNA Manipulation by Interface Movement
2.7. DNA Origami and Control of DNA Folding
2.8. Nanopores and DNA Manipulation
3. Single-Molecule Imaging for Biological Processes
3.1. Fluorescence Labeling of Biomolecules for Single-Molecule Imaging
3.1.1. Fluorescence Staining of Double-Stranded DNA
3.1.2. Fluorescence Labeling of DNA-Binding Proteins
3.1.3. Single-Molecule Imaging by Single-Molecule FRET
3.2. Optical Mapping on Single DNA Molecules
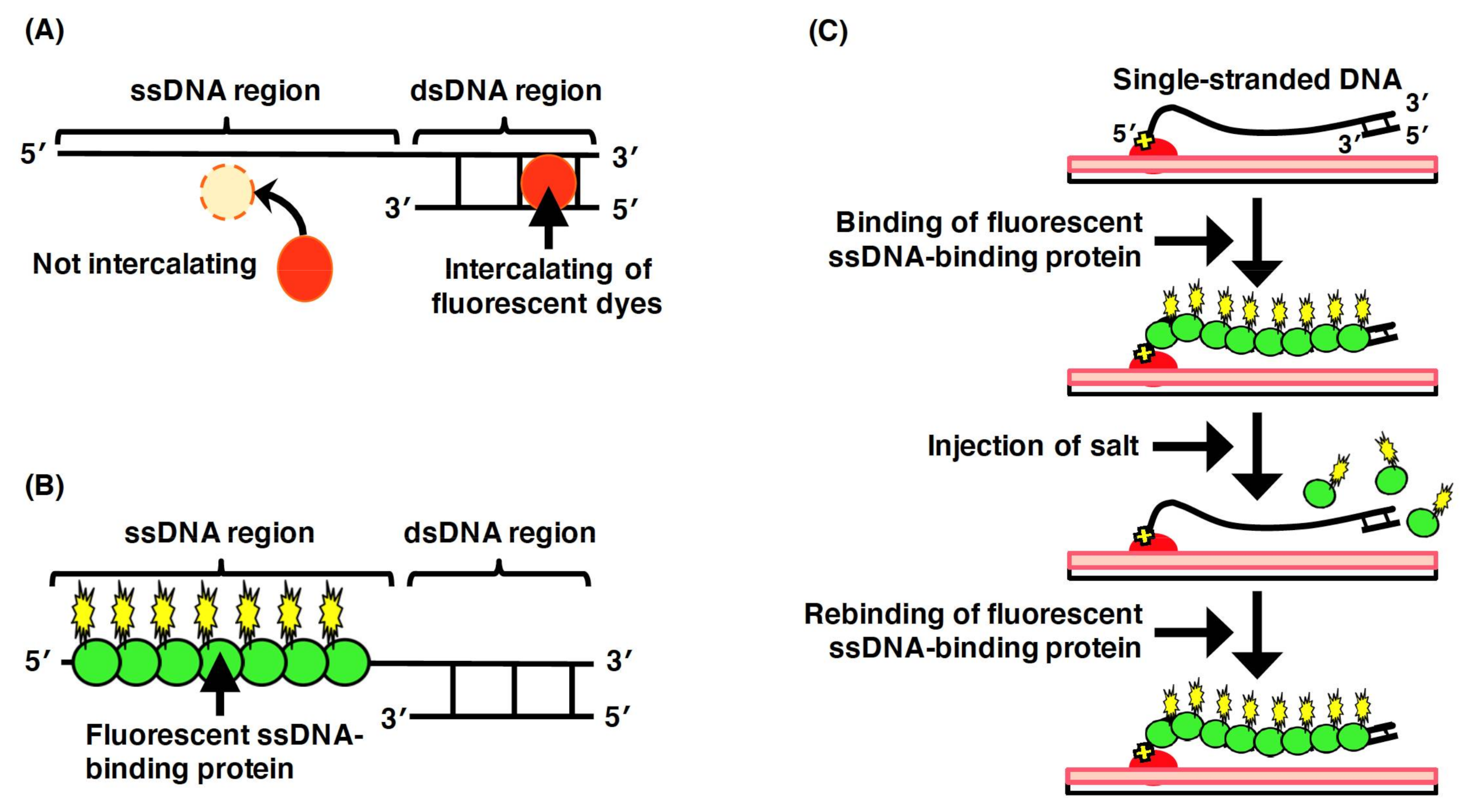
3.3. Single-Molecule Imaging for ssDNA Molecules by Fluorescent ssDNA-Binding Protein
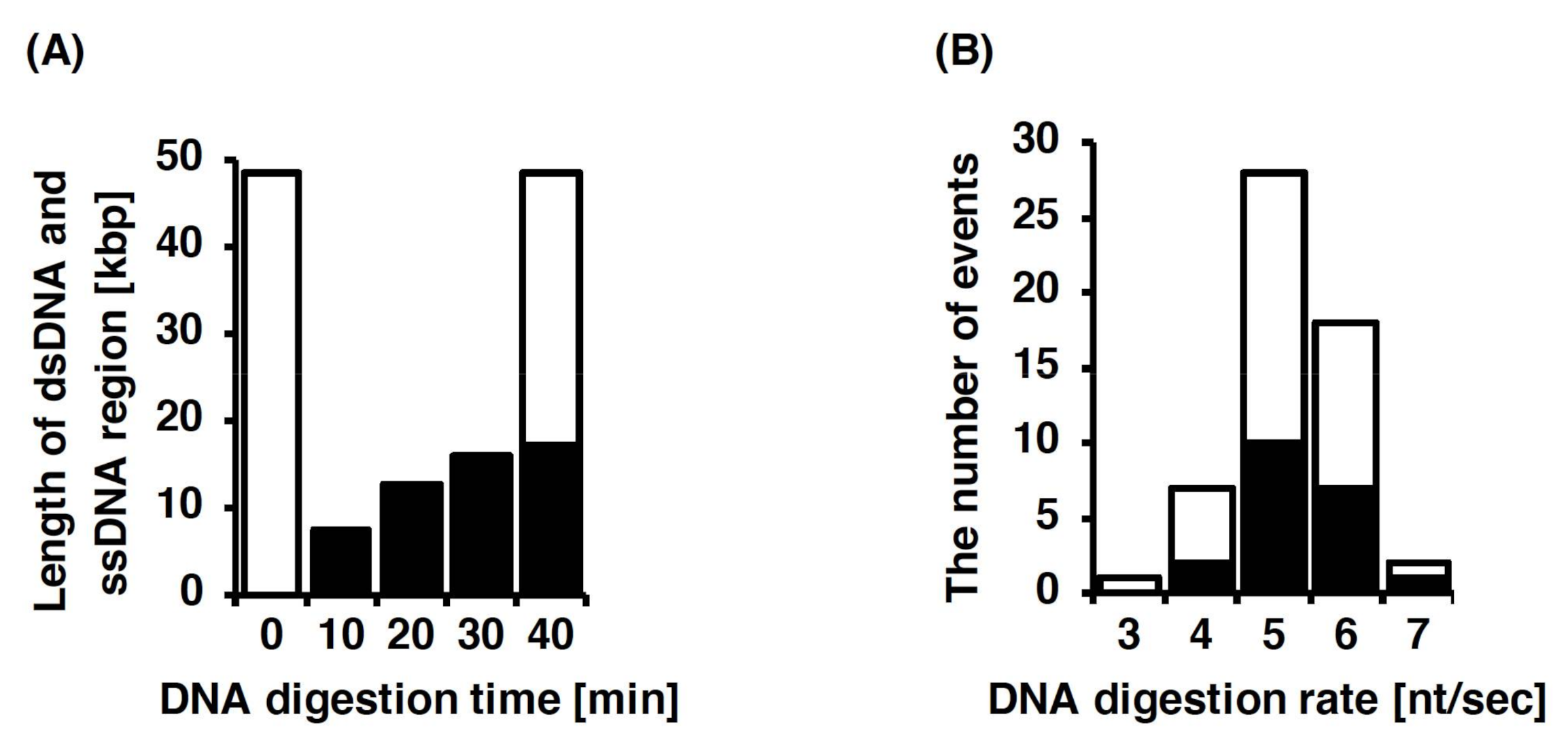
3.4. Single-Molecule Imaging for DNA Digestion by Exonuclease
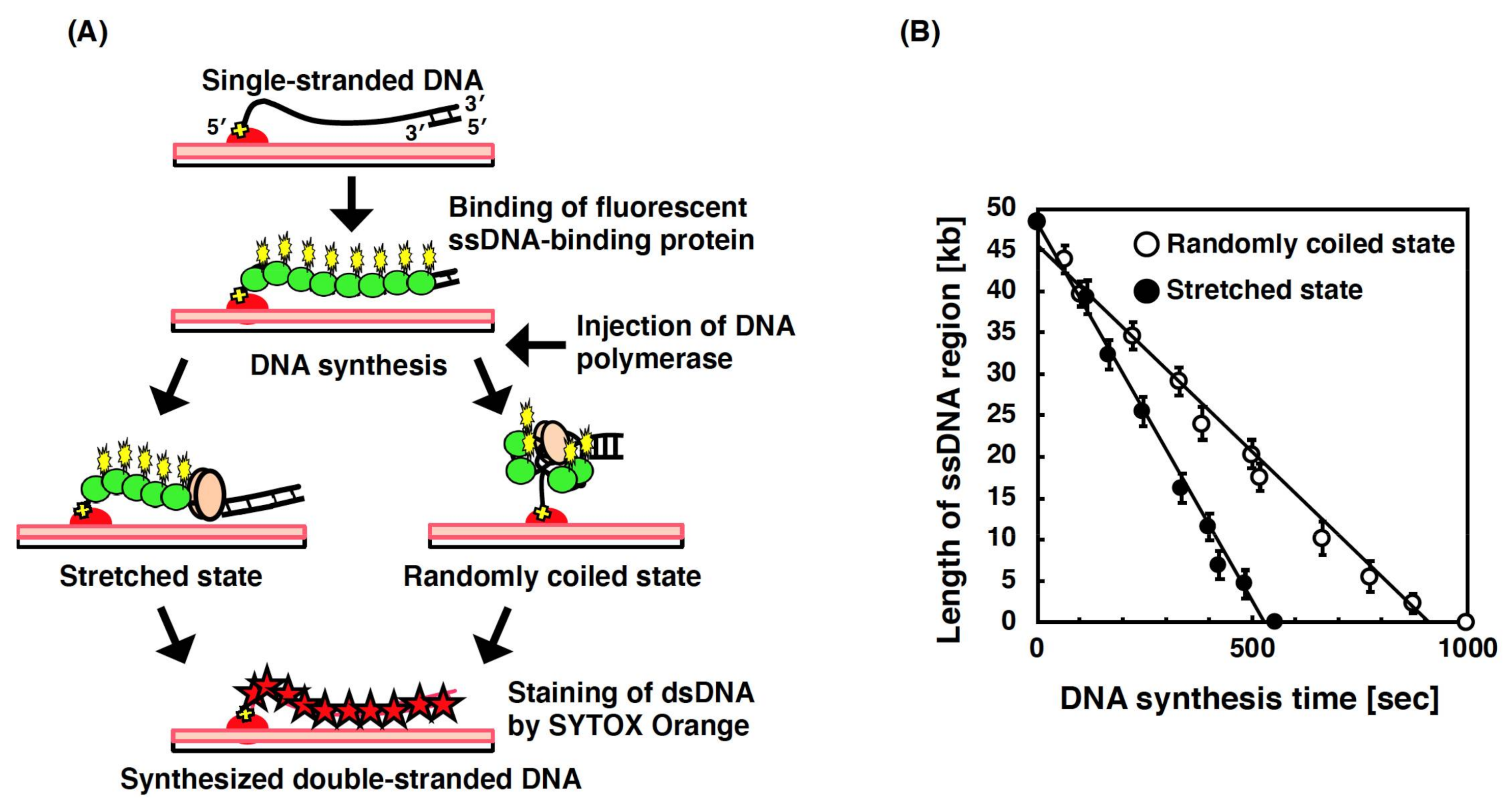
3.5. Single-Molecule Imaging for DNA Synthesis by DNA Polymerase
3.6. Single-Molecule Imaging for Supercoiled DNA and DNA Secondary Structures
3.7. Single-Molecule Imaging for DNA Origami
3.8. Single-Molecule Imaging for the Initiation of DNA Replication
3.9. Single-Molecule Imaging Based on Zero-Mode Waveguides (ZMWs)
4. Conclusions
- DNA manipulation by orientation forces, optical tweezers, fluid flow, and magnetic tweezers;
- Nanopores and DNA manipulation;
- Single-molecule imaging for double- and single-stranded DNA;
- The optical mapping of single DNA molecules;
- Single-molecule imaging for biological processes between DNA and protein;
- Single-molecule imaging for supercoiled DNA and DNA secondary structures;
- Single-molecule imaging for DNA origami and DNA sequencing.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Walter, N.G.; Huang, C.Y.; Manzo, A.J.; Sobhy, M.A. Do-it-yourself guide: How to use the modern single-molecule toolkit. Nat. Methods 2008, 5, 475–489. [Google Scholar] [CrossRef] [PubMed]
- Van Oijen, A.M. Single-molecule approaches to characterizing kinetics of biomolecular interactions. Curr. Opin. Biotechnol. 2011, 22, 75–80. [Google Scholar] [CrossRef]
- Neuman, K.C.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 5, 491–505. [Google Scholar] [CrossRef]
- Kaur, G.; Lewis, J.S.; van Oijen, A.M. Shining a Spotlight on DNA: Single-Molecule Methods to Visualise DNA. Molecules 2019, 24, 491. [Google Scholar] [CrossRef]
- Duzdevich, D.; Redding, S.; Greene, E.C. DNA Dynamics and Single-Molecule Biology. Chem. Rev. 2014, 114, 3072–3086. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.; Huberman, J.A.; Kornberg, A. Electron microscopy of DNA polymerase bound to DNA. J. Mol. Biol. 1971, 55, 209–214. [Google Scholar] [CrossRef]
- Morris, V.J.; Kirby, A.R.; Gunning, A.P. Atomic Force Microscopy for Biologists; Imperial College Press: London, UK, 2010; Volume 81, p. 332. [Google Scholar]
- Watson, J.D.; Crick, F.H. Molecular structure of nucleic acids. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Franklin, R.E.; Gosling, R.G. Molecular configuration in sodium thymonucleate. Nature 1953, 171, 740–741. [Google Scholar] [CrossRef]
- Fang, Y.; Hoh, J.H.; Spisz, T.S. Ethanol-induced structural transitions of DNA on mica. Nucleic Acids Res. 1999, 27, 1943–1949. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.Z.; Bash, R.; Balagurumoorthy, P.; Lohr, D.; Harrington, R.E.; Lindsay, S.M. Conformational transition in DNA on a cold surface. Nucleic Acids Res. 2000, 28, 593–596. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Japaridze, A.; Vobornik, D.; Lipiec, E.; Cerreta, A.; Szczerbinski, J.; Zenobi, R.; Dietler, G. Toward an effective control of DNA’s submolecular conformation on a surface. Macromolecules 2016, 49, 643–652. [Google Scholar] [CrossRef]
- Bustamante, C.; Bryant, Z.; Smith, S.B. Ten years of tension: Single-molecule DNA mechanics. Nature 2003, 421, 423–427. [Google Scholar] [CrossRef]
- Perkins, T.T.; Smith, D.E.; Chu, S. Direct observation of tube-like motion of a single polymer chain. Science 1994, 264, 819–822. [Google Scholar] [CrossRef]
- Smith, D.E.; Perkins, T.T.; Chu, S. Self-diffusion of an entangled DNA molecule by reptation. Phys. Rev. Lett. 1995, 75, 4146–4149. [Google Scholar] [CrossRef]
- Morikawa, K.; Yanagida, M. Visualization of Individual DNA Molecules in Solution by Light Microscopy: DAPI Staining Method. J. Biochem. 1981, 89, 693–696. [Google Scholar] [CrossRef] [PubMed]
- De Gennes, P.G. Reptation of a polymer chain in the presence of fixed obstacles. J. Chem. Phys. 1971, 55, 572–579. [Google Scholar] [CrossRef]
- Doi, M.; Edwards, S. Dynamics of concentrated polymer systems. Part 1. Brownian motion in the equilibriumstate. J. Chem. Soc. Faraday Trans. 1978, 74, 1789–1801. [Google Scholar] [CrossRef]
- Saenger, W. Principles of Nucleic Acid Structure; Springer: New York, NY, USA, 1984. [Google Scholar]
- Paleček, E. Local supercoil-stabilized DNA structure. Crit. Rev. Biochem. Mol. Biol. 1991, 26, 151–226. [Google Scholar] [CrossRef]
- Lyubchenko, Y.L.; Shlyakhtenko, L.S. Early melting of supercoiled DNA. Nucleic Acids Res. 1988, 16, 3269–3281. [Google Scholar] [CrossRef] [PubMed]
- Rich, A.; Wang, A.H.; Nordheim, A. The chemistry and biology of left-handed Z-DNA. Annu. Rev. Biochem. 1984, 53, 791–846. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, D.S.; Wang, J.C. Torsional rigidity of DNA and length dependence of the free energy of DNA supercoiling. J. Mol. Biol. 1984, 173, 75–91. [Google Scholar] [CrossRef]
- Gibson, D.G.; Glass, J.I.; Lartigue, C.; Noskov, V.N.; Chuang, R.Y.; Algire, M.A.; Benders, G.A.; Montague, M.G.; Ma, L.; Moodie, M.M.; et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 2010, 329, 52–56. [Google Scholar] [CrossRef]
- Gibson, D.G.; Benders, G.A.; Andrews-Pfannkoch, C.; Denisova, E.A.; Baden-Tillson, H.; Zaveri, J.; Stockwell, T.B.; Brownley, A.; Thomas, D.W.; Algire, M.A.; et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 2008, 319, 1215–1220. [Google Scholar] [CrossRef] [PubMed]
- Itaya, M.; Fujita, K.; Kuroki, A.; Tsuge, K. Bottom-up genome assembly using the Bacillus subtilis genome vector. Nat. Methods 2008, 5, 41–43. [Google Scholar] [CrossRef] [PubMed]
- Dymond, J.S.; Richardson, S.M.; Coombes, C.E.; Babatz, T.; Muller, H.; Annaluru, N.; Blake, W.J.; Schwerzmann, J.W.; Dai, J.; Lindstrom, D.L.; et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature 2011, 477, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Nawy, T. Yeast 2.0. Nat. Methods 2011, 8, 895. [Google Scholar] [CrossRef]
- Enyeart, P.J.; Ellington, A.D. Synthetic biology: A yeast for all reasons. Nature 2011, 477, 413–414. [Google Scholar] [CrossRef]
- Huey, R.; More, S.C. Condensed State of Nucleic Acids III, Ψ (+) and Ψ (−) Conformational Transitions of DNA Induced by Ethanol and Salt. Biopolymers 1981, 20, 2533–2552. [Google Scholar] [CrossRef]
- Bloomfield, V.A. Condensation of DNA by Multivalent Cations: Considerations on Mechanism. Biopolymers 1991, 31, 1471–1481. [Google Scholar] [CrossRef]
- Lerman, L.S. A Transition to a Compact Form of DNA in Polymer Solutions. Proc. Natl. Acad. Sci. USA 1971, 68, 1886–1890. [Google Scholar] [CrossRef]
- Jordan, C.F.; Lerman, L.S.; Venable, J.H. Structure and Circular Dichroism of DNA in Concentrated Polymer Solutions. Nature 1972, 236, 67–70. [Google Scholar]
- Maniatis, T.; Venable, J.H.J.; Lerman, L.S. Structure of Ψ DNA. J. Mol. Biol. 1974, 84, 37–64. [Google Scholar] [CrossRef]
- Katsura, S.; Yamaguchi, A.; Hirano, K.; Matsuzawa, Y.; Mizuno, A. Manipulation of globular DNA molecules for sizing and separation. Electrophoresis 2000, 21, 171–175. [Google Scholar] [CrossRef]
- Yoshikawa, K.; Matsuzawa, Y. Nucleation and Growth in Single DNA Molecules. J. Am. Chem. Sci. 1996, 118, 929–930. [Google Scholar] [CrossRef]
- Matsuzawa, Y.; Hirano, K.; Mizuno, A.; Ichikawa, M.; Yoshikawa, K. Geometric manipulation of DNA molecules with a laser. Appl. Phys. Lett. 2002, 81, 3494–3496. [Google Scholar] [CrossRef]
- Washizu, M.; Kurosawa, O. Electrostatic Manipulation of DNA in Microfabricated Structures. IEEE Trans. Ind. Appl. 1990, 26, 1165–1172. [Google Scholar] [CrossRef]
- Washizu, M.; Kurosawa, O.; Arai, I.; Suzuki, S.; Shimamoto, N. Applications of electrostatic stretch-and-positioning of DNA. IEEE Trans. Ind. Appl. 1995, 31, 447–456. [Google Scholar] [CrossRef]
- Suzuki, S.; Yamanashi, T.; Tazawa, S.-I.; Kurosawa, O.; Washizu, M. Quantitative analysis of DNA orientation in stationary AC electric fields using fluorescence anisotropy. IEEE Trans. Ind. Appl. 1998, 34, 75–83. [Google Scholar] [CrossRef]
- Kabata, H.; Kurosawa, O.; Arai, I.; Washizu, M.; Margarson, S.A.; Glass, R.E.; Shimamoto, N. Visualization of single molecules of RNA polymerase sliding along DNA. Science 1993, 262, 1561–1563. [Google Scholar] [CrossRef]
- Sung, K.E.; Burns, M.A. Optimization of dielectrophoretic DNA stretching in microfabricated devices. Anal. Chem. 2006, 78, 2939–2947. [Google Scholar] [CrossRef]
- Ashkin, A.; Dziedzic, J.M.; Bjorkholm, J.E.; Chu, S. Observation of a single-beam gradient force optical trap for dielectric particles. Opt. Lett. 1986, 11, 288–290. [Google Scholar] [CrossRef] [PubMed]
- Ashkin, A. Forces of a single-beam gradient laser trap on a dielectric sphere in the ray optics regime. Biophys. J. 1992, 61, 569–582. [Google Scholar] [CrossRef]
- Moffitt, J.R.; Chemla, Y.R.; Smith, S.B.; Bustamante, C. Recent advances in optical tweezers. Annu. Rev. Biochem. 2008, 77, 205. [Google Scholar] [CrossRef] [PubMed]
- Neuman, K.C.; Block, S.M. Optical trapping. Rev. Sci. Instrum. 2004, 75, 2787. [Google Scholar] [CrossRef]
- Brewer, L.R.; Corzett, M.; Balhorn, R. Protamine-induced condensation and decondensation of the same DNA molecule. Science 1999, 286, 120–123. [Google Scholar] [CrossRef]
- Bianco, P.R.; Brewer, L.R.; Corzett, M.; Balhorn, R.; Yeh, Y.; Kowalczykowski, S.C.; Baskin, R.J. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature 2001, 409, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Spies, M.; Bianco, P.R.; Dillingham, M.S.; Handa, N.; Baskin, R.J.; Kowalczykowski, S.C. A molecular throttle: The recombination hotspot chi controls DNA translocation by the RecBCD helicase. Cell 2003, 114, 647–654. [Google Scholar] [CrossRef]
- Kurita, H.; Yasuda, H.; Takashima, K.; Katsura, S.; Mizuno, A. Physical manipulation of single-molecule DNA using microbead and its application to analysis of DNA-protein interaction. J. Magn. Magn. Mater. 2009, 321, 655–658. [Google Scholar] [CrossRef]
- Yin, H.; Wang, M.D.; Svoboda, K.; Landick, R.; Block, S.M.; Gelles, J. Transcription against an applied force. Science 1995, 270, 1653–1657. [Google Scholar] [CrossRef]
- Shaevitz, J.W.; Abbondanzieri, E.A.; Landick, R.; Block, S.M. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature 2003, 426, 684. [Google Scholar] [CrossRef]
- Van den Broek, B.; Noom, M.C.; Wuite, G.J.L. DNA-tension dependence of restriction enzyme activity reveals mechanochemical properties of the reaction pathway. Nucleic Acids Res. 2005, 33, 2676–2684. [Google Scholar] [CrossRef]
- Heller, I.; Hoekstra, T.P.; King, G.A.; Peterman, E.J.; Wuite, G.J. Optical tweezers analysis of DNA–protein complexes. Chem. Rev. 2014, 114, 3087–3119. [Google Scholar] [CrossRef] [PubMed]
- Katsura, S.; Hirano, K.; Matsuzawa, Y.; Mizuno, A.; Yoshikawa, K. Direct laser trapping of single DNA molecules in the globular state. Nucleic Acids Res. 1998, 26, 4943–4945. [Google Scholar] [CrossRef]
- Matsuura, S.-I.; Komatsu, J.; Hirano, K.; Yasuda, H.; Takashima, K.; Katsura, S.; Mizuno, A. Real-time observation of a single DNA digestion by λ exonuclease under a fluorescence microscope field. Nucleic Acids Res. 2001, 29, e79. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Matsuura, S.-I.; Kurita, H.; Nakano, M.; Komatsu, J.; Takashima, K.; Katsura, S.; Mizuno, A. One-end immobilization of individual DNA molecules on a functional hydrophobic glass surface. J. Biomol. Struct. Dyn. 2002, 20, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Graneli, A.; Yeykal, C.C.; Prasad, T.K.; Greene, E.C. Organized arrays of individual DNA molecules tethered to supported lipid bilayers. Langmuir 2006, 22, 292–299. [Google Scholar] [CrossRef]
- Fazio, T.; Visnapuu, M.L.; Wind, S.; Greene, E.C. DNA curtains and nanoscale curtain rods: High-throughput tools for single molecule imaging. Langmuir 2008, 24, 10524–10531. [Google Scholar] [CrossRef]
- Greene, E.C.; Wind, S.; Fazio, T.; Gorman, J.; Visnapuu, M.L. DNA curtains for high-throughput single-molecule optical imaging. Methods Enzymol. 2010, 472, 293. [Google Scholar]
- Finkelstein, I.J.; Greene, E.C. Supported lipid bilayers and DNA curtains for high-throughput single-molecule studies. Methods Mol. Biol. 2011, 745, 447. [Google Scholar]
- Robison, A.D.; Finkelstein, I.J. Rapid Prototyping of Multichannel Microfluidic Devices for Single-Molecule DNA Curtain Imaging. Anal. Chem. 2014, 86, 4157–4163. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, I.F.; Pasupathy, P.; Brown, M.; Manhart, C.M.; Neikirk, D.P.; Alani, E.; Finkelstein, I.J. High-throughput universal DNA curtain arrays for single-molecule fluorescence imaging. Langmuir 2015, 31, 10310–10317. [Google Scholar] [CrossRef] [PubMed]
- Strick, T.R.; Allemand, J.F.; Bensimon, D.; Bensimon, A.; Croquette, V. The elasticity of a single supercoiled DNA molecule. Science 1996, 271, 1835–1837. [Google Scholar] [CrossRef]
- Strick, T.R.; Allemand, J.F.; Bensimon, D.; Croquette, V. Behavior of supercoiled DNA. Biophys. J. 1998, 74, 2016–2028. [Google Scholar] [CrossRef]
- Strick, T.R.; Croquette, V.; Bensimon, D. Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature 2000, 404, 901–904. [Google Scholar] [CrossRef]
- Koster, D.A.; Croquette, V.; Dekker, C.; Shuman, S.; Dekker, N.H. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature 2005, 434, 671–674. [Google Scholar] [CrossRef]
- Gore, J.; Bryant, Z.; Stone, M.D.; Nollmann, M.N.; Cozzarelli, N.R.; Bustamante, C. Mechanochemical analysis of DNA gyrase using rotor bead tracking. Nature 2006, 439, 100–104. [Google Scholar] [CrossRef]
- Koster, D.A.; Crut, A.; Shuman, S.; Bjornsti, M.A.; Dekker, N.H. Cellular strategies for regulating DNA supercoiling: A single-molecule perspective. Cell 2010, 142, 519–530. [Google Scholar] [CrossRef]
- Van Loenhout, M.T.; de Grunt, M.V.; Dekker, C. Dynamics of DNA supercoils. Science 2012, 338, 94–97. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Motooka, S.; Usui, T.; Kawasaki, S.; Miyata, H.; Kurita, H.; Mizuno, T.; Matsuura, S.I.; Mizuno, A.; Oshige, M.; et al. Direct single-molecule observations of local denaturation of a DNA double helix under a negative supercoil state. Anal. Chem. 2015, 87, 3490–3497. [Google Scholar] [CrossRef]
- Bensimon, A.; Simon, A.; Chiffaudel, A.; Croquette, V.; Heslot, F.; Bensimon, D. Alignment and sensitive detection of DNA by a moving interface. Science 1994, 265, 2096–2098. [Google Scholar] [CrossRef] [PubMed]
- Giri, D.; Li, Z.; Ashraf, K.M.; Collinson, M.M.; Higgins, D.A. Molecular combing of λ-DNA using self-propelled water droplets on wettability gradient surfaces. ACS Appl. Mater. Interfaces 2016, 8, 24265–24272. [Google Scholar] [CrossRef] [PubMed]
- Michalet, X.; Ekong, R.; Fougerousse, F.; Rousseaux, S.; Schurra, C.; Hornigold, N.; van Slegtenhorst, M.; Wolfe, J.; Povey, S.; Beckmann, J.S.; et al. Dynamic molecular combing: Stretching the whole human genome for high-resolution studies. Science 1997, 277, 1518–1523. [Google Scholar] [CrossRef] [PubMed]
- Varapula, D.; LaBouff, E.; Raseley, K.; Uppuluri, L.; Ehrlich, G.D.; Noh, M.; Xiao, M. A micropatterned substrate for on-surface enzymatic labelling of linearized long DNA molecules. Sci. Rep. 2019, 9, 15059 . [Google Scholar] [CrossRef] [PubMed]
- Allemand, J.F.; Bensimon, D.; Jullien, L.; Bensimon, A.; Croquette, V. pH-dependent specific binding and combing of DNA. Biophys. J. 1997, 73, 2064–2070. [Google Scholar] [CrossRef]
- Deng, Z.; Mao, C. DNA-Templated Fabrication of 1D Parallel and 2D Crossed Metallic Nanowire Arrays. Nano Lett. 2003, 3, 1545–1548. [Google Scholar] [CrossRef]
- Zhang, J.; Ma, Y.; Stachura, S.; He, H. Assembly of highly aligned DNA strands onto Si chips. Langmuir 2005, 21, 4180–4184. [Google Scholar] [CrossRef] [PubMed]
- Yoda, S.; Han, S.P.; Kudo, H.; Kwak, K.J.; Fujihira, M. Deoxyribonucleic acid interaction with silanized surfaces. Jpn. J. Appl. Phys. 2004, 43, 6297–6304. [Google Scholar] [CrossRef]
- Han, S.P.; Yoda, S.; Kwak, K.J.; Suga, K.; Fujihira, M. Interpretation of DNA adsorption on silanized surfaces by measuring interaction forces at various pHs using atomic force microscopy. Ultramicroscopy 2005, 105, 148–154. [Google Scholar] [CrossRef]
- Oshige, M.; Yamaguchi, K.; Matsuura, S.-I.; Kurita, H.; Mizuno, A.; Katsura, S. A new DNA combing method for biochemical analysis. Anal. Biochem. 2010, 400, 145–147. [Google Scholar] [CrossRef]
- Oshige, M.; Kawasaki, S.; Takano, H.; Yamaguchi, K.; Kurita, H.; Mizuno, T.; Matsuura, S.-I.; Mizuno, A.; Katsura, S. Direct observation method of individual single-stranded DNA molecules using fluorescent replication protein A. J. Fluoresc. 2011, 21, 1189–1194. [Google Scholar] [CrossRef]
- Seeman, N.C. DNA in a material world. Nature 2003, 421, 427–431. [Google Scholar] [CrossRef]
- Aldaye, F.A.; Palmer, A.L.; Sleiman, H.F. Assembling materials with DNA as the guide. Science 2008, 321, 1795–1799. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Yao, C.; Zhu, Y.; Yang, L.; Luo, D.; Yang, D. DNA functional materials assembled from branched DNA: Design, synthesis, and applications. Chem. Rev. 2020, 120, 9420–9481. [Google Scholar] [CrossRef]
- Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef]
- Castro, C.E.; Kilchherr, F.; Kim, D.N.; Shiao, E.L.; Wauer, T.; Wortmann, P.; Bathe, M.; Dietz, H. A primer to scaffolded DNA origami. Nat. Methods 2011, 8, 221. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Katsuda, Y.; Hidaka, K.; Sugiyama, H. A versatile DNA nanochip for direct analysis of DNA base-excision repair. Angew. Chem. Int. Ed. 2010, 49, 9412–9416. [Google Scholar] [CrossRef]
- Suzuki, Y.; Endo, M.; Katsuda, Y.; Ou, K.; Hidaka, K.; Sugiyama, H. DNA origami based visualization system for studying site-specific recombination events. J. Am. Chem. Soc. 2014, 136, 211–218. [Google Scholar] [CrossRef]
- Samanta, A.; Medintz, I.L. Nanoparticles and DNA—A powerful and growing functional combination in bionanotechnology. Nanoscale 2016, 8, 9037–9095. [Google Scholar] [CrossRef]
- Urban, M.J.; Dutta, P.K.; Wang, P.; Duan, X.; Shen, X.; Ding, B.; Ke, Y.; Liu, N. Plasmonic toroidal metamolecules assembled by DNA origami. J. Am. Chem. Soc. 2016, 138, 5495–5498. [Google Scholar] [CrossRef]
- Teschome, B.; Facsko, S.; Gothelf, K.V.; Keller, A. Alignment of gold nanoparticle-decorated DNA origami nanotubes: Substrate prepatterning versus molecular combing. Langmuir 2015, 31, 12823–12829. [Google Scholar] [CrossRef] [PubMed]
- Kuzuya, A.; Sakai, Y.; Yamazaki, T.; Xu, Y.; Komiyama, M. Nanomechanical DNA origami ‘single-molecule beacons’ directly imaged by atomic force microscopy. Nat. Commun. 2011, 2, 449 . [Google Scholar] [CrossRef]
- Wang, S.; Zhou, Z.; Ma, N.; Yang, S.; Li, K.; Teng, C.; Ke, Y.; Tian, Y. DNA Origami-Enabled Biosensors. Sensors 2020, 20, 6899. [Google Scholar] [CrossRef] [PubMed]
- Bell, N.A.; Engst, C.R.; Ablay, M.; Divitini, G.; Ducati, C.; Liedl, T.; Keyser, U.F. DNA origami nanopores. Nano Lett. 2012, 12, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Martin, T.G.; Rant, U.; Dietz, H. DNA origami gatekeepers for solid-state nanopores. Angew. Chem. Int. Ed. 2012, 124, 4948–4951. [Google Scholar] [CrossRef]
- Jiang, Q.; Liu, S.; Liu, J.; Wang, Z.G.; Ding, B. Rationally designed DNA-origami nanomaterials for drug delivery in vivo. Adv. Mater. 2019, 31, 1804785. [Google Scholar] [CrossRef]
- Zhao, Y.X.; Shaw, A.; Zeng, X.; Benson, E.; Nystrom, A.M.; Hogberg, B. DNA origami delivery system for cancer therapy with tunable release properties. ACS Nano 2012, 6, 8684–8691. [Google Scholar] [CrossRef]
- Crampton, N.; Yokokawa, M.; Dryden, D.T.; Edwardson, J.M.; Rao, D.N.; Takeyasu, K.; Yoshimura, S.H.; Henderson, R.M. Fast-scan atomic force microscopy reveals that the type III restriction enzyme EcoP15I is capable of DNA translocation and looping. Proc. Natl. Acad. Sci. USA 2007, 104, 12755–12760. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, J.L.; Suzuki, Y.; Tamulaitis, G.; Siksnys, V.; Takeyasu, K.; Lyubchenko, Y.L. Single-Molecule Dynamics of the DNA−EcoRII Protein Complexes Revealed with High-Speed Atomic Force Microscopy. Biochemistry 2009, 48, 10492–10498. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Katsuda, Y.; Hidaka, K.; Sugiyama, H. Regulation of DNA methylation using different tensions of double strands constructed in a defined DNA nanostructure. J. Am. Chem. Soc. 2010, 132, 1592–1597. [Google Scholar] [CrossRef]
- Endo, M. AFM-based single-molecule observation of the conformational changes of DNA structures. Methods 2019, 169, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Xing, X.; Zhou, X.; Emura, T.; Hidaka, K.; Tuesuwan, B.; Sugiyama, H. Single-Molecule manipulation of the duplex formation and dissociation at the G-Quadruplex/i-Motif site in the DNA nanostructure. ACS Nano 2015, 9, 9922–9929. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, A.; Endo, M.; Hidaka, K.; Lan Thao Tran, P.; Mergny, J.L.; Sugiyama, H. Controlling the stoichiometry and strand polarity of a tetramolecular G-quadruplex structure by using a DNA origami frame. Nucleic Acids Res. 2013, 41, 8738–8747. [Google Scholar] [CrossRef] [PubMed]
- Räz, M.H.; Hidaka, K.; Sturla, S.J.; Sugiyama, H.; Endo, M. Torsional constraints of DNA substrates impact Cas9 cleavage. J. Am. Chem. Soc. 2016, 138, 13842–13845. [Google Scholar] [CrossRef] [PubMed]
- Howorka, S.; Siwy, Z. Nanopore analytics: Sensing of single molecules. Chem. Soc. Rev. 2009, 38, 2360–2384. [Google Scholar] [CrossRef]
- Meller, A.; Nivon, L.; Branton, D. Voltage-driven DNA translocations through a nanopore. Phys. Rev. Lett. 2001, 86, 3435. [Google Scholar] [CrossRef] [PubMed]
- Deamer, D.; Akeson, M.; Branton, D. Three decades of nanopore sequencing. Nat. Biotechnol. 2016, 34, 518–524. [Google Scholar] [CrossRef]
- Yuan, Z.; Wang, C.; Yi, X.; Ni, Z.; Chen, Y.; Li, T. Solid-state nanopore. Nanoscale Res. Lett. 2018, 13, 56. [Google Scholar] [CrossRef]
- Keyser, U.F.; Koeleman, B.N.; Van Dorp, S.; Krapf, D.; Smeets, R.M.; Lemay, S.G.; Dekker, N.H.; Dekker, C. Direct force measurements on DNA in a solid-state nanopore. Nat. Phys. 2006, 2, 473–477. [Google Scholar] [CrossRef]
- Peng, H.; Ling, X.S. Reverse DNA translocation through a solid-state nanopore by magnetic tweezers. Nanotechnology 2009, 20, 185101. [Google Scholar] [CrossRef]
- Nelson, E.M.; Li, H.; Timp, G. Direct, concurrent measurements of the forces and currents affecting DNA in a nanopore with comparable topography. ACS Nano 2014, 8, 5484–5493. [Google Scholar] [CrossRef]
- Zhu, L.; Zhang, Z.; Liu, Q. Deformation-mediated translocation of DNA origami nanoplates through a narrow solid-state nanopore. Anal. Chem. 2020, 92, 13238–13245. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, R.; Pal, S.; Joshi, H.; Rao, A.; Naik, A.; Varma, M.; Chakraborty, B.; Maiti, P.K. DNA Translocation through Hybrid Bilayer Nanopores. J. Phys. Chem. C 2019, 123, 11908–11916. [Google Scholar] [CrossRef] [PubMed]
- Barati Farimani, A.; Dibaeinia, P.; Aluru, N.R. DNA Origami–Graphene Hybrid Nanopore for DNA Detection. ACS Appl. Mater. Interfaces 2017, 9, 92–100. [Google Scholar] [CrossRef]
- Ding, T.; Yang, J.; Pan, V.; Zhao, N.; Lu, Z.; Ke, Y.; Zhang, C. DNA nanotechnology assisted nanopore-based analysis. Nucleic Acids Res. 2020, 48, 2791–2806. [Google Scholar] [CrossRef]
- Shen, B.; Piskunen, P.; Nummelin, S.; Liu, Q.; Kostiainen, M.A.; Linko, V. Advanced DNA Nanopore Technologies. ACS Appl. Bio Mater. 2020, 3, 5606–5619. [Google Scholar] [CrossRef]
- Murade, C.U.; Subramaniam, V.; Otto, C.; Bennink, M.L. Interaction of oxazole yellow dyes with DNA studied with hybrid optical tweezers and fluorescence microscopy. Biophys. J. 2009, 97, 835–843. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Martin, R.M.; Leonhardt, H.; Cardoso, M.C. DNA labeling in living cells. Cytom. Part A 2005, 67, 45–52. [Google Scholar] [CrossRef]
- Bakajin, O.B.; Duke, T.A.J.; Chou, C.F.; Chan, S.S.; Austin, R.H.; Cox, E.C. Electrohydrodynamic stretching of DNA in confined environments. Phys. Rev. Lett. 1998, 80, 2737. [Google Scholar] [CrossRef]
- Dragan, A.I.; Casas-Finet, J.R.; Bishop, E.S.; Strouse, R.J.; Schenerman, M.A.; Geddes, C.D. Characterization of PicoGreen interaction with dsDNA and the origin of its fluorescence enhancement upon binding. Biophys. J. 2010, 99, 3010–3019. [Google Scholar] [CrossRef]
- Gudnason, H.; Dufva, M.; Bang, D.D.; Wolff, A. Comparison of multiple DNA dyes for real-time PCR: Effects of dye concentration and sequence composition on DNA amplification and melting temperature. Nucleic Acids Res. 2007, 35, e127. [Google Scholar] [CrossRef]
- Gurrieri, S.; Wells, K.S.; Johnson, I.D.; Bustamante, C. Direct visualization of individual DNA molecules by fluorescence microscopy: Characterization of the factors affecting signal/background and optimization of imaging conditions using YOYO. Anal. Biochem. 1997, 249, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Reuter, M.; Dryden, D.T. The kinetics of YOYO-1 intercalation into single molecules of double-stranded DNA. Biochem. Biophys. Res. Commun. 2010, 403, 225–229. [Google Scholar] [CrossRef]
- Yan, X.; Habbersett, R.C.; Yoshida, T.M.; Nolan, J.P.; Jett, J.H.; Marrone, B.L. Probing the kinetics of SYTOX Orange stain binding to double-stranded DNA with implications for DNA analysis. Anal. Chem. 2005, 77, 3554–3562. [Google Scholar] [CrossRef]
- Biebricher, A.S.; Heller, I.; Roijmans, R.F.; Hoekstra, T.P.; Peterman, E.J.; Wuite, G.J. The impact of DNA intercalators on DNA and DNA-processing enzymes elucidated through force-dependent binding kinetics. Nat. Commun. 2015, 6, 7304. [Google Scholar] [CrossRef] [PubMed]
- Günther, K.; Mertig, M.; Seidel, R. Mechanical and structural properties of YOYO-1 complexed DNA. Nucleic Acids Res. 2010, 38, 6526–6532. [Google Scholar] [CrossRef] [PubMed]
- Japaridze, A.; Benke, A.; Renevey, S.; Benadiba, C.; Dietler, G. Influence of DNA binding dyes on bare DNA structure studied with atomic force microscopy. Macromolecules 2015, 48, 1860–1865. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, A.; Baker, B.; Sun, L.; Howard, A.; Buswell, J.; Maurel, D.; Masharina, A.; Johnsson, K.; Noren, C.J.; et al. Development of SNAP-tag fluorogenic probes for wash-free fluorescence imaging. ChemBioChem 2011, 12, 2217–2226. [Google Scholar] [CrossRef]
- Los, G.V.; Encell, L.P.; McDougall, M.G.; Hartzell, D.D.; Karassina, N.; Zimprich, C.; Wood, M.G.; Learish, R.; Ohana, R.F.; Urh, M.; et al. HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 2008, 3, 373–382. [Google Scholar] [CrossRef]
- Rusinova, E.; Tretyachenko-Ladokhina, V.; Vele, O.E.; Senear, D.F.; Ross, J.A. Alexa and Oregon Green dyes as fluorescence anisotropy probes for measuring protein–protein and protein–nucleic acid interactions. Anal. Biochem. 2002, 308, 18–25. [Google Scholar] [CrossRef]
- Toseland, C.P. Fluorescent labeling and modification of proteins. J. Chem. Biol. 2013, 6, 85–95. [Google Scholar] [CrossRef]
- Selvin, P.R. The renaissance of fluorescence resonance energy transfer. Nat. Struct. Mol. Biol. 2000, 7, 730–734. [Google Scholar] [CrossRef]
- Clegg, R.M. Fluorescence resonance energy transfer. Curr. Opin. Biotech. 1995, 6, 103–110. [Google Scholar] [CrossRef]
- Roy, R.; Hohng, S.; Ha, T. A practical guide to single-molecule FRET. Nat. Methods 2008, 5, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Hohng, S.; Lee, S.; Lee, J.; Jo, M.H. Maximizing information content of single-molecule FRET experiments: Multi-color FRET and FRET combined with force or torque. Chem. Soc. Rev. 2014, 43, 1007–1013. [Google Scholar] [CrossRef]
- Marklund, E.; van Oosten, B.; Mao, G.; Amselem, E.; Kipper, K.; Sabantsev, A.; Emmerich, A.; Globisch, D.; Zheng, X.; Lehmann, L.C.; et al. DNA surface exploration and operator bypassing during target search. Nature 2020, 583, 858–861. [Google Scholar] [CrossRef]
- Fijen, C.; Silva, A.M.; Hochkoeppler, A.; Hohlbein, J. A single-molecule FRET sensor for monitoring DNA synthesis in real time. Phys. Chem. Chem. Phys. 2017, 19, 4222–4230. [Google Scholar] [CrossRef]
- Grimme, J.M.; Honda, M.; Wright, R.; Okuno, Y.; Rothenberg, E.; Mazin, A.V.; Ha, T.; Spies, M. Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52–ssDNA complexes. Nucleic Acids Res. 2010, 38, 2917–2930. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Kim, S.H.; Hong, S.C. Minute negative superhelicity is sufficient to induce the BZ transition in the presence of low tension. Proc. Natl. Acad. Sci. USA 2010, 107, 4985–4990. [Google Scholar] [CrossRef]
- Long, X.; Parks, J.W.; Bagshaw, C.R.; Stone, M.D. Mechanical unfolding of human telomere G-quadruplex DNA probed by integrated fluorescence and magnetic tweezers spectroscopy. Nucleic Acids Res. 2013, 41, 2746–2755. [Google Scholar] [CrossRef]
- Chapados, B.R.; Hosfield, D.J.; Han, S.; Qiu, J.; Yelent, B.; Shen, B.; Tainer, J.A. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell 2004, 116, 39–50. [Google Scholar] [CrossRef]
- Collins, T.R.; Hammes, G.G.; Hsieh, T.S. Analysis of the eukaryotic topoisomerase II DNA gate: A single-molecule FRET and structural perspective. Nucleic Acids Res. 2009, 37, 712–720. [Google Scholar] [CrossRef]
- Ticau, S.; Friedman, L.J.; Champasa, K.; Corrêa, I.R., Jr.; Gelles, J.; Bell, S.P. Mechanism and timing of Mcm2–7 ring closure during DNA replication origin licensing. Nat. Struct. Mol. Biol. 2017, 24, 309. [Google Scholar] [CrossRef]
- Wu, W.Q.; Hou, X.M.; Zhang, B.; Fossé, P.; René, B.; Mauffret, O.; Li, M.; Dou, S.X.; Xi, X.G. Single-molecule studies reveal reciprocating of WRN helicase core along ssDNA during DNA unwinding. Sci. Rep. 2017, 7, 43954. [Google Scholar] [CrossRef]
- Washizu, M.; Yamamoto, T.; Kurosawa, O.; Suzuki, S.; Shimamoto, N. Moleculer Surgery of DNA Using Enzyme-Immobilized Particles. IEEJ Trans. Sens. Micromach. 1996, 116, 196–202. [Google Scholar] [CrossRef][Green Version]
- Meng, X.; Benson, K.; Chada, K.; Huff, E.J.; Schwartz, D.C. Optical mapping of lambda bacteriophage clones using restriction endonucleases. Nat. Genet. 1995, 9, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Wei, F.; Nguyen, J.; Bechner, M.; Potamousis, K.; Goldstein, S.; Pape, L.; Mehan, M.R.; Churas, C.; Pasternak, S.; et al. A single molecule scaffold for the maize genome. PLoS Genet. 2009, 5, e1000711. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.W.; Schramm, T.M.; Zhou, S.; Schwartz, D.C.; Talaat, A.M. Optical mapping of the Mycobacterium avium subspecies paratuberculosis genome. BMC Genom. 2009, 10, 25. [Google Scholar] [CrossRef]
- Mizuno, A. Manipulation of Single DNA Molecules, Micromachines as Tools for Nanotechnology; Fujita, H., Ed.; Springer: Berlin/Heidelberg, Germany, 2003; pp. 45–82. [Google Scholar]
- Katsura, S.; Harada, N.; Maeda, Y.; Komatsu, J.; Matsuura, S.-I.; Takashima, K.; Mizuno, A. Activation of restriction enzyme by electrochemically released magnesium ion. J. Biosci. Bioeng. 2004, 98, 293–297. [Google Scholar] [CrossRef]
- Kim, S.; Gottfried, A.; Lin, R.R.; Dertinger, T.; Kim, A.S.; Chung, S.; Colyer, R.A.; Weinhold, E.; Weiss, S.; Ebenstein, Y. Enzymatically Incorporated Genomic Tags for Optical Mapping of DNA-Binding Proteins. Angew. Chem. Int. Ed. 2012, 51, 3578–3581. [Google Scholar] [CrossRef]
- Ananiev, G.E.; Goldstein, S.; Runnheim, R.; Forrest, D.K.; Zhou, S.; Potamousis, K.; Churas, C.P.; Bergendahl, V.; Thomson, J.A.; Schwartz, D.C. Optical mapping discerns genome wide DNA methylation profiles. BMC Mol. Biol. 2008, 9, 68. [Google Scholar] [CrossRef] [PubMed]
- Neely, R.K.; Deen, J.; Hofkens, J. Optical mapping of DNA: Single-molecule-based methods for mapping genomes. Biopolymers 2011, 95, 298–311. [Google Scholar] [CrossRef]
- Cheng, X.; Roberts, R.J. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001, 29, 3784–3795. [Google Scholar] [CrossRef] [PubMed]
- Klimašauskas, S.; Weinhold, E. A new tool for biotechnology: AdoMet-dependent methyltransferases. Trends Biotechnol. 2007, 25, 99–104. [Google Scholar] [CrossRef]
- Lukinavicius, G.; Lapiene, V.; Stasevskij, Z.; Dalhoff, C.; Weinhold, E.; Klimasauskas, S. Targeted labeling of DNA by methyltransferase-directed transfer of activated groups (mTAG). J. Am. Chem. Soc. 2007, 129, 2758–2759. [Google Scholar] [CrossRef]
- Neely, R.K.; Dedecker, P.; Hotta, J.I.; Urbanavičiūtė, G.; Klimašauskas, S.; Hofkens, J. DNA fluorocode: A single molecule, optical map of DNA with nanometre resolution. Chem. Sci. 2010, 1, 453–460. [Google Scholar] [CrossRef]
- Grunwald, A.; Dahan, M.; Giesbertz, A.; Nilsson, A.; Nyberg, L.K.; Weinhold, E.; Ambjörnsson, T.; Westerlund, F.; Ebenstein, Y. Bacteriophage strain typing by rapid single molecule analysis. Nucleic Acids Res. 2015, 43, e117. [Google Scholar] [CrossRef]
- Marie, R.; Pedersen, J.N.; Bærlocher, L.; Koprowska, K.; Pødenphant, M.; Sabatel, C.; Zalkovskij, M.; Mironov, A.; Bilenberg, B.; Ashley, N.; et al. Single-molecule DNA-mapping and whole-genome sequencing of individual cells. Proc. Natl. Acad. Sci. USA 2018, 115, 11192–11197. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Xie, M.; Jiang, Y.; Xiao, N.; Du, X.; Zhang, W.; Tosser-Klopp, G.; Wang, J.; Yang, S.; Liang, J.; et al. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat. Biotechnol. 2013, 31, 135–141. [Google Scholar] [CrossRef]
- Chan, E.K.; Cameron, D.L.; Petersen, D.C.; Lyons, R.J.; Baldi, B.F.; Papenfuss, A.T.; Thomas, D.M.; Hayes, V.M. Optical mapping reveals a higher level of genomic architecture of chained fusions in cancer. Genome Res. 2018, 28, 726–738. [Google Scholar] [CrossRef] [PubMed]
- Gibb, B.; Silverstein, T.D.; Finkelstein, I.J.; Greene, E.C. Single-stranded DNA curtains for real-time single-molecule visualization of protein–nucleic acid interactions. Anal. Chem. 2012, 84, 7607–7612. [Google Scholar] [CrossRef]
- Sakaguchi, K.; Ishibashi, T.; Uchiyama, Y.; Iwabata, K. The multi-replication protein A (RPA) system—A new perspective. FEBS J. 2009, 276, 943–963. [Google Scholar] [CrossRef]
- Fanning, E.; Klimovich, V.; Nager, A.R. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006, 34, 4126–4137. [Google Scholar] [CrossRef]
- Takahashi, S.; Kawasaki, S.; Yamaguchi, K.; Miyata, H.; Kurita, H.; Mizuno, T.; Matsuura, S.-I.; Mizuno, A.; Oshige, M.; Katsura, S. Direct observation of fluorescently labeled single-stranded λDNA molecules in a micro-flow channel. J. Fluoresc. 2013, 23, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Gibb, B.; Ling, F.Y.; Gergoudis, S.C.; Kwon, Y.; Niu, H.; Sung, P.; Greene, E.C. Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLoS ONE 2014, 9, e87922. [Google Scholar] [CrossRef]
- Shevelev, I.V.; Hübscher, U. The 3′-5′ exonucleases. Nat. Rev. Mol. Cell Biol. 2002, 3, 364–376. [Google Scholar] [CrossRef] [PubMed]
- Nimonkar, A.V.; Genschel, J.; Kinoshita, E.; Polaczek, P.; Campbell, J.L.; Wyman, C.; Modrich, P.; Kowalczykowski, S.C. BLM–DNA2–RPA–MRN and EXO1–BLM–RPA–MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011, 25, 350–362. [Google Scholar] [CrossRef]
- Reha-Krantz, L.J. DNA polymerase proofreading: Multiple roles maintain genome stability. Biochim. Biophys. Acta Proteins Proteom. 2010, 1804, 1049–1063. [Google Scholar] [CrossRef]
- Van Oijen, A.M.; Blainey, P.C.; Crampton, D.J.; Richardson, C.C.; Ellenberger, T.; Xie, X.S. Single-molecule kinetics of λ exonuclease reveal base dependence and dynamic disorder. Science 2003, 301, 1235–1238. [Google Scholar] [CrossRef]
- Kurita, H.; Inaishi, K.-I.; Torii, K.; Urisu, M.; Nakano, M.; Katsura, S.; Mizuno, A. Real-time direct observation of single-molecule DNA hydrolysis by exonuclease III. J. Biomol. Struct. Dyn. 2008, 25, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Kurita, H.; Torii, K.; Yasuda, H.; Takashima, K.; Katsura, S.; Mizuno, A. The Effect of Physical Form of DNA on Exonuclease III Activity Revealed by Single-molecule Observations. J. Fluoresc. 2009, 19, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Usui, T.; Kawasaki, S.; Miyata, H.; Kurita, H.; Matsuura, S.-I.; Mizuno, A.; Oshige, M.; Katsura, S. Real-time single-molecule observations of T7 Exonuclease activity in a microflow channel. Anal. Biochem. 2014, 457, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Fijalkowska, I.J.; Schaaper, R.M.; Jonczyk, P. DNA replication fidelity in Escherichia coli: A multi-DNA polymerase affair. FEMS Microbiol. Rev. 2012, 36, 1105–1121. [Google Scholar] [CrossRef]
- Lange, S.S.; Takata, K.-I.; Wood, R.D. DNA polymerases and cancer. Nat. Rev. Cancer 2011, 11, 96–110. [Google Scholar] [CrossRef]
- Tanner, N.A.; Loparo, J.J.; Hamdan, S.M.; Jergic, S.; Dixon, N.E.; van Oijen, A.M. Real-time single-molecule observation of rolling-circle DNA replication. Nucleic Acids Res. 2009, 37, e27. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.E.; Marians, K.J.; Kowalczykowski, S.C. Independent and stochastic action of DNA polymerases in the replisome. Cell 2017, 169, 1201–1213. [Google Scholar] [CrossRef]
- Lewis, J.S.; Spenkelink, L.M.; Jergic, S.; Wood, E.A.; Monachino, E.; Horan, N.P.; Duderstadt, K.E.; Cox, M.M.; Robinson, A.; Dixon, N.E. Single-molecule visualization of fast polymerase turnover in the bacterial replisome. eLife 2017, 6, 23932. [Google Scholar] [CrossRef] [PubMed]
- Duderstadt, K.E.; Geertsema, H.J.; Stratmann, S.A.; Punter, C.M.; Kulczyk, A.W.; Richardson, C.C.; van Oijen, A.M. Simultaneous real-time imaging of leading and lagging strand synthesis reveals the coordination dynamics of single replisomes. Mol. Cell. 2016, 64, 1035–1047. [Google Scholar] [CrossRef]
- Derbyshire, V.; Freemont, P.S.; Sanderson, M.R.; Beese, L.; Friedman, J.M.; Joyce, C.M.; Steitz, T.A. Genetic and crystallographic studies of the 3′,5′-exonucleolytic site of DNA polymerase I. Science 1988, 240, 199–201. [Google Scholar] [CrossRef]
- Bebenek, K.; Joyce, C.M.; Fitzgerald, M.P.; Kunkel, T.A. The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J. Biol. Chem. 1990, 265, 13878–13887. [Google Scholar] [CrossRef]
- Takahashi, S.; Kawasaki, S.; Miyata, H.; Kurita, H.; Mizuno, T.; Matsuura, S.-I.; Mizuno, A.; Oshige, M.; Katsura, S. A new direct single-molecule observation method for DNA synthesis reaction using fluorescent replication protein A. Sensors 2014, 14, 5174–5182. [Google Scholar] [CrossRef]
- Gemmen, G.J.; Millin, R.; Smith, D.E. Tension-dependent DNA cleavage by restriction endonucleases: Two-site enzymes are “switched off” at low force. Proc. Natl. Acad. Sci. USA 2006, 103, 11555–11560. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Ganji, M.; Kim, E.; van der Torre, J.; Abbondanzieri, E.; Dekker, C. DNA sequence encodes the position of DNA supercoils. eLife 2018, 7, e36557. [Google Scholar] [CrossRef] [PubMed]
- Jeppsson, K.; Kanno, T.; Shirahige, K.; Sjögren, C. The maintenance of chromosome structure: Positioning and functioning of SMC complexes. Nat. Rev. Mol. Cell Biol. 2014, 15, 601–614. [Google Scholar] [CrossRef]
- Datta, S.; Lecomte, L.; Haering, C.H. Structural insights into DNA loop extrusion by SMC protein complexes. Curr. Opin. Struct. Biol. 2020, 65, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Ganji, M.; Shaltiel, I.A.; Bisht, S.; Kim, E.; Kalichava, A.; Haering, C.H.; Dekker, C. Real-time imaging of DNA loop extrusion by condensin. Science 2018, 360, 102–105. [Google Scholar] [CrossRef]
- Hong, F.; Zhang, F.; Liu, Y.; Yan, H. DNA origami: Scaffolds for creating higher order structures. Chem. Rev. 2017, 117, 12584–12640. [Google Scholar] [CrossRef]
- Ando, T. High-speed AFM imaging. Curr. Opin. Struct. Biol. 2014, 28, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Ando, T. High-speed atomic force microscopy and its future prospects. Biophys. Rev. 2018, 10, 285–292. [Google Scholar] [CrossRef]
- Ando, T.; Uchihashi, T.; Kodera, N. High-speed AFM and applications to biomolecular systems. Annu. Rev. Biophys. 2013, 42, 393–414. [Google Scholar] [CrossRef]
- Katan, A.J.; Dekker, C. High-speed AFM reveals the dynamics of single biomolecules at the nanometer scale. Cell 2011, 147, 979–982. [Google Scholar] [CrossRef]
- Casuso, I.; Redondo-Morata, L.; Rico, F. Biological physics by high-speed atomic force microscopy. Philos. Trans. R. Soc. A Math Phys. Eng. Sci. 2020, 378, 20190604. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Shen, B.; Kostiainen, M.A.; Grundmeier, G.; Keller, A.; Linko, V. Real-Time Observation of Superstructure-Dependent DNA Origami Digestion by DNase I Using High-Speed Atomic Force Microscopy. ChemBioChem 2019, 20, 2818–2823. [Google Scholar] [CrossRef] [PubMed]
- Sannohe, Y.; Endo, M.; Katsuda, Y.; Hidaka, K.; Sugiyama, H. Visualization of dynamic conformational switching of the G-quadruplex in a DNA nanostructure. J. Am. Chem. Soc. 2010, 132, 16311–16313. [Google Scholar] [CrossRef]
- Shashkova, S.; Leake, M.C. Single-molecule fluorescence microscopy review: Shedding new light on old problems. Biosci. Rep. 2017, 37, BSR20170031. [Google Scholar] [CrossRef]
- Huang, B.; Bates, M.; Zhuang, X. Super-resolution fluorescence microscopy. Annu. Rev. Biochem. 2009, 78, 993–1016. [Google Scholar] [CrossRef]
- Li, H.; Vaughan, J.C. Switchable fluorophores for single-molecule localization microscopy. Chem. Rev. 2018, 118, 9412–9454. [Google Scholar] [CrossRef]
- Wu, Y.L.; Tschanz, A.; Krupnik, L.; Ries, J. Quantitative Data Analysis in Single-Molecule Localization Microscopy. Trends Cell Biol. 2020, 30, 837–885. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Clowsley, A.H.; Lutz, T.; Baddeley, D.; Soeller, C. 3D super-resolution microscopy performance and quantitative analysis assessment using DNA-PAINT and DNA origami test samples. Methods 2020, 174, 56–71. [Google Scholar] [CrossRef] [PubMed]
- Kuzyk, A.; Jungmann, R.; Acuna, G.P.; Liu, N. DNA origami route for nanophotonics. ACS Photonics 2018, 5, 1151–1163. [Google Scholar] [CrossRef]
- Nieves, D.J.; Gaus, K.; Baker, M.A. DNA-based super-resolution microscopy: DNA-PAINT. Genes 2018, 9, 621. [Google Scholar] [CrossRef]
- Schnitzbauer, J.; Strauss, M.T.; Schlichthaerle, T.; Schueder, F.; Jungmann, R. Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 2017, 12, 1198–1228. [Google Scholar] [CrossRef] [PubMed]
- Filius, M.; Cui, T.J.; Ananth, A.N.; Docter, M.W.; Hegge, J.W.; Van Der Oost, J.; Joo, C. High-Speed Super-Resolution Imaging Using Protein-Assisted DNA-PAINT. Nano Lett. 2020, 20, 2264–2270. [Google Scholar] [CrossRef] [PubMed]
- Strauss, S.; Jungmann, R. Up to 100-fold speed-up and multiplexing in optimized DNA-PAINT. Nat. Methods 2020, 17, 789–791. [Google Scholar] [CrossRef] [PubMed]
- Wickham, S.F.; Auer, A.; Min, J.; Ponnuswamy, N.; Woehrstein, J.B.; Schueder, F.; Strauss, M.T.; Schnitzbauer, J.; Nathwani, B.; Zhao, Z.; et al. Complex multicomponent patterns rendered on a 3D DNA-barrel pegboard. Nat. Commun. 2020, 11, 5768. [Google Scholar] [CrossRef]
- Scheckenbach, M.; Bauer, J.; Zähringer, J.; Selbach, F.; Tinnefeld, P. DNA origami nanorulers and emerging reference structures. APL Mater. 2020, 8, 110902. [Google Scholar] [CrossRef]
- Scheckenbach, J.M.; Tinnefeld, P.; Glembockyte, V.; Schubert, T.; Forthmann, C. Self-Regeneration and Self-Healing in DNA Origami Nanostructures. Angew. Chem. Int. Ed. 2020. [Google Scholar] [CrossRef]
- Bramhill, D.; Kornberg, A. A model for initiation at origins of DNA replication. Cell 1988, 54, 915–918. [Google Scholar] [CrossRef]
- Crooke, E.; Hwang, D.S.; Skarstad, K.; Thöny, B.; Kornberg, A. E. coli minichromosome replication: Regulation of initiation at oriC. Res. Microbiol. 1991, 142, 127–130. [Google Scholar] [CrossRef]
- Masai, H.; Matsumoto, S.; You, Z.; Yoshizawa-Sugata, N.; Oda, M. Eukaryotic chromosome DNA replication: Where, when, and how? Annu. Rev. Biochem. 2010, 79, 89–130. [Google Scholar] [CrossRef]
- Méchali, M. Eukaryotic DNA replication origins: Many choices for appropriate answers. Nat. Rev. Mol. Cell Biol. 2010, 11, 728–738. [Google Scholar] [CrossRef]
- Fuller, R.S.; Kornberg, A. Purified dnaA protein in initiation of replication at the Escherichia coli chromosomal origin of replication. Proc. Natl. Acad. Sci. USA 1983, 80, 5817–5821. [Google Scholar] [CrossRef]
- Funnell, B.E.; Baker, T.A.; Kornberg, A. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J. Biol. Chem. 1987, 262, 10327–10334. [Google Scholar] [CrossRef]
- Asai, T.; Chen, C.P.; Nagata, T.; Takanami, M.; Imai, M. Transcription in vivo within the replication origin of the Escherichia coli chromosome: A mechanism for activating initiation of replication. Mol. Gen. Genet. 1992, 231, 169–178. [Google Scholar] [CrossRef]
- Corless, S.; Gilbert, N. Effects of DNA supercoiling on chromatin architecture. Biophys. Rev. 2016, 8, 245–258. [Google Scholar] [CrossRef]
- Rampakakis, E.; Gkogkas, C.; Di Paola, D.; ZannisHadjopoulos, M. Replication initiation and DNA topology: The twisted life of the origin. J. Cell. Biochem. 2010, 110, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Roca, J. The torsional state of DNA within the chromosome. Chromosoma 2011, 120, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Fanning, E.; Zhao, K. SV40 DNA replication: From the A gene to a nanomachine. Virology 2009, 384, 352–359. [Google Scholar] [CrossRef]
- Yardimci, H.; Wang, X.; Loveland, A.B.; Tappin, I.; Rudner, D.Z.; Hurwitz, J.; van Oijen, A.M.; Walter, J.C. Bypass of a protein barrier by a replicative DNA helicase. Nature 2012, 492, 205–209. [Google Scholar] [CrossRef]
- Douglas, M.E.; Ali, F.A.; Costa, A.; Diffley, J.F.X. The mechanism of eukaryotic CMG helicase activation. Nature 2018, 555, 265–268. [Google Scholar] [CrossRef]
- Kose, H.B.; Larsen, N.B.; Duxin, J.P.; Yardimci, H. Dynamics of the eukaryotic replicative helicase at lagging-strand protein barriers support the steric exclusion model. Cell Rep. 2019, 26, 2113–2125. [Google Scholar] [CrossRef]
- Burnham, D.R.; Kose, H.B.; Hoyle, R.B.; Yardimci, H. The mechanism of DNA unwinding by the eukaryotic replicative helicase. Nat. Commun. 2019, 10, 2159. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Motooka, S.; Kawasaki, S.; Kurita, H.; Mizuno, T.; Matsuura, S.-I.; Hanaoka, F.; Mizuno, A.; Oshige, M.; Katsura, S. Direct single-molecule observations of DNA unwinding by SV40 large tumor antigen under a negative DNA supercoil state. J. Biomol. Struct. Dyn. 2018, 36, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Holzmeister, P.; Acuna, G.P.; Grohmann, D.; Tinnefeld, P. Breaking the concentration limit of optical single-molecule detection. Chem. Soc. Rev. 2014, 43, 1014–1028. [Google Scholar] [CrossRef]
- Crouch, G.M.; Han, D.; Bohn, P.W. Zero-mode waveguide nanophotonic structures for single molecule characterization. J. Phys. D Appl. Phys. 2018, 51, 193001. [Google Scholar] [CrossRef]
- Klimov, V.V. Smart Design of Zero-Mode Waveguide Nanodevices for Effective Detection of Single-Molecule Fluorescence. Phys. Rev. Appl. 2019, 12, 014049. [Google Scholar] [CrossRef]
- Zhu, P.; Craighead, H.G. Zero-Mode Waveguides for Single-Molecule Analysis. Annu. Rev. Biophys. 2012, 41, 269–293. [Google Scholar] [CrossRef] [PubMed]
- Levene, M.J.; Korlach, J.; Turner, S.W.; Foquet, M.; Craighead, H.G.; Webb, W.W. Zero-mode waveguides for single-molecule analysis at high concentrations. Science 2003, 299, 682–686. [Google Scholar] [CrossRef]
- Jiao, X.; Peterson, E.M.; Harris, J.M.; Blair, S. UV Fluorescence Lifetime Modification by Aluminum Nanoapertures. ACS Photonics 2014, 1, 1270–1277. [Google Scholar] [CrossRef]
- Pibiri, E.; Holzmeister, P.; Lalkens, B.; Acuna, G.P.; Tinnefeld, P. Single-Molecule Positioning in Zeromode Waveguides by DNA Origami Nanoadapters. Nano Lett. 2014, 14, 3499–3503. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.S.; Karim, F.; Zhao, C. Single-molecule detection at high concentrations with optical aperture nanoantennas. Nanoscale 2016, 8, 9480–9487. [Google Scholar] [CrossRef]
- Eid, J.; Fehr, A.; Gray, J.; Luong, K.; Lyle, J.; Otto, G.; Peluso, P.; Rank, D.; Baybayan, P.; Bettman, B.; et al. Real-Time DNA Sequencing from Single Polymerase Molecules. Science 2009, 323, 133–138. [Google Scholar] [CrossRef]
- Larkin, J.; Henley, R.Y.; Jadhav, V.; Korlach, J.; Wanunu, M. Length-independent DNA packing into nanopore zero-mode waveguides for low-input DNA sequencing. Nat. Nanotechnol. 2017, 12, 1169–1175. [Google Scholar] [CrossRef]
- Wenger, J.; Gerard, D.; Aouani, H.; Rigneault, H.; Lowder, B.; Blair, S.; Devaux, E.; Ebbesen, T.W. Nanoaperture-Enhanced Signal-to-Noise Ratio in Fluorescence Correlation Spectroscopy. Angew. Chem. Int. Ed. 2009, 81, 834–839. [Google Scholar] [CrossRef] [PubMed]
- Goldschen-Ohm, M.P.; White, D.S.; Klenchin, V.A.; Chanda, B.; Goldsmith, R.H. Observing Single-Molecule Dynamics at Millimolar Concentrations. Angew. Chem. Int. Ed. 2017, 129, 2439–2442. [Google Scholar] [CrossRef]
- Sandén, T.; Wyss, R.; Santschi, C.; Hassaïne, G.; Deluz, C.; Martin, O.J.; Wennmalm, S.; Vogel, H. A Zeptoliter Volume Meter for Analysis of Single Protein Molecules. Nano Lett. 2012, 12, 370–375. [Google Scholar]
- Chen, J.; Dalal, R.V.; Petrov, A.N.; Tsai, A.; O’Leary, S.E.; Chapin, K.; Cheng, J.; Ewan, M.; Hsiung, P.L.; Lundquist, P.; et al. High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence. Proc. Natl. Acad. Sci. USA 2014, 111, 664–669. [Google Scholar] [CrossRef]
- Jadhav, V.; Hoogerheide, D.P.; Korlach, J.; Wanunu, M. Porous Zero-Mode Waveguides for Picogram-Level DNA Capture. Nano Lett. 2019, 19, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Verschueren, D.V.; Pud, S.; Shi, X.; De Angelis, L.; Kuipers, L.; Dekker, C. Label-Free Optical Detection of DNA Translocations through Plasmonic Nanopores. ACS Nano 2019, 13, 61–70. [Google Scholar] [CrossRef]
- Baibakov, M.; Patra, S.; Claude, J.B.; Moreau, A.; Lumeau, J.; Wenger, J. Extending Single-Molecule Förster Resonance Energy Transfer (FRET) Range beyond 10 Nanometers in Zero-Mode Waveguides. ACS Nano 2019, 13, 8469–8480. [Google Scholar] [CrossRef]
- Zambrana-Puyalto, X.; Ponzellini, P.; Maccaferri, N.; Garoli, D. Förster-Resonance Energy Transfer between Diffusing Molecules and a Functionalized Plasmonic Nanopore. Phys. Rev. Appl. 2020, 14, 054065. [Google Scholar] [CrossRef]
- Baibakov, M.; Patra, S.; Claude, J.B.; Wenger, J. Long-Range Single-Molecule Förster Resonance Energy Transfer between Alexa Dyes in Zero-Mode Waveguides. ACS Omega 2020, 5, 6947–6955. [Google Scholar] [CrossRef] [PubMed]
- Lou, F.; Song, N.; Han, Z.; Gao, T. Single-molecule real-time (SMRT) sequencing facilitates Tachypleus tridentatus genome annotation. Int. J. Biol. Macromol. 2020, 147, 89–97. [Google Scholar] [CrossRef]
- Ardui, S.; Ameur, A.; Vermeesch, J.R.; Hestand, M.S. Single molecule real-time (SMRT) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 2018, 46, 2159–2168. [Google Scholar] [CrossRef]
- Rhoads, A.; Au, K.F. PacBio sequencing and its applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.J.; Carneiro, M.O.; Schatz, M.C. The advantages of SMRT sequencing. Genome Biol. 2013, 14, 405. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, S.; Oshige, M.; Katsura, S. DNA Manipulation and Single-Molecule Imaging. Molecules 2021, 26, 1050. https://doi.org/10.3390/molecules26041050
Takahashi S, Oshige M, Katsura S. DNA Manipulation and Single-Molecule Imaging. Molecules. 2021; 26(4):1050. https://doi.org/10.3390/molecules26041050
Chicago/Turabian StyleTakahashi, Shunsuke, Masahiko Oshige, and Shinji Katsura. 2021. "DNA Manipulation and Single-Molecule Imaging" Molecules 26, no. 4: 1050. https://doi.org/10.3390/molecules26041050
APA StyleTakahashi, S., Oshige, M., & Katsura, S. (2021). DNA Manipulation and Single-Molecule Imaging. Molecules, 26(4), 1050. https://doi.org/10.3390/molecules26041050





