Polyphyllin D Shows Anticancer Effect through a Selective Inhibition of Src Homology Region 2-Containing Protein Tyrosine Phosphatase-2 (SHP2)
Abstract
1. Introduction
2. Results and Discussions
2.1. Expression, Purification, and Kinetic Characterization of SHP2
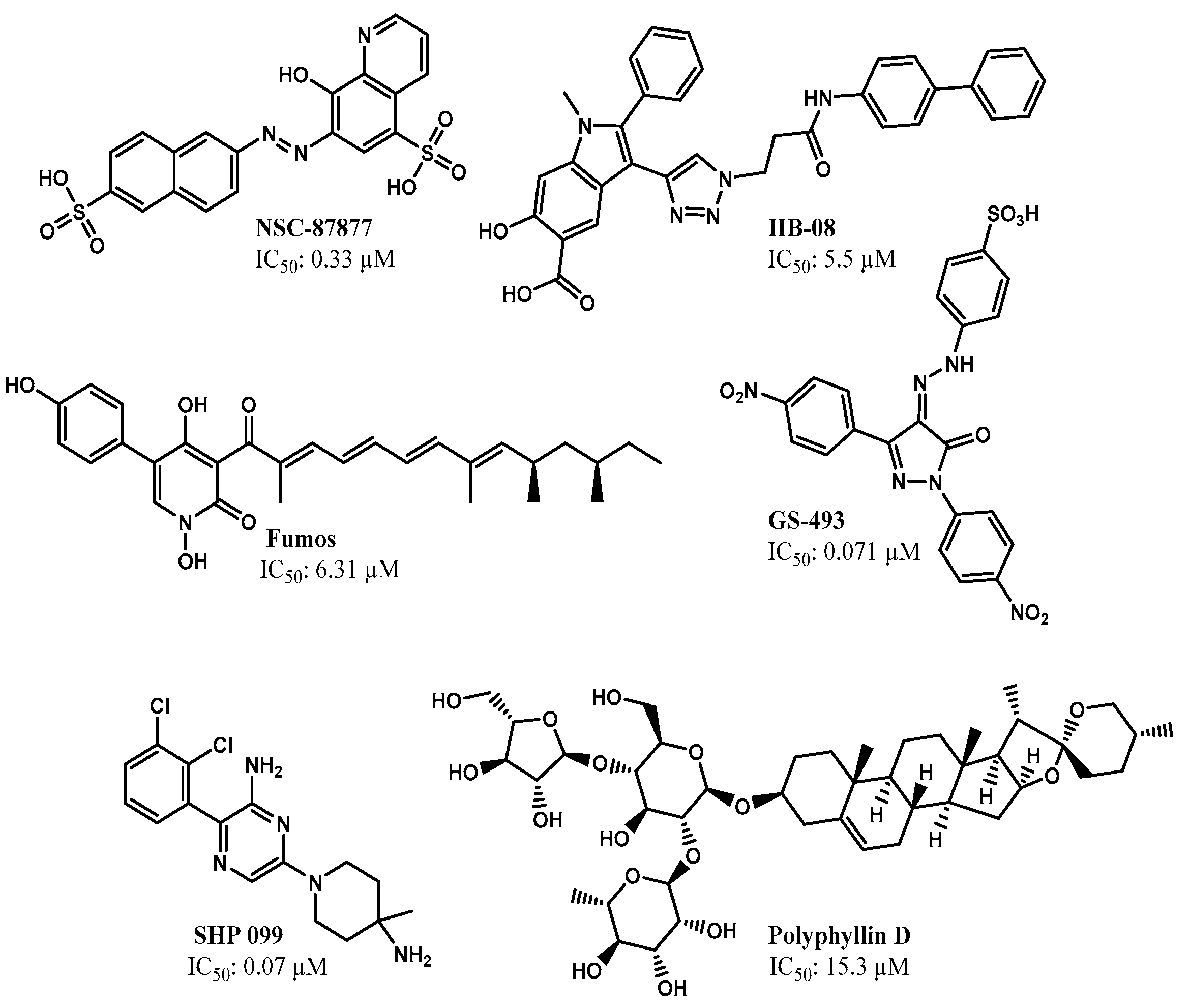
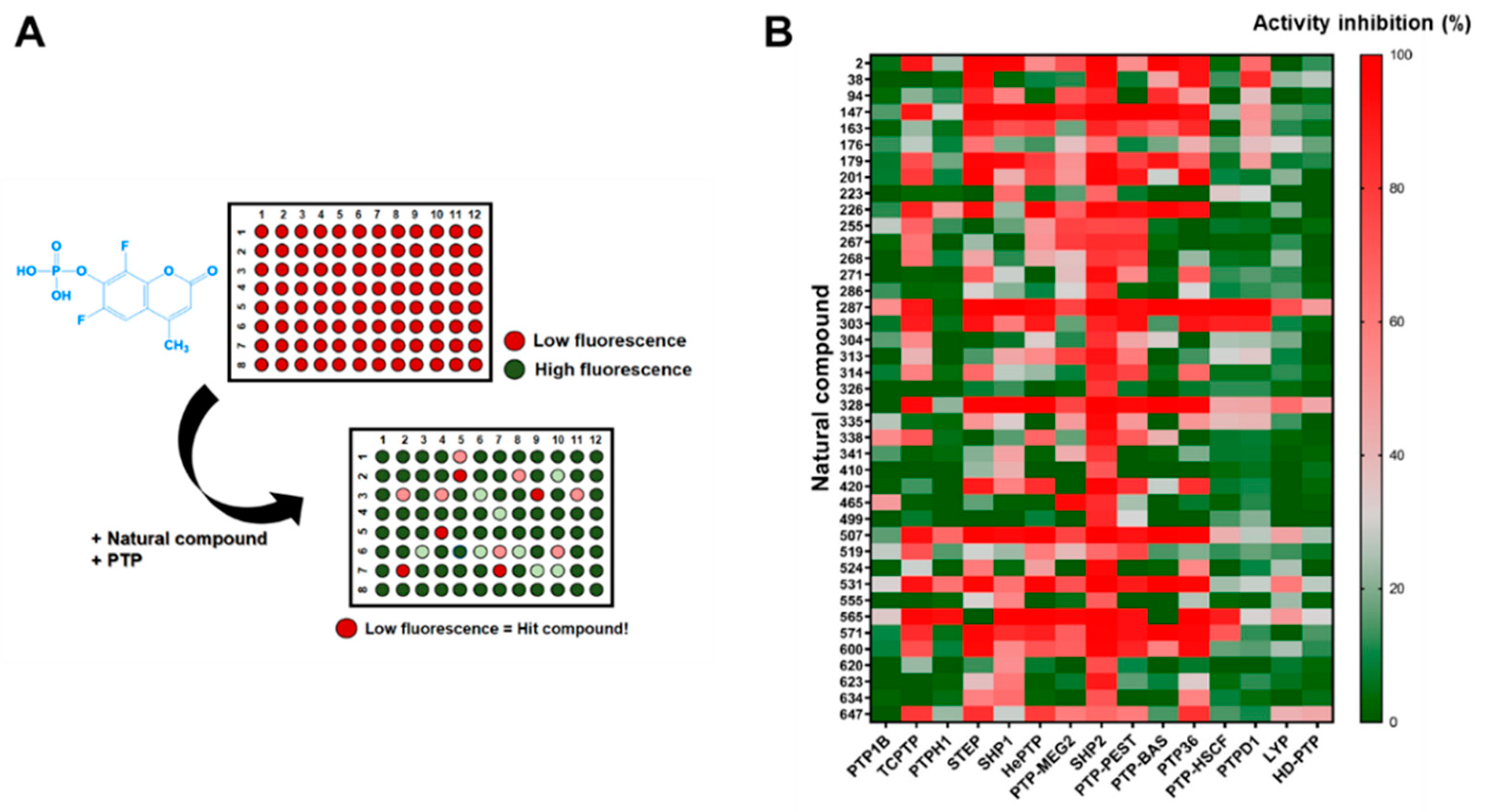
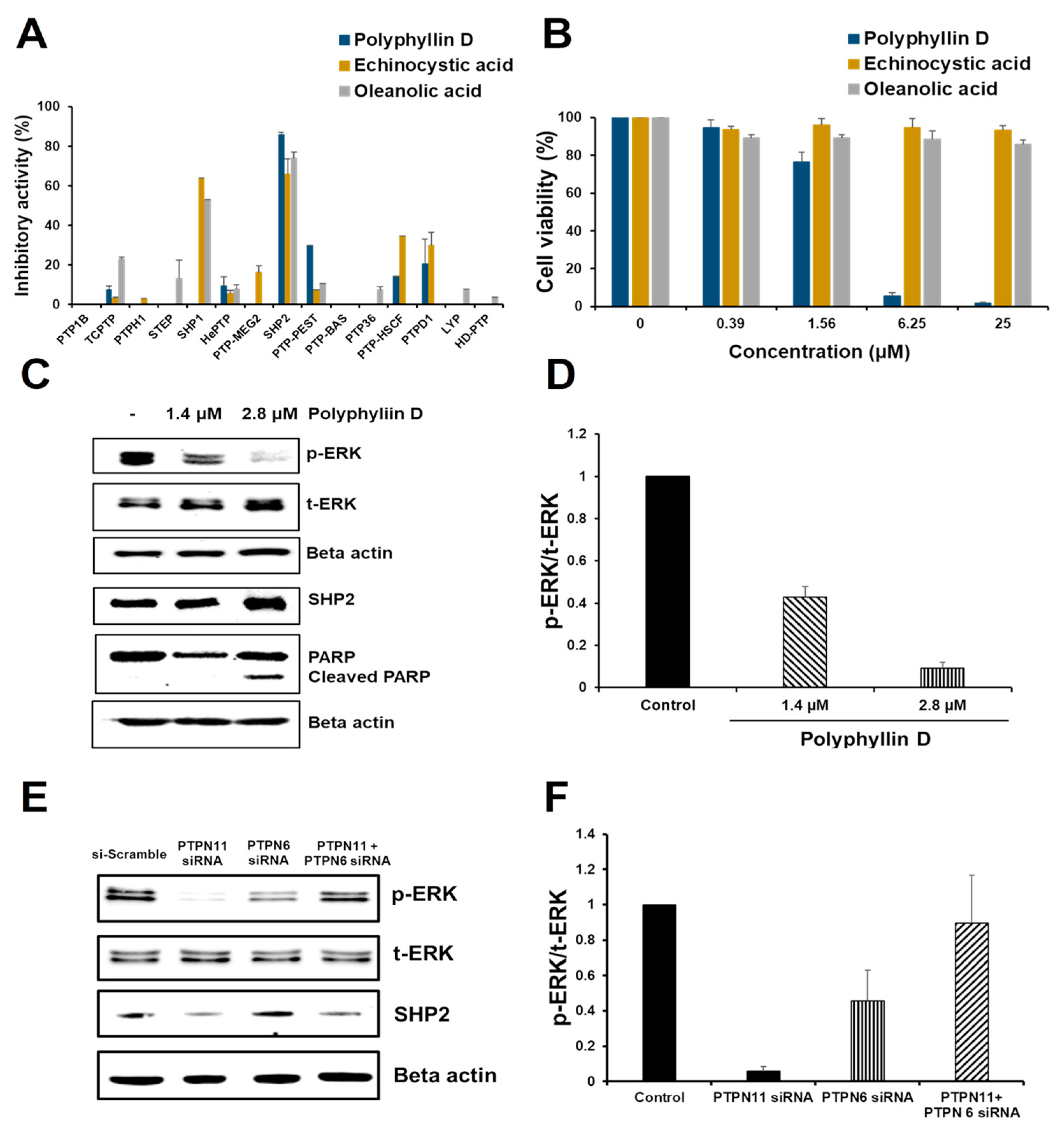
2.2. Identification of SHP2 Inhibitors from Natural Products
2.3. Effect of Polyphyllin D for the Proliferation and Signaling of Jurkat Cells
2.4. siRNA Knockdown Study of PTPN6 and PTPN11 in Jurkat Cells
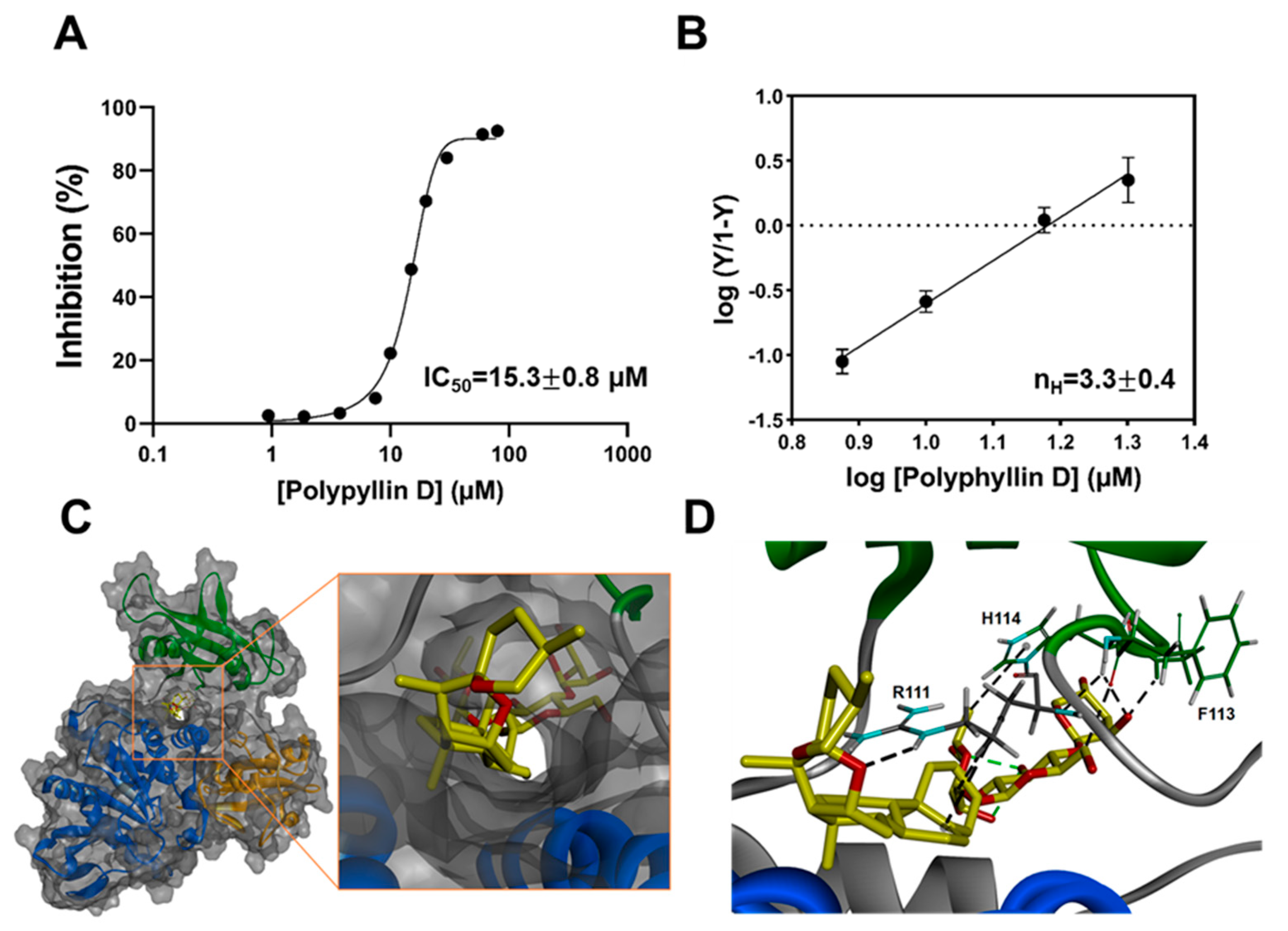
2.5. Kinetic Study of Polyphyllin D for SHP2 Inhibition
3. Material and Methods
3.1. Materials
3.2. Construction of SHP2 Overexpression Vectors
3.3. Expression and Purification of Recombinant SHP2
3.4. Kinetic Characterization of SHP2 Enzyme Activity
3.5. Screening of Natural Compound Library for SHP2 Inhibition
3.6. Assessment of Antitumor Activity
3.7. Western Blot Analysis
3.8. RNA Interference and Quantitative Real Time-Polymerase Chain Reaction (qRT-PCR)
3.9. Docking Study of Polyphyllin D into SHP2
3.10. Statistical Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Neel, B.G.; Tonks, N.K. Protein tyrosine phosphatases in signal transduction. Curr. Opin. Cell Biol. 1997, 9, 193–204. [Google Scholar] [CrossRef]
- Wee, P.; Wang, Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef]
- Normanno, N.; De Luca, A.; Bianco, C.; Strizzi, L.; Mancino, M.; Maiello, M.R.; Carotenuto, A.; De Feo, G.; Caponigro, F.; Salomon, D.S. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006, 366, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, M.; Martinelli, S.; Cazzaniga, G.; Cordeddu, V.; Iavarone, I.; Spinelli, M.; Palmi, C.; Carta, C.; Pession, A.; Aricò, M.; et al. Genetic evidence for lineage-related and differentiation stage–related contribution of somatic PTPN11 mutations to leukemogenesis in childhood acute leukemia. Blood 2004, 104, 307–313. [Google Scholar] [CrossRef]
- Cai, T.; Nishida, K.; Hirano, T.; Khavari, P.A. Gab1 and SHP-2 promote Ras/MAPK regulation of epidermal growth and differentiation. J. Cell Biol. 2002, 159, 103–112. [Google Scholar] [CrossRef]
- Hof, P.; Pluskey, S.; Dhe-Paganon, S.; Eck, M.J.; Shoelson, S.E. Crystal structure of the tyrosine phosphatase SHP-2. Cell 1998, 92, 441–450. [Google Scholar] [CrossRef]
- Huang, W.Q.; Lin, Q.; Zhuang, X.; Cai, L.L.; Ruan, R.S.; Lu, Z.X.; Tzeng, C.M. Structure, function, and pathogenesis of SHP2 in developmental disorders and tumorigenesis. Curr. Cancer Drug Targets 2014, 14, 567–588. [Google Scholar] [CrossRef]
- Shi, Z.Q.; Yu, D.H.; Park, M.; Marshall, M.; Feng, G.S. Molecular mechanism for the Shp-2 tyrosine phosphatase function in promoting growth factor stimulation of Erk activity. Mol. Cell. Biol. 2000, 20, 1526–1536. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Martin, E.; Matalkah, F.; Shah, N.; Ivanov, A.; Ruppert, J.M.; Lockman, P.R.; Agazie, Y.M. Conditional knockout of SHP2 in ErbB2 transgenic mice or inhibition in HER2-amplified breast cancer cell lines blocks oncogene expression and tumorigenesis. Oncogene 2019, 38, 2275–2290. [Google Scholar] [CrossRef]
- Chen, X.; Fu, X.; Zhao, W.; Ho, W.-T.T.; Xing, S.; Zhao, Z.J. Loss of tyrosine phosphatase SHP2 activity promotes growth of colorectal carcinoma HCT-116 cells. Signal Transduct. Target. Ther. 2020, 5, 83. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Bu, H.; Zhou, J.; Yang, C.-Y.; Zhang, H. Recent advances of SHP2 inhibitors in cancer therapy: Current development and clinical application. J. Med. Chem. 2020, 63, 11368–11396. [Google Scholar] [CrossRef]
- Chen, Y.N.; LaMarche, M.J.; Chan, H.M.; Fekkes, P.; Garcia-Fortanet, J.; Acker, M.G.; Antonakos, B.; Chen, C.H.; Chen, Z.; Cooke, V.G.; et al. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature 2016, 535, 148–152. [Google Scholar] [CrossRef]
- Chen, L.; Sung, S.S.; Yip, M.L.; Lawrence, H.R.; Ren, Y.; Guida, W.C.; Sebti, S.M.; Lawrence, N.J.; Wu, J. Discovery of a novel shp2 protein tyrosine phosphatase inhibitor. Mol. Pharmacol. 2006, 70, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Cao, M.; Zhu, S.; Wang, C.; Liang, F.; Yan, L.; Luo, D. Discovery of a Novel Inhibitor of the Protein Tyrosine Phosphatase Shp2. Sci. Rep. 2015, 5, 17626. [Google Scholar] [CrossRef] [PubMed]
- Grosskopf, S.; Eckert, C.; Arkona, C.; Radetzki, S.; Böhm, K.; Heinemann, U.; Wolber, G.; von Kries, J.-P.; Birchmeier, W.; Rademann, J. Selective Inhibitors of the Protein Tyrosine Phosphatase SHP2 Block Cellular Motility and Growth of Cancer Cells in vitro and in vivo. ChemMedChem 2015, 10, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; He, Y.; Liu, S.; Yu, Z.; Jiang, Z.X.; Yang, Z.; Dong, Y.; Nabinger, S.C.; Wu, L.; Gunawan, A.M.; et al. Salicylic acid based small molecule inhibitor for the oncogenic Src homology-2 domain containing protein tyrosine phosphatase-2 (SHP2). J. Med. Chem. 2010, 53, 2482–2493. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Zeng, L.-F.; He, Y.; Zhang, S.; Zhang, Z.-Y. Small molecule tools for functional interrogation of protein tyrosine phosphatases. FEBS J. 2013, 280, 731–750. [Google Scholar] [CrossRef]
- Hellmuth, K.; Grosskopf, S.; Lum, C.T.; Würtele, M.; Röder, N.; von Kries, J.P.; Rosario, M.; Rademann, J.; Birchmeier, W. Specific inhibitors of the protein tyrosine phosphatase Shp2 identified by high-throughput docking. Proc. Natl. Acad. Sci. USA 2008, 105, 7275–7280. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.N.; Mortensen, O.H.; Peters, G.H.; Drake, P.G.; Iversen, L.F.; Olsen, O.H.; Jansen, P.G.; Andersen, H.S.; Tonks, N.K.; Møller, N.P. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol. Cell. Biol. 2001, 21, 7117–7136. [Google Scholar] [CrossRef] [PubMed]
- Varone, A.; Spano, D.; Corda, D. Shp1 in Solid Cancers and Their Therapy. Front. Oncol. 2020, 10, 935. [Google Scholar] [CrossRef] [PubMed]
- Fodor, M.; Price, E.; Wang, P.; Lu, H.; Argintaru, A.; Chen, Z.; Glick, M.; Hao, H.-X.; Kato, M.; Koenig, R.; et al. Dual Allosteric Inhibition of SHP2 Phosphatase. ACS Chem. Biol. 2018, 13, 647–656. [Google Scholar] [CrossRef]
- Yoon, S.-Y.; Kang, H.J.; Ahn, D.; Hwang, J.Y.; Kwon, S.J.; Chung, S.J. Identification of chebulinic acid as a dual targeting inhibitor of protein tyrosine phosphatases relevant to insulin resistance. Bioorg. Chem. 2019, 90, 103087. [Google Scholar] [CrossRef]
- Yoon, S.-Y.; Lee, J.H.; Kwon, S.J.; Kang, H.J.; Chung, S.J. Ginkgolic acid as a dual-targeting inhibitor for protein tyrosine phosphatases relevant to insulin resistance. Bioorg. Chem. 2018, 81, 264–269. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, W.; Lee, Y.G.; Kang, H.J.; Lee, S.-H.; Park, S.Y.; Min, J.-K.; Lee, S.-R.; Chung, S.J. Identification of sennoside A as a novel inhibitor of the slingshot (SSH) family proteins related to cancer metastasis. Pharmacol. Res. 2017, 119, 422–430. [Google Scholar] [CrossRef]
- Jeong, M.S.; Kim, E.; Kang, H.J.; Choi, E.J.; Cho, A.R.; Chung, S.J.; Park, S.B. A selective Seoul-Fluor-based bioprobe, SfBP, for vaccinia H1-related phosphatase--a dual-specific protein tyrosine phosphatase. Chem. Commun. (Camb.) 2012, 48, 6553–6555. [Google Scholar] [CrossRef]
- Xu, R.; Yu, Y.; Zheng, S.; Zhao, X.; Dong, Q.; He, Z.; Liang, Y.; Lu, Q.; Fang, Y.; Gan, X.; et al. Overexpression of Shp2 tyrosine phosphatase is implicated in leukemogenesis in adult human leukemia. Blood 2005, 106, 3142–3149. [Google Scholar] [CrossRef]
- Morales, L.D.; Casillas Pavón, E.A.; Shin, J.W.; Garcia, A.; Capetillo, M.; Kim, D.J.; Lieman, J.H. Protein Tyrosine Phosphatases PTP-1B, SHP-2, and PTEN Facilitate Rb/E2F-Associated Apoptotic Signaling. PLoS ONE 2014, 9, e97104. [Google Scholar] [CrossRef]
- Lu, H.; Liu, C.; Huynh, H.; Le, T.B.U.; LaMarche, M.J.; Mohseni, M.; Engelman, J.A.; Hammerman, P.S.; Caponigro, G.; Hao, H.-X. Resistance to allosteric SHP2 inhibition in FGFR-driven cancers through rapid feedback activation of FGFR. Oncotarget 2020, 11, 265–281. [Google Scholar] [CrossRef]
- Lorenz, U. SHP-1 and SHP-2 in T cells: Two phosphatases functioning at many levels. Immunol. Rev. 2009, 228, 342–359. [Google Scholar] [CrossRef]
- Lee, M.-S.; Chan, J.Y.-W.; Kong, S.-K.; Yu, B.; Eng-Choon, V.O.; Nai-Ching, H.W.; Mak Chung-Wai, T.; Fung, K.-P. Effects of polyphyllin D, a steroidal saponin in Paris Polyphylla, in growth inhibition of human breast cancer cells and in xenograft. Cancer Biol. Ther. 2005, 4, 1248–1254. [Google Scholar] [CrossRef]
- Cheung, J.Y.; Ong, R.C.; Suen, Y.K.; Ooi, V.; Wong, H.N.; Mak, T.C.; Fung, K.P.; Yu, B.; Kong, S.K. Polyphyllin D is a potent apoptosis inducer in drug-resistant HepG2 cells. Cancer Lett. 2005, 217, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Liu, W.; Chen, Z.; Gu, Y.; Peng, S.; Shen, L.; Shen, Y.; Wang, X.; Feng, G.-S.; Sun, Y.; et al. Tyrosine phosphatase SHP2 negatively regulates NLRP3 inflammasome activation via ANT1-dependent mitochondrial homeostasis. Nat. Commun. 2017, 8, 2168. [Google Scholar] [CrossRef]
- Chang, J.; Li, Y.; Wang, X.; Hu, S.; Wang, H.; Shi, Q.; Wang, Y.; Yang, Y. Polyphyllin I suppresses human osteosarcoma growth by inactivation of Wnt/β-catenin pathway in vitro and in vivo. Sci. Rep. 2017, 7, 7605. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwon, S.J.; Ahn, D.; Yang, H.-M.; Kang, H.J.; Chung, S.J. Polyphyllin D Shows Anticancer Effect through a Selective Inhibition of Src Homology Region 2-Containing Protein Tyrosine Phosphatase-2 (SHP2). Molecules 2021, 26, 848. https://doi.org/10.3390/molecules26040848
Kwon SJ, Ahn D, Yang H-M, Kang HJ, Chung SJ. Polyphyllin D Shows Anticancer Effect through a Selective Inhibition of Src Homology Region 2-Containing Protein Tyrosine Phosphatase-2 (SHP2). Molecules. 2021; 26(4):848. https://doi.org/10.3390/molecules26040848
Chicago/Turabian StyleKwon, Se Jeong, Dohee Ahn, Hyun-Mo Yang, Hyo Jin Kang, and Sang J. Chung. 2021. "Polyphyllin D Shows Anticancer Effect through a Selective Inhibition of Src Homology Region 2-Containing Protein Tyrosine Phosphatase-2 (SHP2)" Molecules 26, no. 4: 848. https://doi.org/10.3390/molecules26040848
APA StyleKwon, S. J., Ahn, D., Yang, H.-M., Kang, H. J., & Chung, S. J. (2021). Polyphyllin D Shows Anticancer Effect through a Selective Inhibition of Src Homology Region 2-Containing Protein Tyrosine Phosphatase-2 (SHP2). Molecules, 26(4), 848. https://doi.org/10.3390/molecules26040848






