Transcriptome Changes Reveal the Molecular Mechanisms of Humic Acid-Induced Salt Stress Tolerance in Arabidopsis
Abstract
1. Introduction
2. Results and Discussion
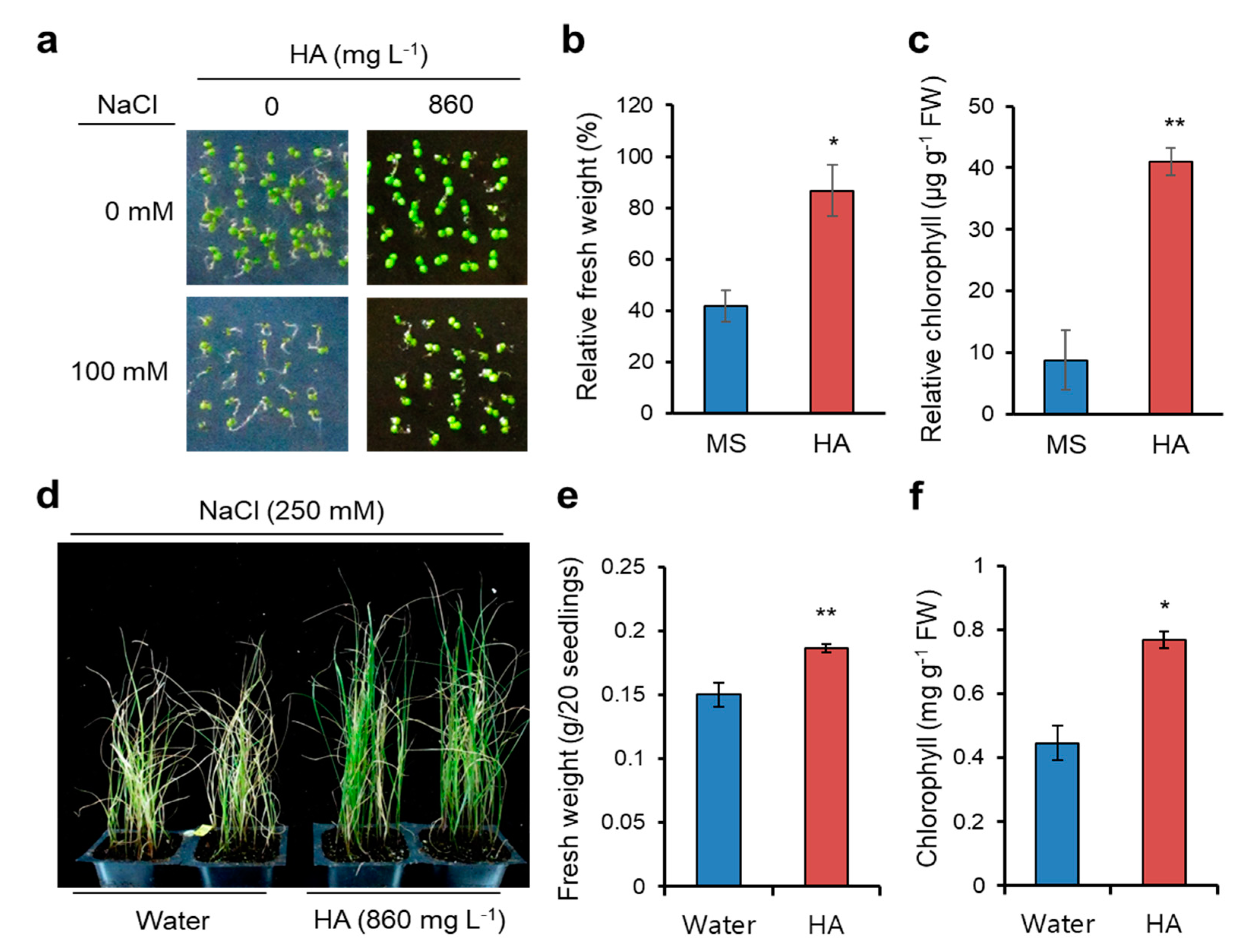
2.1. HA Application Confers Salt Stress Yolerance in Arabidopsis and Italian Ryegrass
2.2. Illumina Sequencing, Mapping Sequence Reads, and Total Differentially Expressed Genes (DEGs)
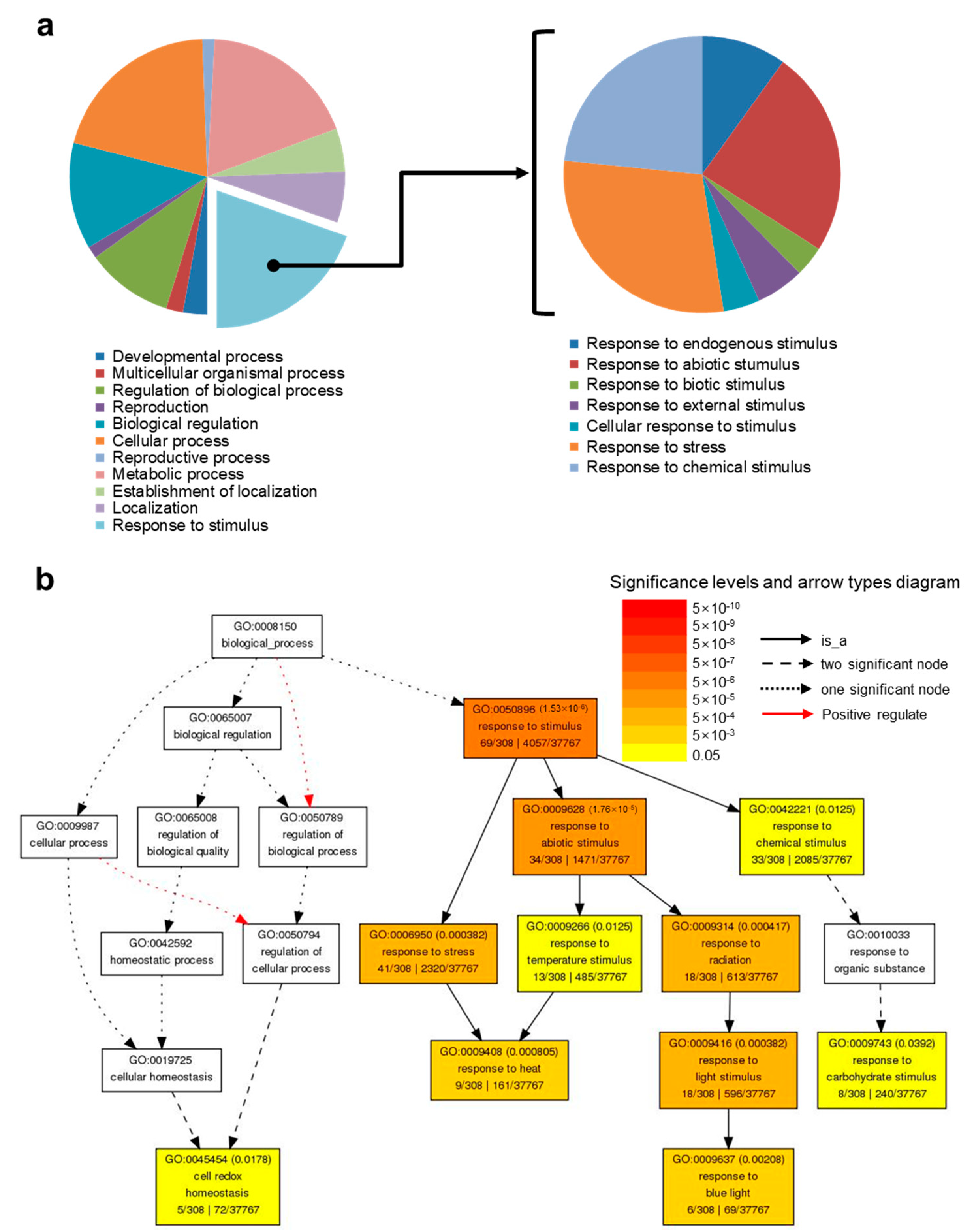
2.3. Up-Regulated Genes by HA under Salt Stress (DEG #3)
2.3.1. Up-regulated Genes Involved in Response to Light Stimulus
2.3.2. Up-Regulated Genes Involved in Response to Heat
2.3.3. Up-Regulated Genes Involved in Response to Cell Redox Homeostasis
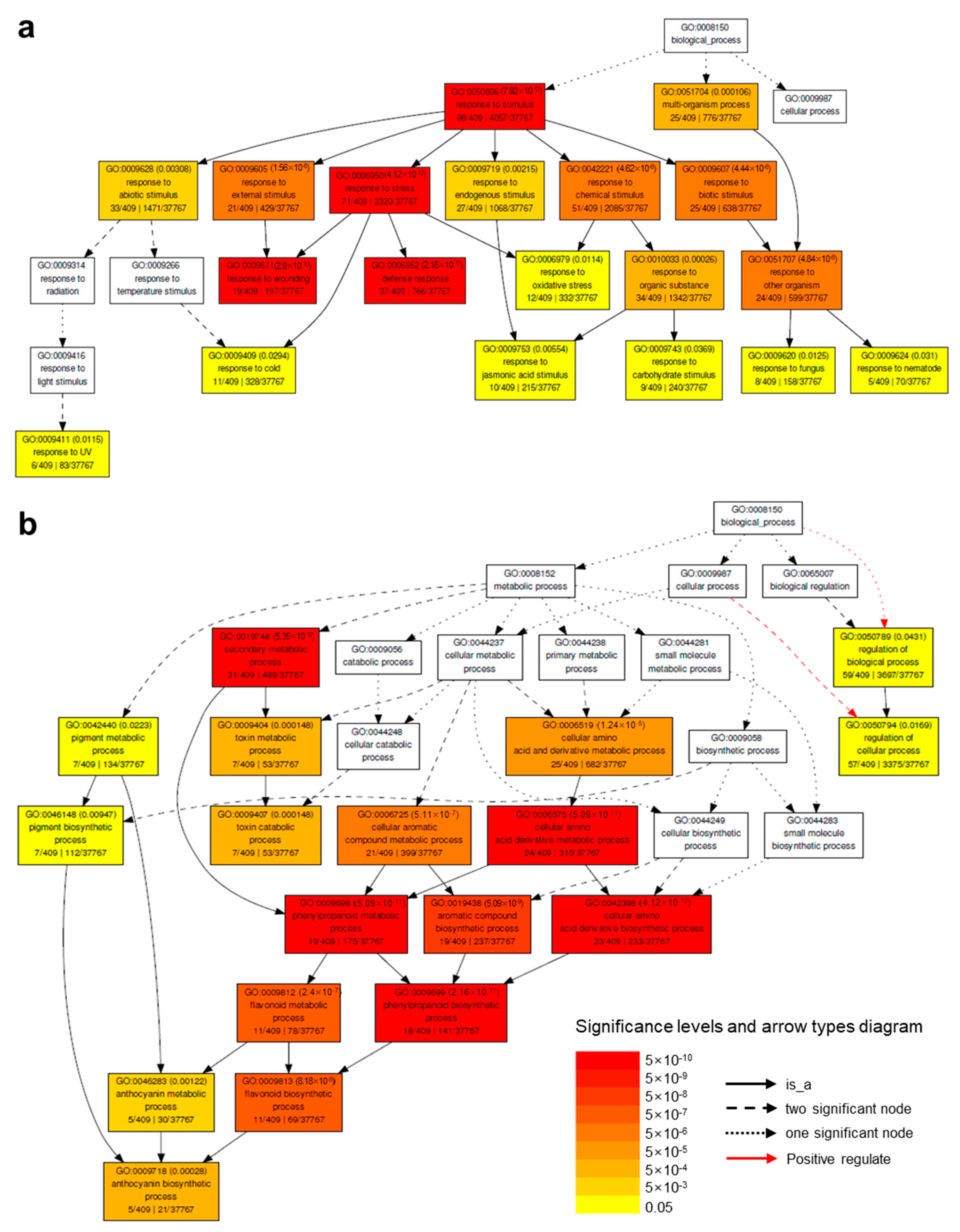
2.4. Down-Regulated Genes by HA under Salt Stress (DEG #3)
2.4.1. Down-Regulated Genes Involved in Toxin Catabolic Process
2.4.2. Down-Regulated Genes Involved in the Metabolic Process
2.5. Validation of DEGs by Quantitative RT-PCR (qRT-PCR)
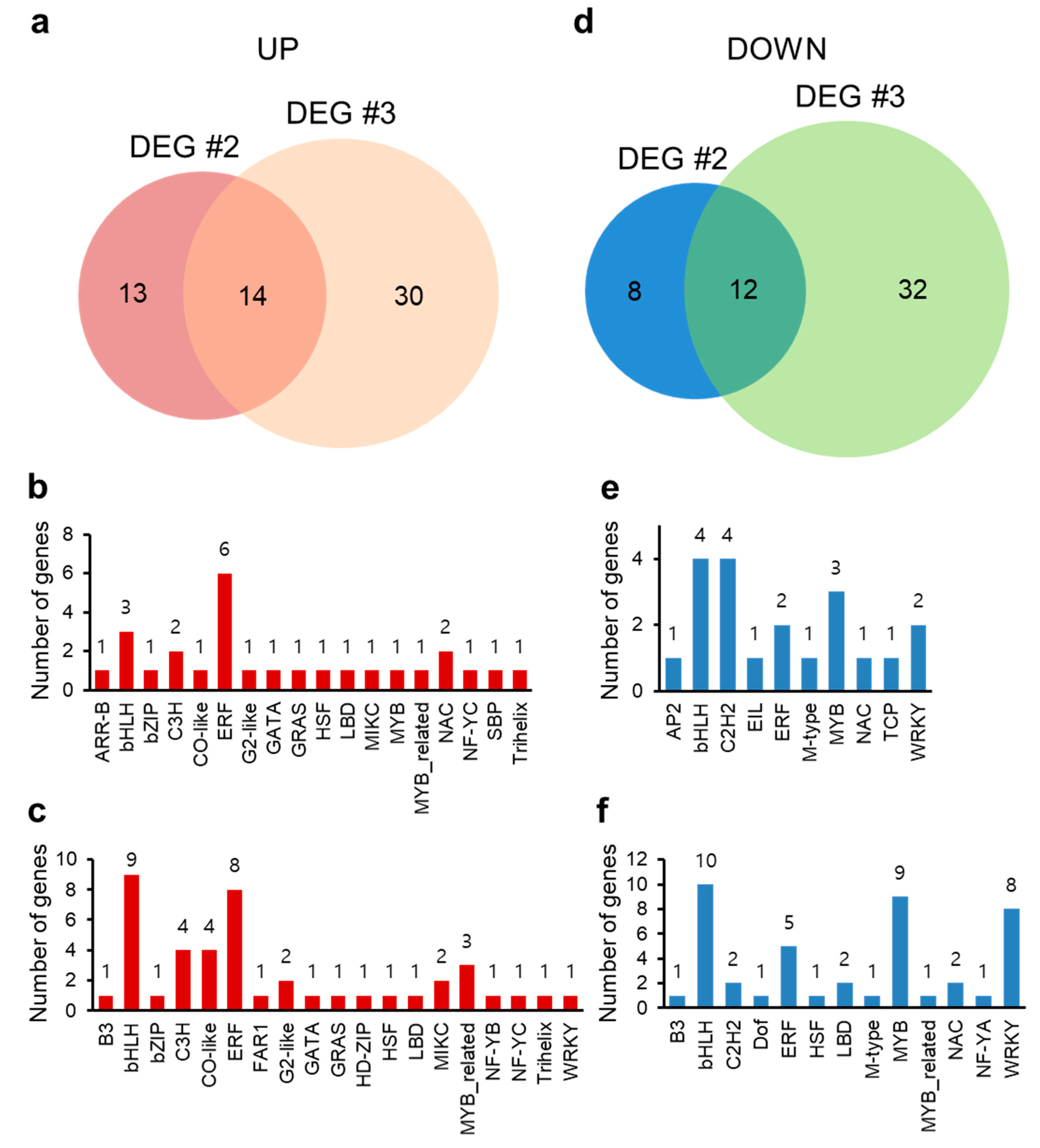
2.6. Transcripts Regulated in Common between DEG #2 (MS vs. HA) and DEG #3 (Salt vs. Salt + HA)
2.7. Transcription Factors Regulated in Common between DEG #2 (MS vs. HA) and DEG #3 (Salt vs. Salt + HA)
3. Materials and Methods
3.1. Plant Materials, Growth Conditions and Treatments
3.2. Chlorophyll Content
3.3. RNA Extraction, Illumina RNA-Seq and Analysis of RNA-Seq Data
3.4. DEG and GO Analysis
3.5. Validation by Quantitative Reverse-Transcription PCR (qRT-PCR)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Orsi, M. Molecular dynamics simulation of humic substances. Chem. Biol. Technol. Agric. 2014, 1, 10. [Google Scholar] [CrossRef]
- Keeler, C.; Maciel, G.E. Quantitation in the Solid-State13C NMR Analysis of Soil and Organic Soil Fractions. Anal. Chem. 2003, 75, 2421–2432. [Google Scholar] [CrossRef] [PubMed]
- Muscolo, A.; Sidari, M.; Nardi, S. Humic substance: Relationship between structure and activity. Deeper information suggests univocal findings. J. Geochem. Explor. 2013, 129, 57–63. [Google Scholar] [CrossRef]
- Nikbakht, A.; Kafi, M.; Babalar, M.; Xia, Y.P.; Luo, A.; Etemadi, N.-A. Effect of Humic Acid on Plant Growth, Nutrient Uptake, and Postharvest Life of Gerbera. J. Plant Nutr. 2008, 31, 2155–2167. [Google Scholar] [CrossRef]
- Trevisan, S.; Francioso, O.; Quaggiotti, S.; Nardi, S. Humic substances biological activity at the plant-soil interface. Plant Signal. Behav. 2010, 5, 635–643. [Google Scholar] [CrossRef]
- Kulikova, N.A.; Abroskin, D.P.; Badun, G.A.; Chernysheva, M.G.; Korobkov, V.I.; Beer, A.S.; Tsvetkova, E.A.; Senik, S.V.; Klein, O.I.; Perminova, I.V. Label Distribution in Tissues of Wheat Seedlings Cultivated with Tritium-Labeled Leonardite Humic Acid. Sci. Rep. 2016, 6, 28869. [Google Scholar] [CrossRef]
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef]
- Cheong, M.S.; Yun, D.-J. Salt-stress signaling. J. Plant Biol. 2007, 50, 148–155. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef]
- Liang, W.; Ma, X.; Wan, P.; Liu, L. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018, 495, 286–291. [Google Scholar] [CrossRef]
- Khaleda, L.; Park, H.J.; Yun, D.-J.; Jeon, J.-R.; Kim, M.G.; Cha, J.-Y.; Kim, W.-Y. Humic Acid Confers HIGH-AFFINITY K+ TRANSPORTER 1-Mediated Salinity Stress Tolerance in Arabidopsis. Mol. Cells 2017, 40, 966–975. [Google Scholar] [PubMed]
- Cha, J.-Y.; Kang, S.-H.; Ali, I.; Lee, S.C.; Ji, M.G.; Jeong, S.Y.; Shin, G.-I.; Kim, M.G.; Jeon, J.-R.; Kim, W.-Y. Humic acid enhances heat stress tolerance via transcriptional activation of Heat-Shock Proteins in Arabidopsis. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.-Y.; Kim, T.-W.; Choi, J.H.; Jang, K.-S.; Khaleda, L.; Kim, W.-Y.; Jeon, J.-R. Fungal Laccase-Catalyzed Oxidation of Naturally Occurring Phenols for Enhanced Germination and Salt Tolerance ofArabidopsis thaliana: A Green Route for Synthesizing Humic-like Fertilizers. J. Agric. Food Chem. 2017, 65, 1167–1177. [Google Scholar] [CrossRef]
- Saleki, R.; Young, P.G.; Lefebvre, D.D. Mutants of Arabidopsis thaliana Capable of Germination under Saline Conditions. Plant Physiol. 1993, 101, 839–845. [Google Scholar] [CrossRef]
- Laila Khaleda; Min Gab Kim; Woe-Yeon Kim; Jong-Rok Jeon; Joon-Yung Cha Humic Acid and Synthesized Humic Mimic Promote the Growth of Italian Ryegrass. J. Korean Soc. Grassl. Forage Sci. 2017, 37, 242–247. [CrossRef]
- Pinton, R.; Cesco, S.; Iacolettig, G.; Astolfi, S.; Varanini, Z. Modulation of NO3- uptake by water-extractable humic substances: Involvement of root plasma membrane H+ATPase. Plant Soil 1999, 215, 155–161. [Google Scholar] [CrossRef]
- Canellas, L.P.; Olivares, F.L.; Okorokova-Façanha, A.L.; Façanha, A.R. Humic Acids Isolated from Earthworm Compost Enhance Root Elongation, Lateral Root Emergence, and Plasma Membrane H+-ATPase Activity in Maize Roots. Plant Physiol. 2002, 130, 1951–1957. [Google Scholar] [CrossRef]
- Quaggiotti, S.; Ruperti, B.; Pizzeghello, D.; Francioso, O.; Tugnoli, V.; Nardi, S. Effect of low molecular size humic substances on nitrate uptake and expression of genes involved in nitrate transport in maize (Zea mays L.). J. Exp. Bot. 2004, 55, 803–813. [Google Scholar] [CrossRef]
- Russell, L.; Stokes, A.R.; Macdonald, H.; Muscolo, A.; Nardi, S. Stomatal Responses to Humic Substances and Auxin are Sensitive to Inhibitors of Phospholipase A2. Plant Soil 2006, 283, 175–185. [Google Scholar] [CrossRef]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef] [PubMed]
- Casal, J.J.; Candia, A.N.; Sellaro, R. Light perception and signalling by phytochrome A. J. Exp. Bot. 2013, 65, 2835–2845. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.W.; Jang, I.-C.; Henriques, R.; Chua, N. FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE Associate with the Arabidopsis Transcription Factors LAF1 and HFR1 to Transmit Phytochrome A Signals for Inhibition of Hypocotyl Elongation. Plant Cell 2009, 21, 1341–1359. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Ji, Q.; Huang, Y.; Jiang, Z.; Bao, M.; Wang, H.; Lin, R. FAR-RED ELONGATED HYPOCOTYL3 and FAR-RED IMPAIRED RESPONSE1 Transcription Factors Integrate Light and Abscisic Acid Signaling in Arabidopsis. Plant Physiol. 2013, 163, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Schepens, I.; Boccalandro, H.E.; Kami, C.; Casal, J.J.; Fankhauser, C. Phytochrome kinase Substrate4 modulates phytochrome-mediated control of hypocotyl growth orientation. Plant Physiol. 2008, 147, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Nortes, T.; Pérez-Pérez, J.M.; Sarmiento-Mañús, R.; Candela, H.; Micol, J.L. Deficient glutamate biosynthesis triggers a concerted upregulation of ribosomal protein genes in Arabidopsis. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Xu, Y.-H.; Liu, R.; Yan, L.; Liu, Z.-Q.; Jiang, S.-C.; Shen, Y.-Y.; Wang, X.-F.; Zhang, D.-P. Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 2011, 63, 1095–1106. [Google Scholar] [CrossRef]
- Koini, M.A.; Alvey, L.; Allen, T.; Tilley, C.A.; Harberd, N.P.; Whitelam, G.C.; Franklin, K.A. High Temperature-Mediated Adaptations in Plant Architecture Require the bHLH Transcription Factor PIF4. Curr. Biol. 2009, 19, 408–413. [Google Scholar] [CrossRef]
- Gururani, M.A.; Venkatesh, J.; Tran, L.P. Regulation of Photosynthesis during Abiotic Stress-Induced Photoinhibition. Mol. Plant 2015, 8, 1304–1320. [Google Scholar] [CrossRef]
- Morrow, G.; Hightower, L.E.; Tanguay, R.M. Small heat shock proteins: Big folding machines. Cell Stress Chaperones 2014, 20, 207–212. [Google Scholar] [CrossRef]
- Sun, W.; Bernard, C.; Van De Cotte, B.; Van Montagu, M.; Verbruggen, N. At-HSP17.6A, encoding a small heat-shock protein in Arabidopsis, can enhance osmotolerance upon overexpression. Plant J. 2001, 27, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Network of Plant Heat Stress Response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Sakuma, Y.; Todaka, D.; Maruyama, K.; Qin, F.; Mizoi, J.; Kidokoro, S.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem. Biophys. Res. Commun. 2008, 368, 515–521. [Google Scholar] [CrossRef]
- Chi, W.-T.; Fung, R.W.M.; Liu, H.-C.; Hsu, C.-C.; Charng, Y.-Y. Temperature-induced lipocalin is required for basal and acquired thermotolerance inArabidopsis. Plant Cell Environ. 2009, 32, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Abo-Ogiala, A.; Carsjens, C.; Diekmann, H.; Fayyaz, P.; Herrfurth, C.; Feussner, I.; Polle, A. Temperature-induced lipocalin (TIL) is translocated under salt stress and protects chloroplasts from ion toxicity. J. Plant Physiol. 2014, 171, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Huang, Y.; Sun, S.; Sun, J.; Cao, H.; Shabala, S.; Bie, Z. Root respiratory burst oxidase homologue-dependent H2O2 production confers salt tolerance on a grafted cucumber by controlling Na+ exclusion and stomatal closure. J. Exp. Bot. 2018, 69, 3465–3476. [Google Scholar] [CrossRef] [PubMed]
- Meyer, Y.; Belin, C.; Delorme-Hinoux, V.; Reichheld, J.-P.; Riondet, C. Thioredoxin and Glutaredoxin Systems in Plants: Molecular Mechanisms, Crosstalks, and Functional Significance. Antioxid. Redox Signal. 2012, 17, 1124–1160. [Google Scholar] [CrossRef]
- Patterson, K.; Walters, L.A.; Cooper, A.M.; Olvera, J.G.; Rosas, M.A.; Rasmusson, A.G.; Escobar, M.A. Nitrate-Regulated Glutaredoxins Control Arabidopsis Primary Root Growth. Plant Physiol. 2015, 170, 989–999. [Google Scholar] [CrossRef]
- Jung, J.-Y.; Ahn, J.H.; Schachtman, D.P. CC-type glutaredoxins mediate plant response and signaling under nitrate starvation in Arabidopsis. BMC Plant Biol. 2018, 18, 281. [Google Scholar] [CrossRef]
- Deavall, D.G.; Martin, E.A.; Horner, J.M.; Roberts, R.A. Drug-Induced Oxidative Stress and Toxicity. J. Toxicol. 2012, 2012, 1–13. [Google Scholar] [CrossRef]
- Sappl, P.G.; Carroll, A.J.; Clifton, R.; Lister, R.; Whelan, J.; Millar, A.H.; Singh, K.B. The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. Plant J. 2009, 58, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Trivedi, P.K. Glutathione S-Transferases: Role in Combating Abiotic Stresses Including Arsenic Detoxification in Plants. Front. Plant Sci. 2018, 9, 751. [Google Scholar] [CrossRef] [PubMed]
- Kovinich, N.; Kayanja, G.; Chanoca, A.; Otegui, M.S.; Grotewold, E. Abiotic stresses induce different localizations of anthocyanins in Arabidopsis. Plant Signal. Behav. 2015, 10, e1027850. [Google Scholar] [CrossRef] [PubMed]
- Commisso, M.; Toffali, K.; Strazzer, P.; Stocchero, M.; Ceoldo, S.; Baldan, B.; Levi, M.; Guzzo, F. Impact of Phenylpropanoid Compounds on Heat Stress Tolerance in Carrot Cell Cultures. Front. Plant Sci. 2016, 7, 1439. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.R.; Kim, J.H.; Gil Nam, H.; Lim, P.O. The Delayed Leaf Senescence Mutants of Arabidopsis, ore1, ore3, and ore9 are Tolerant to Oxidative Stress. Plant Cell Physiol. 2004, 45, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Blanvillain, R.; Kim, J.H.; Wu, S.; Lima, A.; Ow, D.W. OXIDATIVE STRESS 3 is a chromatin-associated factor involved in tolerance to heavy metals and oxidative stress. Plant J. 2009, 57, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Volkov, R.A.; Panchuk, I.I.; Mullineaux, P.M.; Schöffl, F. Heat stress-induced H2O2 is required for effective expression of heat shock genes in Arabidopsis. Plant Mol. Biol. 2006, 61, 733–746. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Huang, Y.-P.; Xi, J.; Cao, M.-J.; Ni, W.-S.; Chen, X.; Zhu, J.; Oliver, D.J.; Xiang, C.-B. Functional gene-mining for salt-tolerance genes with the power of Arabidopsis. Plant J. 2008, 56, 653–664. [Google Scholar] [CrossRef]
- Luhua, S.; Ciftci-Yilmaz, S.; Harper, J.; Cushman, J.; Mittler, R. Enhanced Tolerance to Oxidative Stress in Transgenic Arabidopsis Plants Expressing Proteins of Unknown Function. Plant Physiol. 2008, 148, 280–292. [Google Scholar] [CrossRef]
- Jung, Y.J.; Melencion, S.M.B.; Lee, E.S.; Park, J.H.; Alinapon, C.V.; Oh, H.T.; Yun, D.-J.; Chi, Y.H.; Lee, S.Y. Universal Stress Protein Exhibits a Redox-Dependent Chaperone Function in Arabidopsis and Enhances Plant Tolerance to Heat Shock and Oxidative Stress. Front. Plant Sci. 2015, 6, 1141. [Google Scholar] [CrossRef]
- Melencion, S.M.B.; Chi, Y.H.; Pham, T.T.; Paeng, S.K.; Wi, S.D.; Lee, C.; Ryu, S.W.; Koo, S.S.; Lee, S.Y. RNA Chaperone Function of a Universal Stress Protein in Arabidopsis Confers Enhanced Cold Stress Tolerance in Plants. Int. J. Mol. Sci. 2017, 18, 2546. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Liu, W.; Yao, Y.; Wei, Y.; Chan, Z. Alcohol dehydrogenase 1 (ADH1) confers both abiotic and biotic stress resistance in Arabidopsis. Plant Sci. 2017, 262, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Appelhagen, I.; Thiedig, K.; Nordholt, N.; Schmidt, N.; Huep, G.; Sagasser, M.; Weisshaar, B. Update on transparent testa mutants from Arabidopsis thaliana: Characterisation of new alleles from an isogenic collection. Planta 2014, 240, 955–970. [Google Scholar] [CrossRef]
- Sagasser, M.; Lu, G.-H.; Hahlbrock, K.; Weisshaar, B.A. thaliana TRANSPARENT TESTA 1 is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins. Genes Dev. 2002, 16, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Yonekura-Sakakibara, K.; Tohge, T.; Niida, R.; Saito, K. Identification of a Flavonol 7-O-Rhamnosyltransferase Gene Determining Flavonoid Pattern in Arabidopsis by Transcriptome Coexpression Analysis and Reverse Genetics. J. Biol. Chem. 2007, 282, 14932–14941. [Google Scholar] [CrossRef]
- Wang, G.; Ellendorff, U.; Kemp, B.; Mansfield, J.W.; Forsyth, A.; Mitchell, K.; Bastas, K.; Liu, C.-M.; Woods-Tör, A.; Zipfel, C.; et al. A Genome-Wide Functional Investigation into the Roles of Receptor-Like Proteins in Arabidopsis. Plant Physiol. 2008, 147, 503–517. [Google Scholar] [CrossRef]
- Andersson, M.X.; Hamberg, M.; Kourtchenko, O.; Brunnström, Å.; McPhail, K.L.; Gerwick, W.H.; Göbel, C.; Feussner, I.; Ellerström, M. Oxylipin Profiling of the Hypersensitive Response inArabidopsis thaliana. J. Biol. Chem. 2006, 281, 31528–31537. [Google Scholar] [CrossRef]
- Ma, Q.-H. Monocot chimeric jacalins: A novel subfamily of plant lectins. Crit. Rev. Biotechnol. 2014, 34, 300–306. [Google Scholar] [CrossRef]
- Alvarez, A.; Montesano, M.; Schmelz, E.A.; De León, I.P. Activation of Shikimate, Phenylpropanoid, Oxylipins, and Auxin Pathways in Pectobacterium carotovorum Elicitors-Treated Moss. Front. Plant Sci. 2016, 7, 328. [Google Scholar] [CrossRef]
- Iida, K.; Seki, M.; Sakurai, T.; Satou, M.; Akiyama, K.; Toyoda, T.; Konagaya, A.; Shinozaki, K. RARTF: Database and Tools for Complete Sets of Arabidopsis Transcription Factors. DNA Res. 2005, 12, 247–256. [Google Scholar] [CrossRef]
- Han, G.; Wang, M.; Yuan, F.; Sui, N.; Song, J.; Wang, B. The CCCH zinc finger protein gene AtZFP1 improves salt resistance in Arabidopsis thaliana. Plant Mol. Biol. 2014, 86, 237–253. [Google Scholar] [CrossRef]
- Bogamuwa, S.; Jang, J.-C. Plant Tandem CCCH Zinc Finger Proteins Interact with ABA, Drought, and Stress Response Regulators in Processing-Bodies and Stress Granules. PLoS ONE 2016, 11, e0151574. [Google Scholar] [CrossRef]
- Park, H.-Y.; Seok, H.-Y.; Woo, D.-H.; Lee, S.-Y.; Tarte, V.N.; Lee, E.-H.; Lee, C.-H.; Moon, Y.-H. AtERF71/HRE2 transcription factor mediates osmotic stress response as well as hypoxia response in Arabidopsis. Biochem. Biophys. Res. Commun. 2011, 414, 135–141. [Google Scholar] [CrossRef]
- Dai, S.; Wei, X.; Pei, L.; Thompson, R.L.; Liu, Y.; Heard, J.E.; Ruff, T.G.; Beachy, R.N. BROTHER OF LUX ARRHYTHMO Is a Component of the Arabidopsis Circadian Clock. Plant Cell 2011, 23, 961–972. [Google Scholar] [CrossRef]
- Shin, J.M.; Chung, K.; Sakamoto, S.; Kojima, S.; Yeh, C.-M.; Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. The chimeric repressor for the GATA4 transcription factor improves tolerance to nitrogen deficiency in Arabidopsis. Plant Biotechnol. 2017, 34, 151–158. [Google Scholar] [CrossRef]
- Shuai, B.; Reynaga-Peña, C.G.; Springer, P.S. The Lateral Organ Boundaries Gene Defines a Novel, Plant-Specific Gene Family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef]
- Gan, Y.; Bernreiter, A.; Filleur, S.; Abram, B.; Forde, B.G. Overexpressing the ANR1 MADS-Box Gene in Transgenic Plants Provides New Insights into its Role in the Nitrate Regulation of Root Development. Plant Cell Physiol. 2012, 53, 1003–1016. [Google Scholar] [CrossRef]
- Shi, M.-Z. Biosynthesis and Metabolic Engineering of Anthocyanins in Arabidopsis thaliana. Recent Patents Biotechnol. 2014, 8, 47–60. [Google Scholar] [CrossRef]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 2667–2679. [Google Scholar] [CrossRef]
- Liang, Y.S.; Ermawati, N.; Cha, J.-Y.; Jung, M.H.; Su’Udi, M.; Kim, M.G.; Ha, S.-H.; Park, C.-G.; Son, D. Overexpression of an AP2/ERF-type transcription factor CRF5 confers pathogen resistance to arabidopsis plants. J. Korean Soc. Appl. Biol. Chem. 2010, 53, 142–148. [Google Scholar] [CrossRef]
- Raines, T.; Shanks, C.; Cheng, C.-Y.; McPherson, D.; Argueso, C.T.; Kim, H.J.; Franco-Zorrilla, J.M.; López-Vidriero, I.; Solano, R.; Vaňková, R.; et al. The cytokinin response factors modulate root and shoot growth and promote leaf senescence in Arabidopsis. Plant J. 2016, 85, 134–147. [Google Scholar] [CrossRef]
- Malcolm, R.L.; MacCarthy, P. Limitations in the use of commercial humic acids in water and soil research. Environ. Sci. Technol. 1986, 20, 904–911. [Google Scholar] [CrossRef]
- Ni, Z.; Kim, E.-D.; Ha, M.; Lackey, E.; Liu, J.; Zhang, Y.; Sun, Q.; Chen, Z.J. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nat. Cell Biol. 2008, 457, 327–331. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B-Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]
- Fisher, R.A. On the Interpretation of χ2 from Contingency Tables, and the Calculation of P. J. R. Stat. Soc. 1922, 85, 87. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cha, J.-Y.; Kang, S.-H.; Ji, M.G.; Shin, G.-I.; Jeong, S.Y.; Ahn, G.; Kim, M.G.; Jeon, J.-R.; Kim, W.-Y. Transcriptome Changes Reveal the Molecular Mechanisms of Humic Acid-Induced Salt Stress Tolerance in Arabidopsis. Molecules 2021, 26, 782. https://doi.org/10.3390/molecules26040782
Cha J-Y, Kang S-H, Ji MG, Shin G-I, Jeong SY, Ahn G, Kim MG, Jeon J-R, Kim W-Y. Transcriptome Changes Reveal the Molecular Mechanisms of Humic Acid-Induced Salt Stress Tolerance in Arabidopsis. Molecules. 2021; 26(4):782. https://doi.org/10.3390/molecules26040782
Chicago/Turabian StyleCha, Joon-Yung, Sang-Ho Kang, Myung Geun Ji, Gyeong-Im Shin, Song Yi Jeong, Gyeongik Ahn, Min Gab Kim, Jong-Rok Jeon, and Woe-Yeon Kim. 2021. "Transcriptome Changes Reveal the Molecular Mechanisms of Humic Acid-Induced Salt Stress Tolerance in Arabidopsis" Molecules 26, no. 4: 782. https://doi.org/10.3390/molecules26040782
APA StyleCha, J.-Y., Kang, S.-H., Ji, M. G., Shin, G.-I., Jeong, S. Y., Ahn, G., Kim, M. G., Jeon, J.-R., & Kim, W.-Y. (2021). Transcriptome Changes Reveal the Molecular Mechanisms of Humic Acid-Induced Salt Stress Tolerance in Arabidopsis. Molecules, 26(4), 782. https://doi.org/10.3390/molecules26040782





