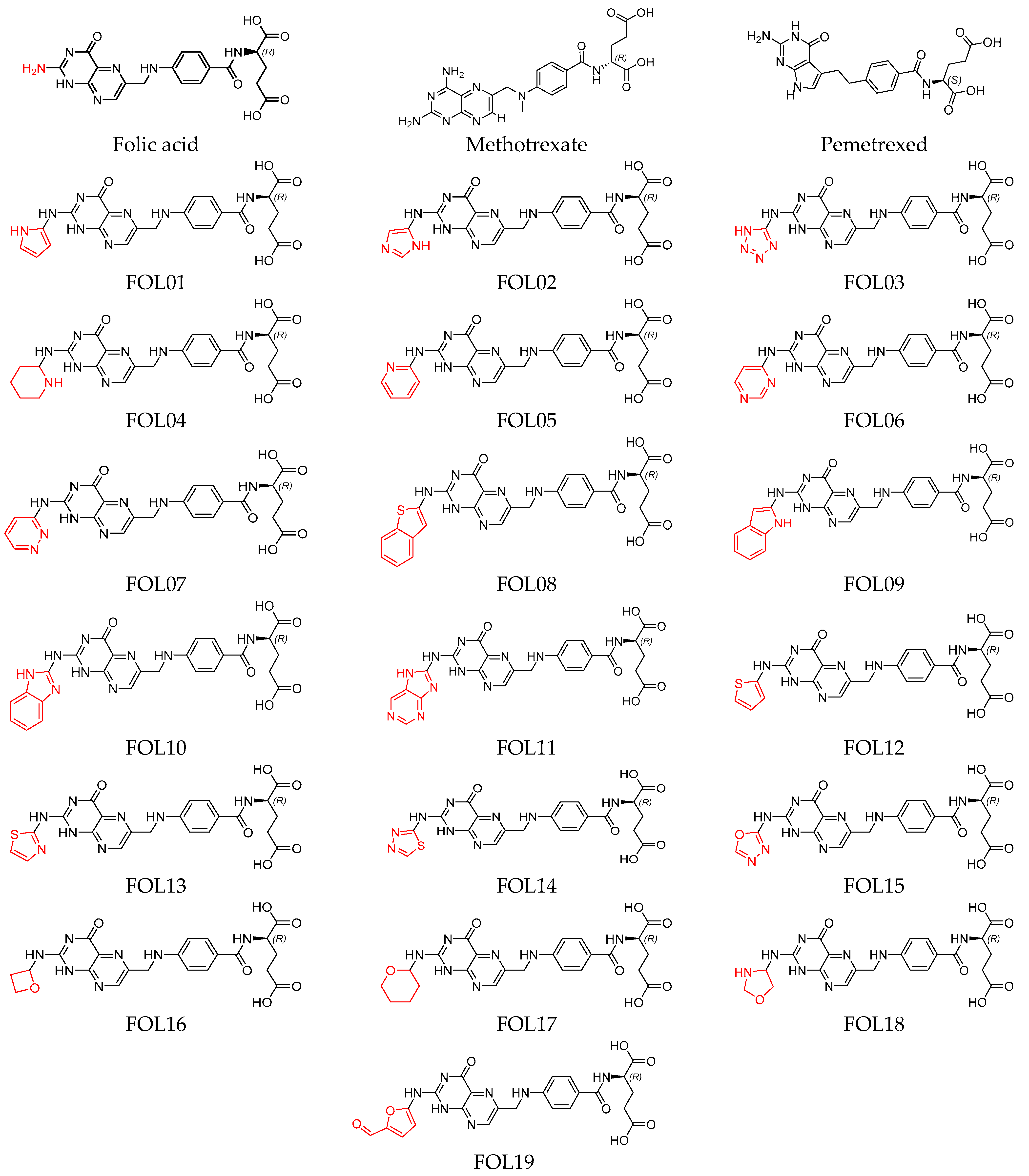
Heterocyclic Substitutions Greatly Improve Affinity and Stability of Folic Acid towards FRα. an In Silico Insight
Abstract
1. Introduction
2. Results and Discussion
2.1. Investigation of FRα Binding Site
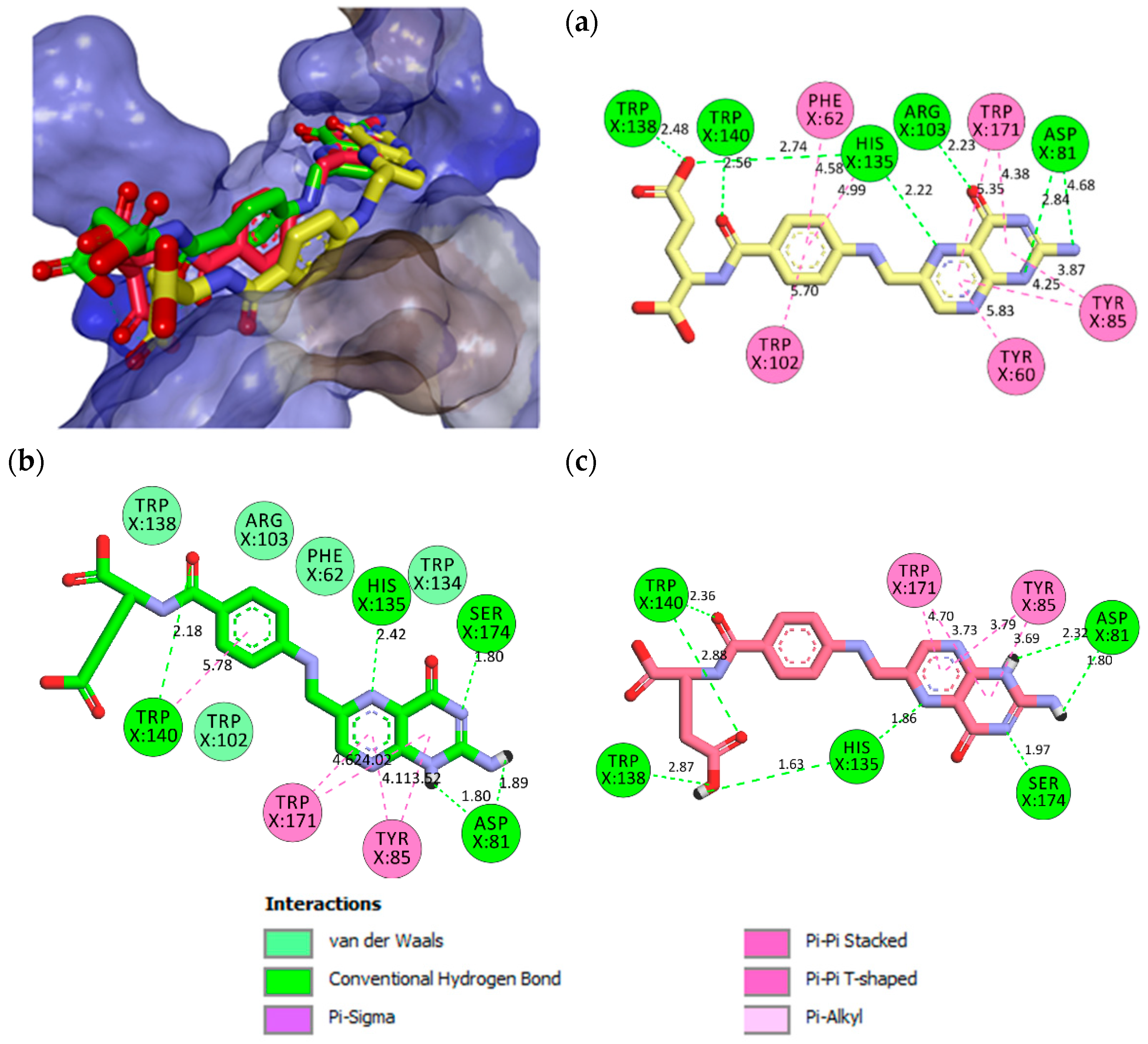
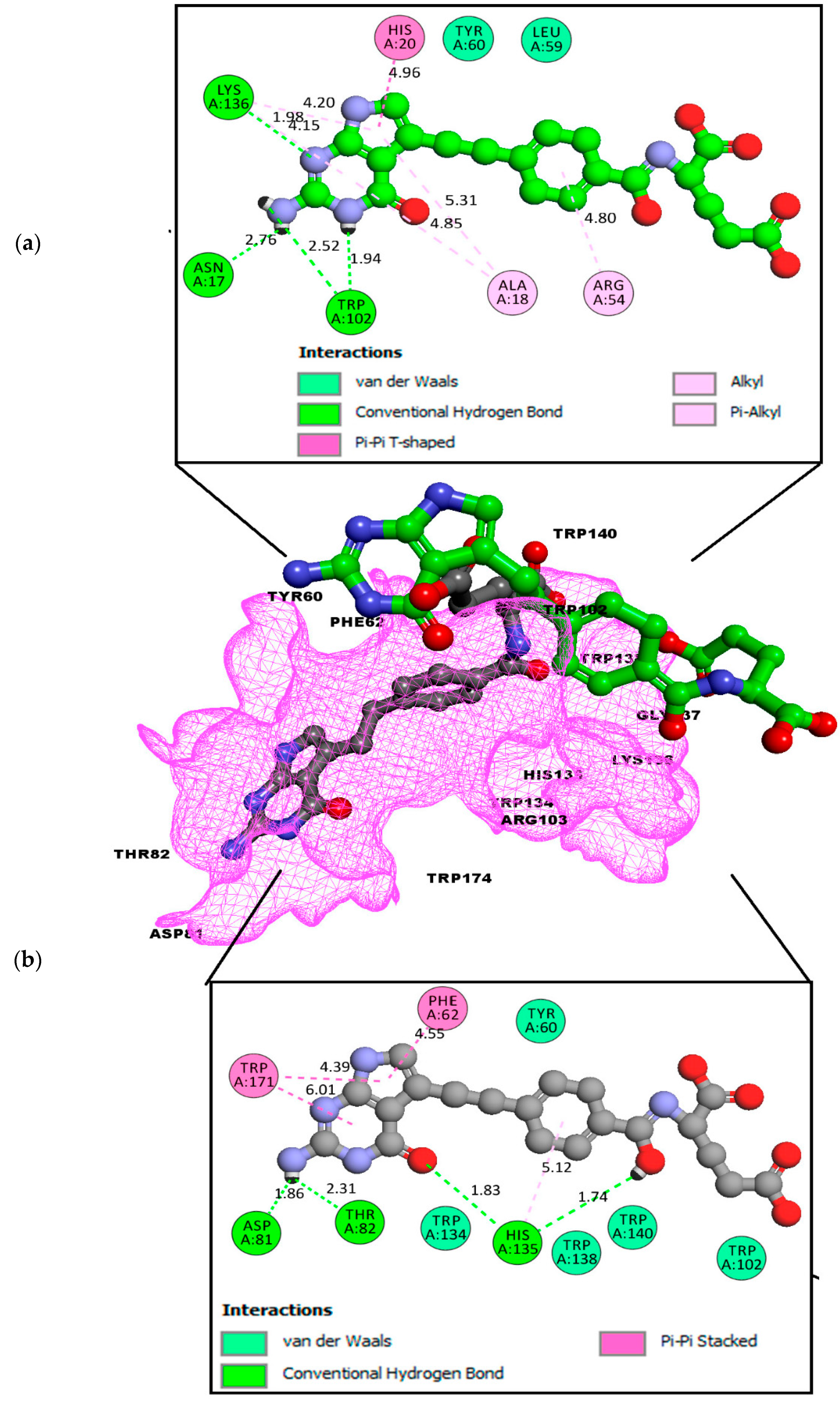
2.2. Molecular Docking
2.3. Molecular Dynamics (MD) Simulation
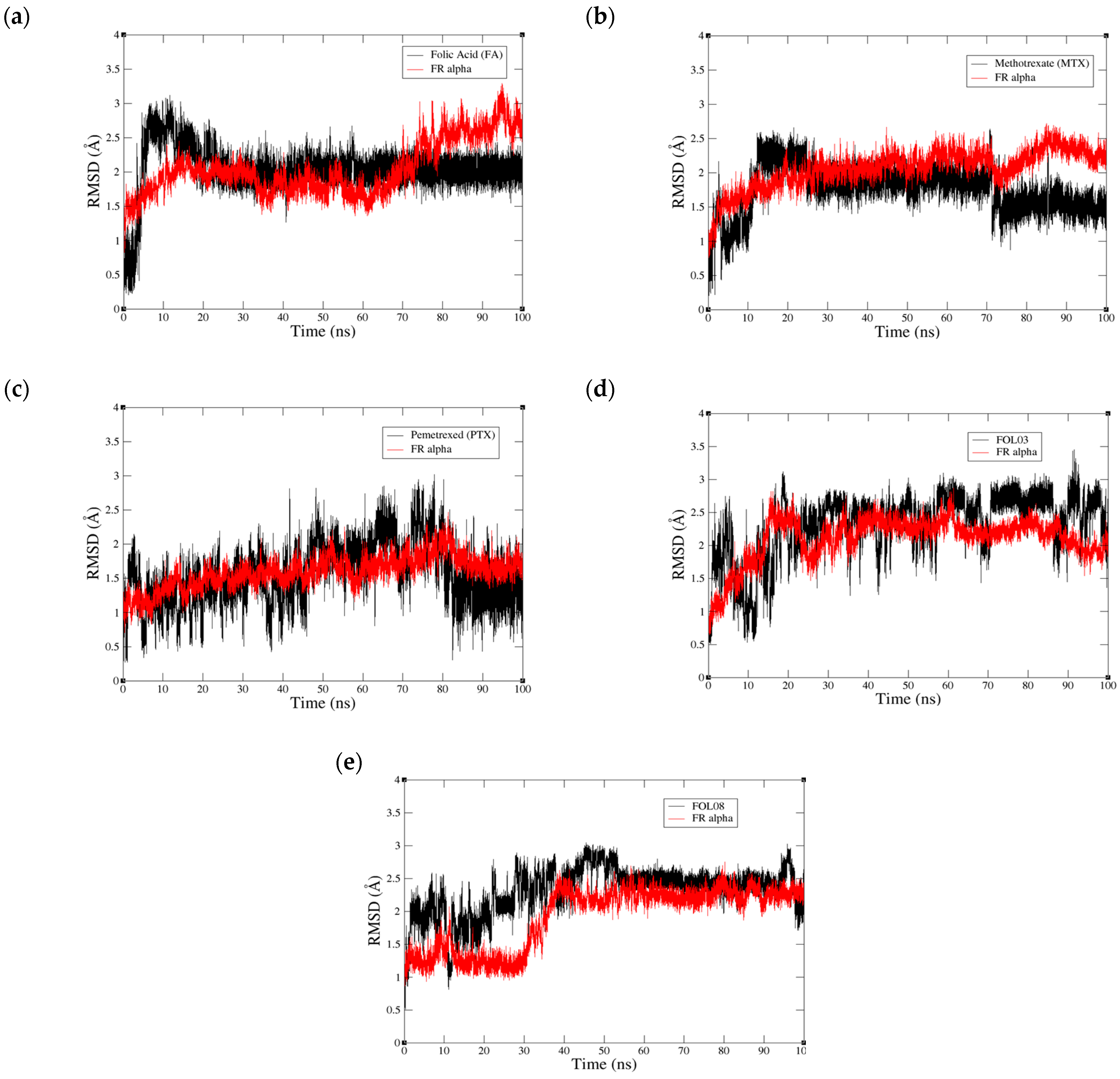
2.3.1. Root Mean Square Deviation (RMSD) Analysis
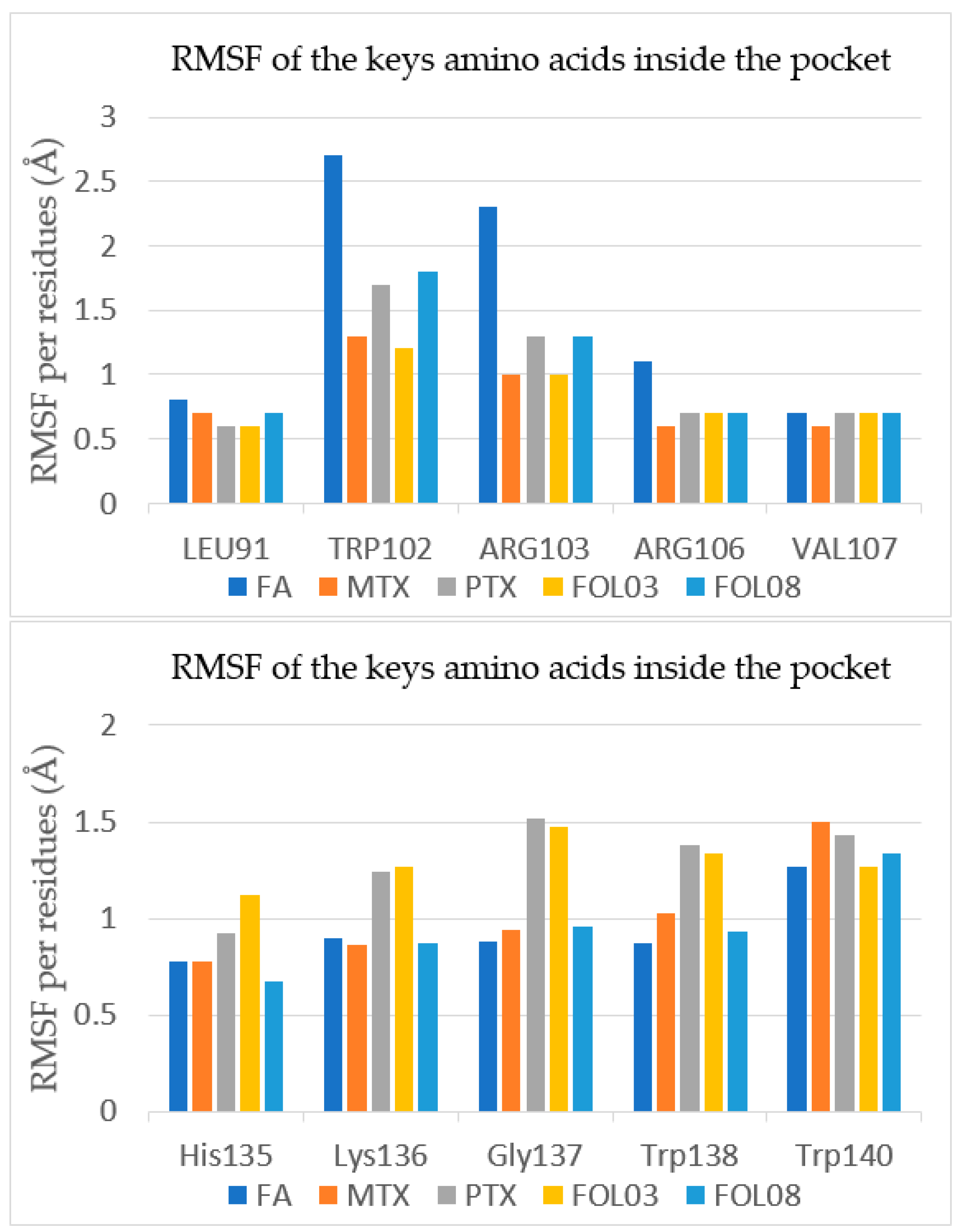
2.3.2. Root Mean Square Fluctuation (RMSF) Analysis
2.3.3. Binding Free Energy Calculation by Molecular Mechanics–Poisson-Boltzmann Surface Accessible (MM-PBSA)
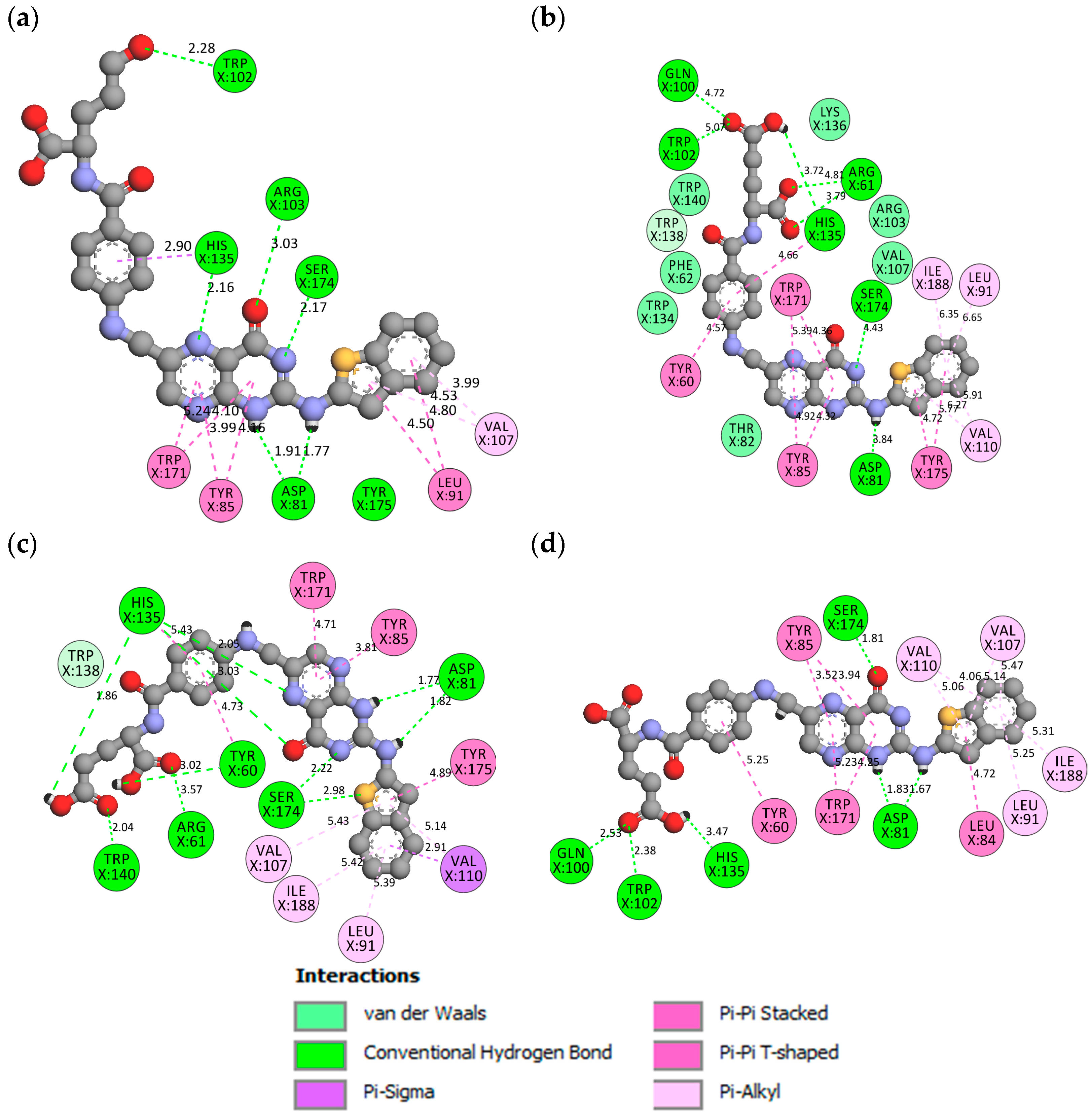
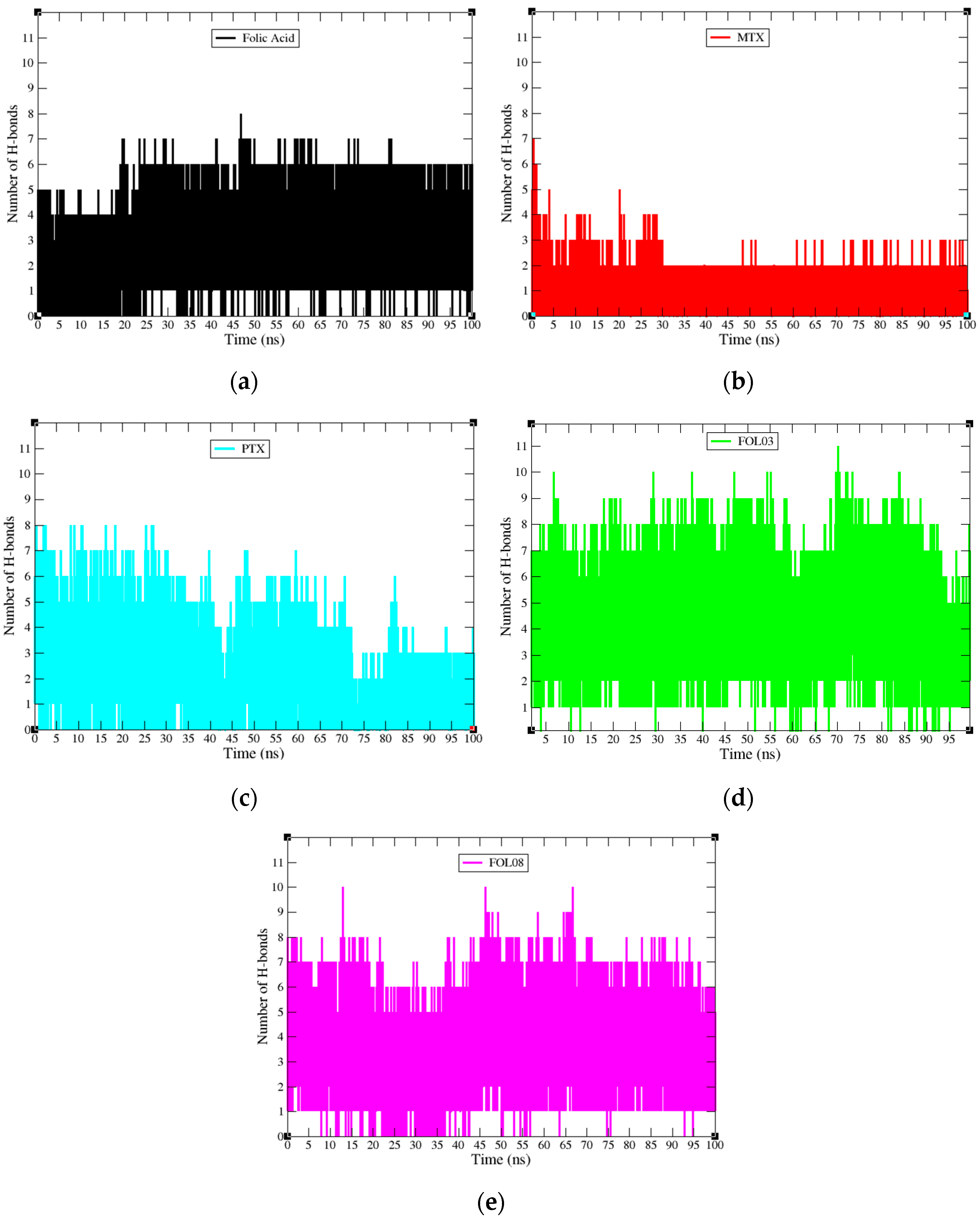
2.3.4. Hydrogen Bond Properties
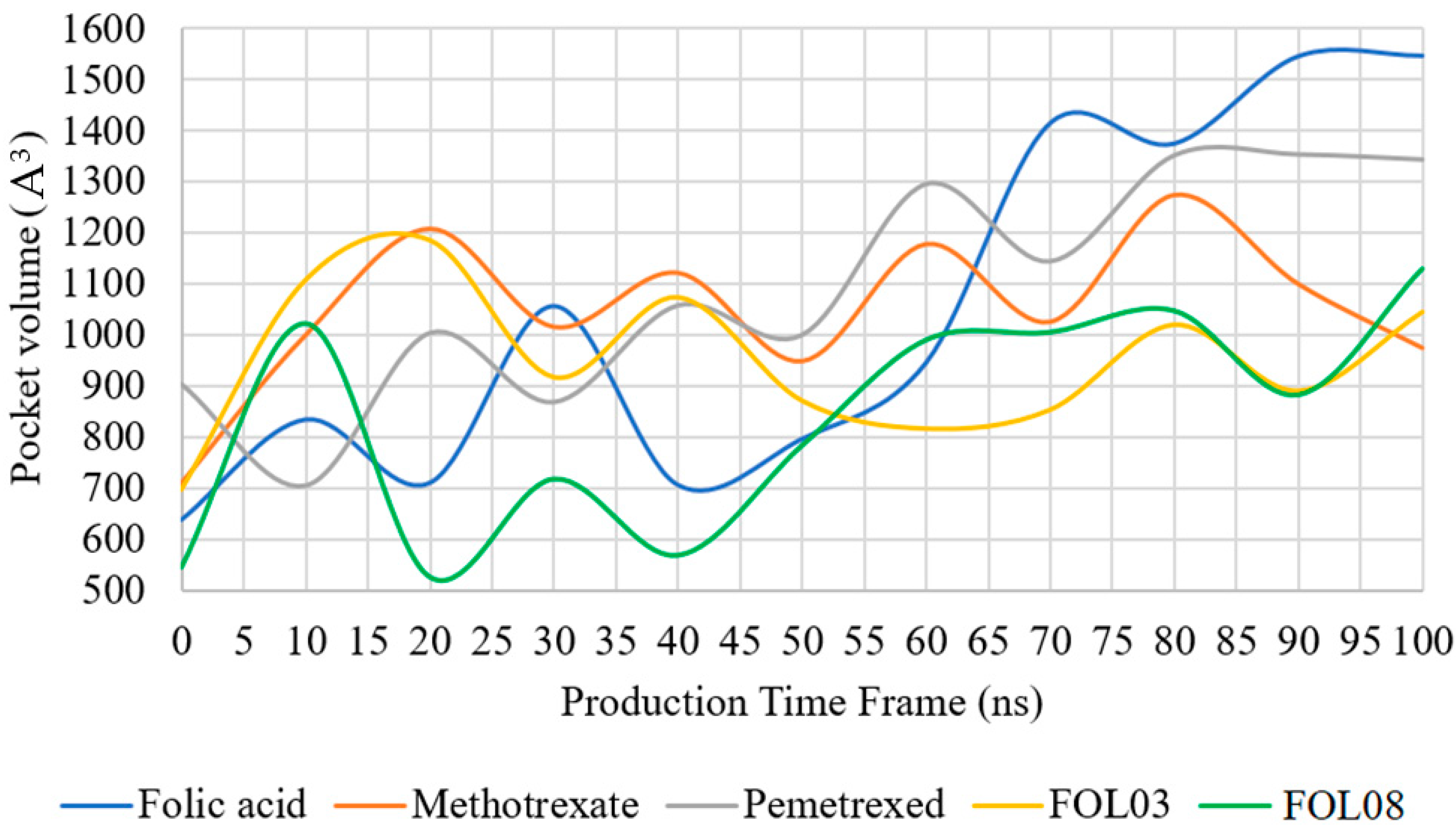
2.3.5. Pocket Volume Calculations
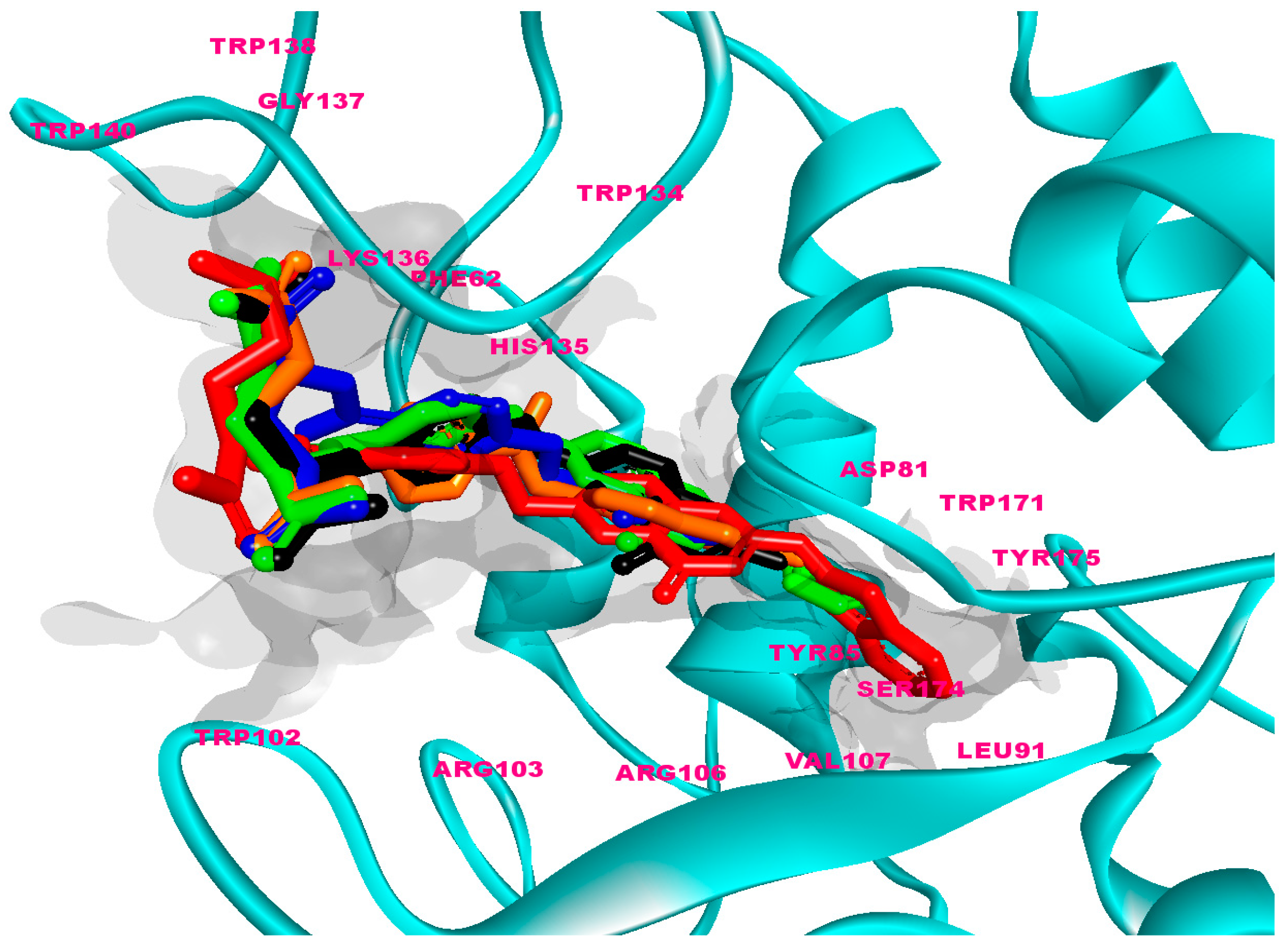
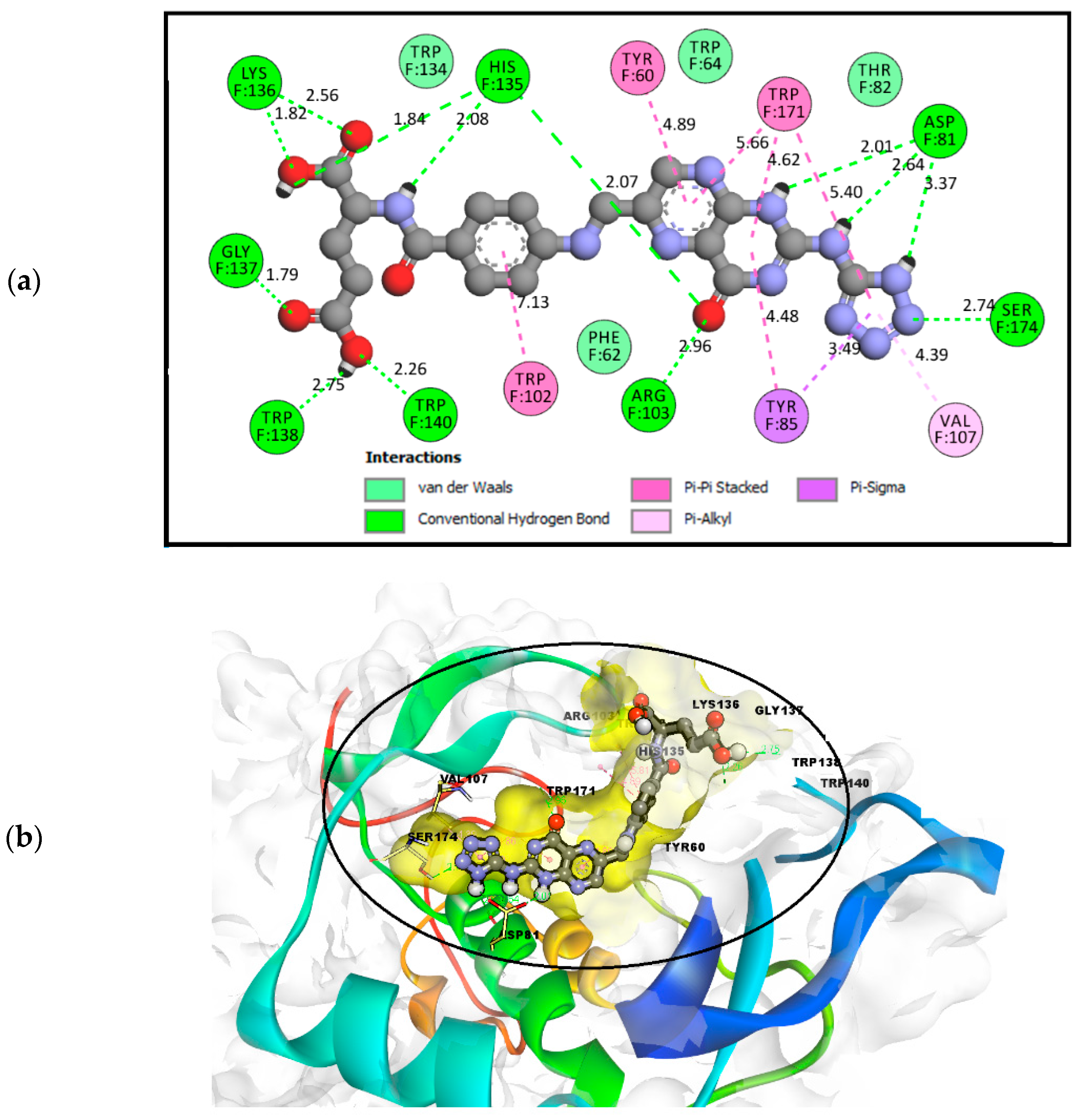
2.3.6. General Effects of the Binding of FOL03 and FOL08 Inside FRα
2.4. ADMET Prediction
3. Materials and Methods
3.1. Determination of the Size of the Binding Site
3.2. Molecular Docking
3.3. Molecular Dynamics (MD) Simulation
3.4. Free Binding Energy Calculation by MM-PBSA
3.5. Pocket Volume (POVME) Algorithm
3.6. ADMET Prediction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Fernández, M.; Javaid, F.; Chudasama, V. Advances in targeting the folate receptor in the treatment/imaging of cancers. Chem. Sci. 2018, 9, 790–810. [Google Scholar] [CrossRef]
- Necela, B.M.; Crozier, J.A.; Andorfer, C.A.; Lewis-Tuffin, L.; Kachergus, J.M.; Geiger, X.J.; Kalari, K.R.; Serie, D.J.; Sun, Z.; Aspita, A.M. Folate receptor-α (FOLR1) expression and function in triple negative tumors. PLoS ONE 2015, 10, e0122209. [Google Scholar]
- Guo, J.; Schlich, M.; Cryan, J.F.; O’Driscoll, C.M. Targeted drug delivery via folate receptors for the treatment of brain cancer: Can the promise deliver? J. Pharm. Sci. 2017, 106, 3413–3420. [Google Scholar] [CrossRef] [PubMed]
- Quici, S.; Casoni, A.; Foschi, F.; Armelao, L.; Bottaro, G.; Seraglia, R.; Bolzati, C.; Salvarese, N.; Carpanese, D.; Rosato, A. Folic acid-conjugated europium complexes as luminescent probes for selective targeting of cancer cells. J. Med. Chem 2015, 58, 2003–2014. [Google Scholar] [CrossRef] [PubMed]
- Assaraf, Y.G.; Leamon, C.P.; Reddy, J.A. The folate receptor as a rational therapeutic target for personalized cancer treatment. Drug Resist. Updat. 2014, 17, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Gattoc, L.; O’Connor, C.; Yang, S.; Wallace-Povirk, A.; George, C.; Orr, S.; Polin, L.; White, K.; Kushner, J. Dual targeting of epithelial ovarian cancer via folate receptor α and the proton-coupled folate transporter with 6-substituted pyrrolo [2,3-d] pyrimidine antifolates. Mol. Cancer Ther. 2017, 16, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Vergote, I.B.; Marth, C.; Coleman, R.L. Role of the folate receptor in ovarian cancer treatment: Evidence, mechanism, and clinical implications. Cancer Metastasis Rev. 2015, 34, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Farran, B.; Pavitra, E.; Kasa, P.; Peela, S.; Raju, G.S.R.; Nagaraju, G.P. Folate-targeted immunotherapies: Passive and active strategies for cancer. Cytokine Growth Factor Rev. 2019, 45, 45–52. [Google Scholar] [CrossRef]
- Patel, N.R.; Piroyan, A.; Nack, A.H.; Galati, C.A.; McHugh, M.; Orosz, S.; Keeler, A.W.; O’Neal, S.; Zamboni, W.C.; Davis, B. Design, synthesis, and characterization of folate-targeted platinum-loaded theranostic nanoemulsions for therapy and imaging of ovarian cancer. Mol. Pharm. 2016, 13, 1996–2009. [Google Scholar] [CrossRef]
- Siwowska, K.; Haller, S.; Bortoli, F.; Benešová, M.; Groehn, V.; Bernhardt, P.; Schibli, R.; Müller, C. Preclinical comparison of albumin-binding radiofolates: Impact of linker entities on the in vitro and in vivo properties. Mol. Pharm. 2017, 14, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Senol, S.; Ceyran, A.B.; Aydin, A.; Zemheri, E.; Ozkanli, S.; Kösemetin, D.; Sehitoglu, I.; Akalin, I. Folate receptor α expression and significance in endometrioid endometrium carcinoma and endometrial hyperplasia. Int. J. Clin. Exp. Pathol. 2015, 8, 5633. [Google Scholar] [PubMed]
- Scaranti, M.; Cojocaru, E.; Banerjee, S.; Banerji, U. Exploiting the folate receptor α in oncology. Nat. Rev. Clin. Oncol. 2020, 17, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, A.; Napoli, A.; Roggiani, F.; Tomassetti, A.; Bagnoli, M.; Mezzanzanica, D. One-carbon metabolism: Biological players in epithelial ovarian cancer. Int. J. Mol. Sci. 2018, 19, 2092. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, Z.; Sun, Y.; Geng, X.; Hu, Y.; Meng, H.; Ge, J.; Qu, L. Synthesis of Luminescent Carbon Dots with Ultrahigh Quantum Yield and Inherent Folate Receptor-Positive Cancer Cell Targetability. Sci. Rep. 2018, 8, 1086. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, X.; Xian, M.; Dong, C.; Shuang, S. Folic acid-conjugated green luminescent carbon dots as a nanoprobe for identifying folate receptor-positive cancer cells. Talanta 2018, 183, 39–47. [Google Scholar] [CrossRef]
- Soe, Z.C.; Thapa, R.K.; Ou, W.; Gautam, M.; Nguyen, H.T.; Jin, S.G.; Ku, S.K.; Oh, K.T.; Choi, H.-G.; Yong, C.S.; et al. Folate receptor-mediated celastrol and irinotecan combination delivery using liposomes for effective chemotherapy. Colloids Surf. B 2018, 170, 718–728. [Google Scholar] [CrossRef]
- Klein, P.M.; Kern, S.; Lee, D.-J.; Schmaus, J.; Höhn, M.; Gorges, J.; Kazmaier, U.; Wagner, E. Folate receptor-directed orthogonal click-functionalization of siRNA lipopolyplexes for tumor cell killing in vivo. Biomaterials 2018, 178, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Allard-Vannier, E.; Hervé-Aubert, K.; Kaaki, K.; Blondy, T.; Shebanova, A.; Shaitan, K.V.; Ignatova, A.A.; Saboungi, M.-L.; Feofanov, A.V.; Chourpa, I. Folic acid-capped PEGylated magnetic nanoparticles enter cancer cells mostly via clathrin-dependent endocytosis. Biochim. Biophys. Acta Gen. Sub. 2017, 1861, 1578–1586. [Google Scholar] [CrossRef]
- He, H.-R.; Liu, P.; He, G.-H.; Dong, W.-H.; Wang, M.-Y.; Dong, Y.-L.; Lu, J. Association between reduced folate carrier G80A polymorphism and methotrexate toxicity in childhood acute lymphoblastic leukemia: A meta-analysis. Leuk. Lymphoma 2014, 55, 2793–2800. [Google Scholar] [CrossRef]
- Kelemen, L.E. The role of folate receptor α in cancer development, progression and treatment: Cause, consequence or innocent bystander? Int. J. Cancer 2006, 119, 243–250. [Google Scholar] [CrossRef]
- Chen, C.; Ke, J.; Zhou, X.E.; Yi, W.; Brunzelle, J.S.; Li, J.; Yong, E.-L.; Xu, H.E.; Melcher, K. Structural basis for molecular recognition of folic acid by folate receptors. Nature 2013, 500, 486. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.K.; Sarkar, A.; Feldmann, D.P.; Hoffmann, P.; Merkel, O.M. Revisiting the value of competition assays in folate receptor-mediated drug delivery. Biomaterials 2017, 138, 35–45. [Google Scholar] [CrossRef]
- O’Shannessy, D.J.; Somers, E.B.; Wang, L.-c.; Wang, H.; Hsu, R. Expression of folate receptors alpha and beta in normal and cancerous gynecologic tissues: Correlation of expression of the beta isoform with macrophage markers. J. Ovarian Res. 2015, 8, 1–9. [Google Scholar] [CrossRef]
- Boshnjaku, V.; Shim, K.-W.; Tsurubuchi, T.; Ichi, S.; Szany, E.V.; Xi, G.; Mania-Farnell, B.; McLone, D.G.; Tomita, T.; Mayanil, C.S. Nuclear localization of folate receptor alpha: A new role as a transcription factor. Sci. Rep. 2012, 2, 980. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, B.; Mohammadnia-Afrouzi, M.; Bakhshaei, P.; Yazdani, Y.; Ghalamfarsa, G.; Yousefi, M.; Sadreddini, S.; Jadidi-Niaragh, F.; Hojjat-Farsangi, M. Folate-conjugated nanoparticles as a potent therapeutic approach in targeted cancer therapy. Tumour Biol. 2015, 36, 5727–5742. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, Y.; Ma, Q.; Cao, Y.; Yu, B. Development and characterization of folic acid-conjugated chitosan nanoparticles for targeted and controlled delivery of gemcitabinein lung cancer therapeutics. Artif. Cells Nanomed. Biotechnol. 2017, 45, 1530–1538. [Google Scholar] [CrossRef]
- Visentin, M.; Zhao, R.; Goldman, I.D. The antifolates. Hematol. Oncol. Clin. 2012, 26, 629–648. [Google Scholar] [CrossRef]
- Wróbel, A.; Drozdowska, D. Recent Design and Structure-Activity Relationship Studies on the Modifications of DHFR Inhibitors as Anticancer Agents. Curr. Med. Chem. 2020. [Google Scholar] [CrossRef]
- Gonen, N.; Assaraf, Y.G. Antifolates in cancer therapy: Structure, activity and mechanisms of drug resistance. Drug Resist. Updat. 2012, 15, 183–210. [Google Scholar] [CrossRef]
- Lutz, R.J. Targeting the folate receptor for the treatment of ovarian cancer. Transl. Cancer Res. 2015, 4, 118–126. [Google Scholar]
- Hagner, N.; Joerger, M. Cancer chemotherapy: Targeting folic acid synthesis. Cancer Manag. Res. 2010, 2, 293. [Google Scholar] [PubMed]
- Lopez, K.; Pina, M.; Alemany, R.; Vögler, O.; Barcelo, F.; Morey, J. Antifolate-modified iron oxide nanoparticles for targeted cancer therapy: Inclusion vs. covalent union. RSC Adv. 2014, 4, 19196–19204. [Google Scholar] [CrossRef]
- Patil, R.; Das, S.; Stanley, A.; Yadav, L.; Sudhakar, A.; Varma, A.K. Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designing. PLoS ONE 2010, 5, e12029. [Google Scholar] [CrossRef]
- Martins, P.; Jesus, J.; Santos, S.; Raposo, L.R.; Roma-Rodrigues, C.; Baptista, P.V.; Fernandes, A.R. Heterocyclic anticancer compounds: Recent advances and the paradigm shift towards the use of nanomedicine’s tool box. Molecules 2015, 20, 16852–16891. [Google Scholar] [CrossRef]
- Afzal, O.; Kumar, S.; Haider, M.R.; Ali, M.R.; Kumar, R.; Jaggi, M.; Bawa, S. A review on anticancer potential of bioactive heterocycle quinoline. Eur. J. Med. Chem. 2015, 97, 871–910. [Google Scholar] [CrossRef]
- Sravanthi, T.; Manju, S. Indoles—a promising scaffold for drug development. Eur. J. Pharm. Sci. 2016, 91, 1–10. [Google Scholar] [CrossRef]
- Huber, R.G.; Margreiter, M.A.; Fuchs, J.E.; von Grafenstein, S.; Tautermann, C.S.; Liedl, K.R.; Fox, T. Heteroaromatic π-stacking energy landscapes. J. Chem. Inf. Model. 2014, 54, 1371–1379. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, M.V.; Randazzo, O.; La Franca, M.; Barone, G.; Vignoni, E.; Rossi, D.; Collina, S. DHFR inhibitors: Reading the past for discovering novel anticancer agents. Molecules 2019, 24, 1140. [Google Scholar] [CrossRef] [PubMed]
- Jendele, L.; Krivak, R.; Skoda, P.; Novotny, M.; Hoksza, D. PrankWeb: A web server for ligand binding site prediction and visualization. Nucleic Acids Res. 2019, 47, W345–W349. [Google Scholar] [CrossRef]
- Anderson, A.C.; Wright, D.L. Antifolate agents: A patent review (2010–2013). Expert Opin. Ther. Pat. 2014, 24, 687–697. [Google Scholar] [CrossRef]
- Krivák, R.; Hoksza, D. P2Rank: Machine learning based tool for rapid and accurate prediction of ligand binding sites from protein structure. J. Cheminform. 2018, 10, 39. [Google Scholar] [CrossRef]
- Tan, K.P.; Nguyen, T.B.; Patel, S.; Varadarajan, R.; Madhusudhan, M.S. Depth: A web server to compute depth, cavity sizes, detect potential small-molecule ligand-binding cavities and predict the pKa of ionizable residues in proteins. Nucleic Acids Res. 2013, 41, W314–W321. [Google Scholar] [CrossRef] [PubMed]
- Della-Longa, S.; Arcovito, A. Structural and functional insights on folate receptor α (FRα) by homology modeling, ligand docking and molecular dynamics. J. Mol. Graph. Model. 2013, 44, 197–207. [Google Scholar] [CrossRef]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef]
- Samar, S.F.; Emad, A.S.; Shaymaa, E.K. CORRIGENDUM A Promising Anti-Cancer and Anti-Oxidant Agents Based on the Pyrrole and Fused Pyrrole: Synthesis, Docking Studies and Biological Evaluation. Anticancer Agents Med. Chem. 2018, 18, 1505. [Google Scholar]
- Taha, M.; Shah, S.A.A.; Afifi, M.; Zulkeflee, M.; Sultan, S.; Wadood, A.; Rahim, F.; Ismail, N.H. Morpholine hydrazone scaffold: Synthesis, anticancer activity and docking studies. Chin. Chem. Lett. 2017, 28, 607–611. [Google Scholar] [CrossRef]
- Popova, E.A.; Protas, A.V.; Trifonov, R.E. Tetrazole derivatives as promising anticancer agents. Anticancer Agents Med. Chem. 2017, 17, 1856–1868. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Zhao, W.; Huang, Z.N.; Yang, S.M.; Wang, L.J.; Jiang, Y.; Zhou, Z.S.; Zheng, M.Y.; Jiang, J.L.; Li, S.H. Evaluation of Novel N-(piperidine-4-yl) benzamide Derivatives as Potential Cell Cycle Inhibitors in HepG2 Cells. Chem. Biol. Drug Des. 2015, 86, 223–231. [Google Scholar] [CrossRef]
- Prachayasittikul, S.; Pingaew, R.; Worachartcheewan, A.; Sinthupoom, N.; Prachayasittikul, V.; Ruchirawat, S.; Prachayasittikul, V. Roles of pyridine and pyrimidine derivatives as privileged scaffolds in anticancer agents. Mini Rev. Med. Chem. 2017, 17, 869–901. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.B. Biological and medicinal significance of pyrimidines: A Review. Int. J. Pharm. Sci. Res. 2018, 9, 44–52. [Google Scholar]
- Jaballah, M.Y.; Serya, R.T.; Abouzid, K. Pyridazine based scaffolds as privileged structures in anti-cancer therapy. Drug Res. 2017, 67, 138–148. [Google Scholar] [CrossRef]
- Keri, R.S.; Chand, K.; Budagumpi, S.; Somappa, S.B.; Patil, S.A.; Nagaraja, B.M. An overview of benzo [b] thiophene-based medicinal chemistry. Eur. J. Med. Chem. 2017, 138, 1002–1033. [Google Scholar] [CrossRef] [PubMed]
- El-sayed, M.T.; Hamdy, N.A.; Osman, D.A.; Ahmed, K.M. Indoles as anticancer agents. Adv. Mod. Oncol. Res. 2015, 1, 20–35. [Google Scholar] [CrossRef]
- Akhtar, M.J.; Yar, M.S.; Sharma, V.K.; Khan, A.A.; Ali, Z.; Haider, M.; Pathak, A. Recent progress of Benzimidazole hybrids for anticancer potential. Curr. Med. Chem. 2020, 27, 5970–6014. [Google Scholar] [CrossRef]
- Camici, M.; Garcia-Gil, M.; Pesi, R.; Allegrini, S.; Tozzi, M.G. Purine-metabolising enzymes and apoptosis in cancer. Cancers 2019, 11, 1354. [Google Scholar] [CrossRef]
- Dos Santos, F.A.; Pereira, M.C.; de Oliveira, T.B.; Mendonça Junior, F.J.B.; de Lima, M.d.C.A.; Pitta, M.G.d.R.; Pitta, I.d.R.; de Melo, R.; Barreto, M.J.; da Rocha, P. Anticancer properties of thiophene derivatives in breast cancer MCF-7 cells. Anticancer Drugs 2018, 29, 157–166. [Google Scholar] [CrossRef]
- Ramos-Inza, S.; Aydillo, C.; Sanmartín, C.; Plano, D. Thiazole Moiety: An Interesting Scaffold for Developing New Antitumoral Compounds. In Heterocycles-Synthesis and Biological Activities; Nandeshwarappa, B.P., Ed.; IntechOpen: London, UK, 2019. [Google Scholar]
- Aliabadi, A. 1,3,4-Thiadiazole based anticancer agents. Anticarcer Agents Med. Chem. 2016, 16, 1301–1314. [Google Scholar] [CrossRef] [PubMed]
- Glomb, T.; Szymankiewicz, K.; Świątek, P. Anti-Cancer Activity of Derivatives of 1, 3, 4-Oxadiazole. Molecules 2018, 23, 3361. [Google Scholar] [CrossRef] [PubMed]
- Bull, J.A.; Croft, R.A.; Davis, O.A.; Doran, R.; Morgan, K.F. Oxetanes: Recent advances in synthesis, reactivity, and medicinal chemistry. Chem. Rev. 2016, 116, 12150–12233. [Google Scholar] [CrossRef]
- Silva, F.; Dantas, B.; Faheina Martins, G.; de Araújo, D.; Vasconcellos, M. Synthesis and Anticancer Activities of Novel Guanylhydrazone and Aminoguanidine Tetrahydropyran Derivatives. Molecules 2016, 21, 671. [Google Scholar] [CrossRef]
- Rodrigues, F.A.; Bomfim, I.d.S.; Cavalcanti, B.C.; Pessoa, C.; Goncalves, R.S.; Wardell, J.L.; Wardell, S.M.; de Souza, M.V. Mefloquine–oxazolidine derivatives: A new class of anticancer agents. Chem. Biol. Drug. Des. 2014, 83, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Murti, Y.; Mishra, P. Synthesis and evaluation of flavanones as anticancer agents. Indian J. Pharm. Sci. 2014, 76, 163. [Google Scholar] [PubMed]
- Jiang, Y.; Wang, C.; Zhang, M.; Fei, X.; Gu, Y. Interacted mechanism of functional groups in ligand targeted with folate receptor via docking, molecular dynamic and MM/PBSA. J. Mol. Graph. Model. 2019, 87, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.-X.; Pang, X. Electrostatic interactions in protein structure, folding, binding, and condensation. Chem. Rev. 2018, 118, 1691–1741. [Google Scholar] [CrossRef]
- Neochoritis, C.G.; Zhao, T.; Dömling, A. Tetrazoles via multicomponent reactions. Chem. Rev. 2019, 119, 1970–2042. [Google Scholar] [CrossRef]
- Della-Longa, S.; Arcovito, A. Intermediate states in the binding process of folic acid to folate receptor α: Insights by molecular dynamics and metadynamics. J. Comput. Aided Mol. Des. 2015, 29, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Yotmanee, P.; Rungrotmongkol, T.; Wichapong, K.; Choi, S.B.; Wahab, H.A.; Kungwan, N.; Hannongbua, S. Binding specificity of polypeptide substrates in NS2B/NS3pro serine protease of dengue virus type 2: A molecular dynamics study. J. Mol. Graph. Model. 2015, 60, 24–33. [Google Scholar] [CrossRef]
- Xie, H.; Li, Y.; Yu, F.; Xie, X.; Qiu, K.; Fu, J. An investigation of molecular docking and molecular dynamic simulation on imidazopyridines as B-Raf kinase inhibitors. Int. J. Mol. Sci. 2015, 16, 27350–27361. [Google Scholar] [CrossRef]
- Shimizu, K.; Yonekawa, T.; Yoshida, M.; Miyazato, M.; Miura, A.; Sakoda, H.; Yamaguchi, H.; Nakazato, M. Conformational change in the ligand-binding pocket via a KISS1R mutation (P147L) leads to isolated gonadotropin-releasing hormone deficiency. JES 2017, 1, 1259–1271. [Google Scholar] [CrossRef][Green Version]
- Gocheva, G.; Ivanova, N.; Iliev, S.; Petrova, J.; Madjarova, G.; Ivanova, A. Characteristics of a folate receptor-α anchored into a multilipid bilayer obtained from atomistic molecular dynamics simulations. J. Chem. Theory Comput. 2019, 16, 749–764. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Lee, T.-S.; Cerutti, D.S.; Mermelstein, D.; Lin, C.; LeGrand, S.; Giese, T.J.; Roitberg, A.; Case, D.A.; Walker, R.C.; York, D.M. GPU-accelerated molecular dynamics and free energy methods in Amber18: Performance enhancements and new features. J. Chem. Inf. Model. 2018, 58, 2043–2050. [Google Scholar] [CrossRef]
- Song, L.F.; Lee, T.-S.; Zhu, C.; York, D.M.; Merz Jr, K.M. Using AMBER18 for Relative Free Energy Calculations. J. Chem. Inf. Model. 2019, 59, 3128–3135. [Google Scholar] [CrossRef] [PubMed]
- Santini, B.L.; Zacharias, M. Rapid in silico design of potential cyclic peptide binders targeting protein-protein interfaces. Front. Chem. 2020, 8, 933. [Google Scholar] [CrossRef]
- Petukh, M.; Li, M.; Alexov, E. Predicting binding free energy change caused by point mutations with knowledge-modified MM/PBSA method. PLoS Comput. Biol. 2015, 11, e1004276. [Google Scholar] [CrossRef]
- Yam, W.K.; Wahab, H.A. Molecular insights into 14-membered macrolides using the MM-PBSA method. J. Chem. Inf. Model. 2009, 49, 1558–1567. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Dassault-Systèmes. Biovia, discovery studio modeling environment; Dassault Systèmes Biovia: San Diego, CA, USA, 2016. [Google Scholar]
- Durrant, J.D.; de Oliveira, C.A.F.; McCammon, J.A. POVME: An algorithm for measuring binding-pocket volumes. J. Mol. Graph. Model. 2011, 29, 773–776. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef] [PubMed]
- Nunes, A.M.V.; de Andrade, F.d.C.P.; Filgueiras, L.A.; de Carvalho Maia, O.A.; Cunha, R.L.; Rodezno, S.V.; Maia Filho, A.L.M.; de Amorim Carvalho, F.A.; Braz, D.C.; Mendes, A.N. preADMET analysis and clinical aspects of dogs treated with the Organotellurium compound RF07: A possible control for canine visceral leishmaniasis? Environ. Toxicol. Pharmacol. 2020, 80, 103470. [Google Scholar] [CrossRef] [PubMed]
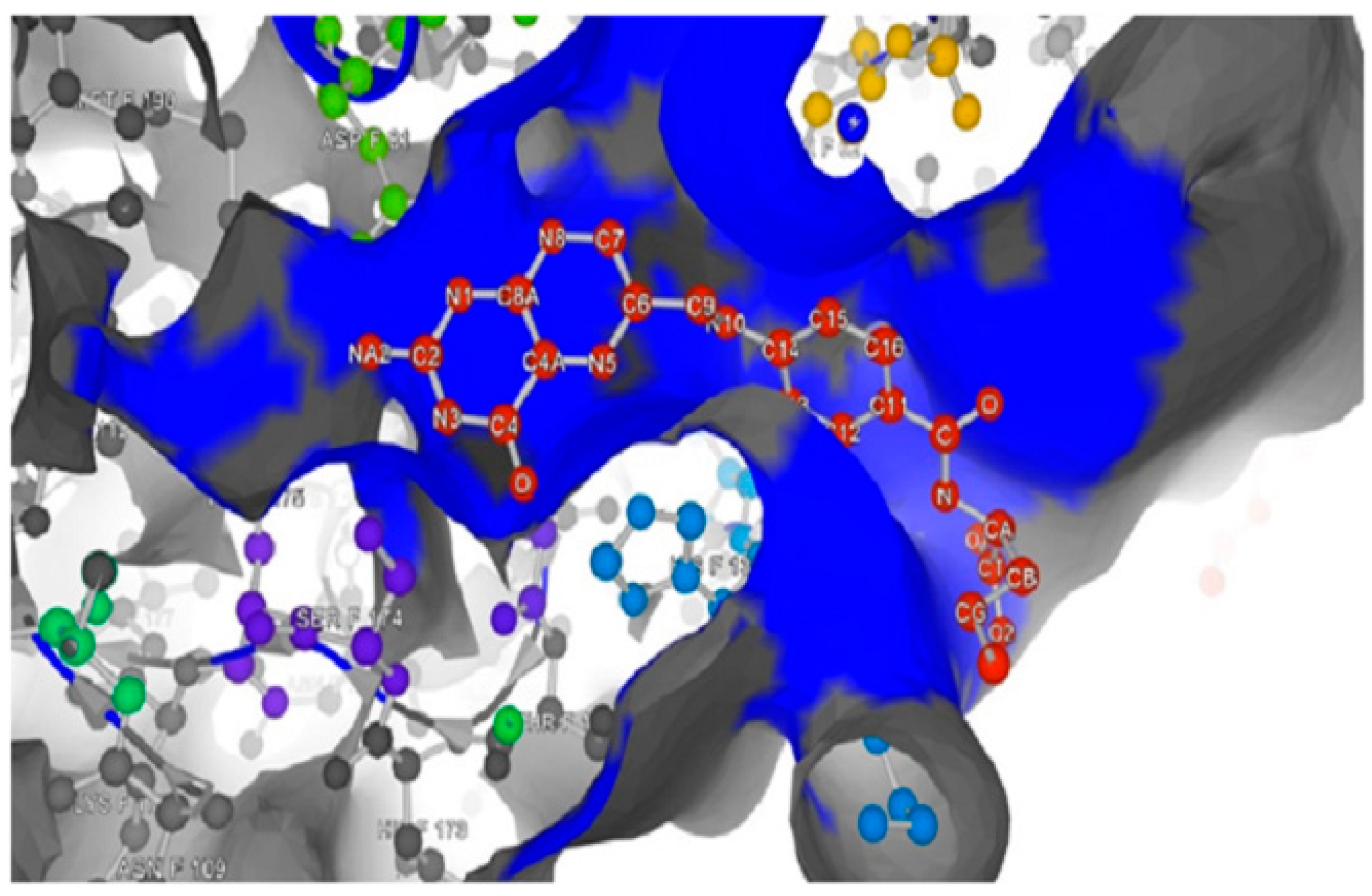
 Unpredictable residues by depth server and confirmed by P2Rank server to form FRα active site. (b) The probability of amino acids, which exist in the binding pocket, forming the binding site.
Unpredictable residues by depth server and confirmed by P2Rank server to form FRα active site. (b) The probability of amino acids, which exist in the binding pocket, forming the binding site.
 Unpredictable residues by depth server and confirmed by P2Rank server to form FRα active site. (b) The probability of amino acids, which exist in the binding pocket, forming the binding site.
Unpredictable residues by depth server and confirmed by P2Rank server to form FRα active site. (b) The probability of amino acids, which exist in the binding pocket, forming the binding site.



| Compound | FBE (kcal/mol) | Ki (Picomolar pM) |
|---|---|---|
| FA | −13.20 | 209.24 |
| MTX | −11.87 | 2000 |
| PTX | −14.05 | 37.88 |
| FOL01 | −15.71 | 3.060 |
| FOL02 | −15.79 | 2.67 |
| FOL03 | −16.83 | 0.460 |
| FOL04 | −15.84 | 2.440 |
| FOL05 | −15.86 | 2.39 |
| FOL06 | −15.88 | 2.28 |
| FOL07 | −15.64 | 3.41 |
| FOL08 | −16.24 | 1.48 |
| FOL09 | −14.34 | 30.88 |
| FOL10 | −15.56 | 3.92 |
| FOL11 | −14.40 | 27.68 |
| FOL12 | −13.84 | 71.24 |
| FOL13 | −14.75 | 15.40 |
| FOL14 | −14.87 | 12.49 |
| FOL15 | −15.71 | 3.06 |
| FOL16 | −14.82 | 13.73 |
| FOL17 | −15.24 | 6.74 |
| FOL18 | −14.81 | 13.84 |
| FOL19 | −14.98 | 10.43 |
| Complex with FRα | kcal/mol | VDWLS kcal/mol | EEL kcal/mol | Gpolar kcal/mol | Gnon-polar kcal/mol | AutoDock kcal/mol |
|---|---|---|---|---|---|---|
| FA | −59.59 ± 0.17 | −55.84 ± 0.15 | −91.97 ± 0.28 | 94.35 ± 0.21 | −6.12 ± 0.01 | −13.20 |
| MTX | −45.12 ± 0.18 | −60.71 ± 0.12 | −48.05 ± 0.39 | 69.98 ± 0.31 | −6.34 ± 0.01 | −11.87 |
| PTX | −30.11 ± 0.36 | −41.38 ± 0.24 | −44.14 ± 0.48 | 60.57 ± 0.40 | −5.16 ± 0.03 | −14.05 |
| FOL03 | −73.62 ± 0.21 | −61.47 ± 0.13 | −134.73 ± 0.40 | 129.16 ± 0.28 | −6.58 ± 0.01 | −16.83 |
| FOL08 | −79.68 ± 0.21 | −75.96 ± 0.15 | −99.95 ± 0.38 | 104.81 ± 0.27 | −8.57 ± 0.01 | −16.24 |
| Code | H-Bond Acceptor (atom@res) | H-Bond Donor (atom@H) | Donor (atom@res) | H-Bond Occupancy (%) | Average Distance (Å) | Average Angle (°) |
|---|---|---|---|---|---|---|
| FA | 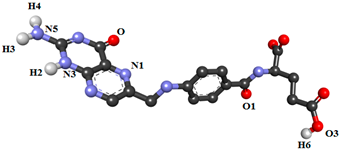 | |||||
| ASP81@OD1 | FA@H2 | FA@N3 | 61.28 | 2.83 | 159.70 | |
| ASP81@OD2 | FA@H4 | FA@N5 | 56.09 | 2.79 | 163.65 | |
| ASP81@OD2 | FA@H3 | FA@N5 | 16.67 | 2.79 | 163.66 | |
| ASP81@OD1 | FA@H3 | FA@N5 | 13.01 | 2.81 | 161.64 | |
| ASP81@OD2 | FA@H2 | FA@N3 | 11.26 | 2.85 | 152.98 | |
| FA@O | ARG103@HH12 | ARG103@NH1 | 17.18 | 2.84 | 149.17 | |
| HIS135@O | FA@H6 | FA@O3 | 56.89 | 2.72 | 156.37 | |
| FA@N1 | HIS135@HE2 | HIS135@NE2 | 21.31 | 2.92 | 162.11 | |
| FA@O1 | TRP140@HE1 | TRP140@NE1 | 43.77 | 2.83 | 148.12 | |
| MTX | 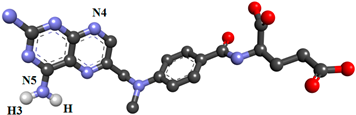 | |||||
| ASP81@OD1 | MTX@H3 | MTX@N5 | 22.89 | 2.82 | 152.41 | |
| ASP81@OD1 | MTX@H | MTX@N5 | 22.15 | 2.81 | 153.59 | |
| ASP81@OD2 | MTX@H | MTX@N5 | 13.03 | 2.83 | 153.19 | |
| MTX@N4 | ARG103@HH12 | ARG103@NH1 | 10.22 | 2.91 | 147.23 | |
| MTX@N4 | HIS135@HE2 | HIS135@NE2 | 10.96 | 2.91 | 158.30 | |
| PTX | 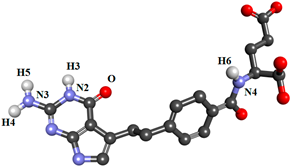 | |||||
| ASP81@OD1 | PTX@H4 | PTX@N3 | 11.46 | 2.81 | 159.08 | |
| PTX@O | HIS135@HE2 | HIS135@NE2 | 45.99 | 2.84 | 160.57 | |
| HIS135@O | PTX@H6 | PTX@N4 | 20.06 | 2.86 | 153.34 | |
| FOL03 | 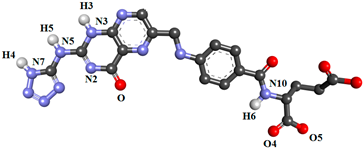 | |||||
| ASP81@OD2 | FOL03@H5 | FOL03@N5 | 75.40 | 2.82 | 157.41 | |
| ASP81@OD2 | FOL03@H3 | FOL03@N3 | 74.93 | 2.76 | 151.43 | |
| ASP81@OD1 | FOL03@H4 | FOL03@N7 | 70.12 | 2.76 | 148.54 | |
| ASP81@OD1 | FOL03@H5 | FOL03@N5 | 28.07 | 2.86 | 147.80 | |
| TYR60@O | FOL03@H6 | FOL03@N10 | 23.44 | 2.89 | 157.28 | |
| FOL03@O4 | ARG61@HE | ARG61@NE | 18.90 | 2.86 | 157.00 | |
| FOL03@O5 | ARG61@HH21 | ARG61@NH2 | 16.70 | 2.88 | 157.43 | |
| FOL03@O FOL03@O FOL03@N2 | ARG107@HH11 ARG107@HH21 ARG107@HH21 | ARG107@NH1 ARG107@NH2 ARG107@NH2 | 26.63 18.56 13.63 | 2.84 2.85 2.92 | 149.99 147.16 156.63 | |
| FOL08 | 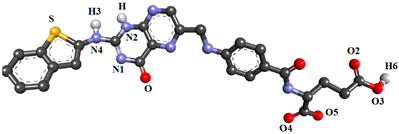 | |||||
| ASP81@OD1 | FOL08@H3 | FOL08@N4 | 63.39 | 2.79 | 163.29 | |
| ASP81@OD2 | FOL08@H | FOL08@N2 | 44.73 | 2.83 | 152.71 | |
| ASP81@OD1 | FOL08@H | FOL08@N2 | 41.74 | 2.83 | 150.17 | |
| ASP81@OD2 | FOL08@H3 | FOL08@N4 | 32.20 | 2.77 | 162.82 | |
| FOL08@O4 | ARG61@HH21 | ARG61@NH2 | 15.71 | 2.84 | 158.32 | |
| HIS135@O | FOL08@H6 | FOL08@O3 | 33.81 | 2.71 | 158.35 | |
| FOL08@N | HIS135@HE2 | HIS135@NE2 | 18.73 | 2.92 | 160.57 | |
| FOL08@O2 | TRP140@HE1 | TRP140@NE1 | 16.08 | 2.86 | 156.43 | |
| FOL08@N1 | SER174@HG | SER174@OG | 32.52 | 2.89 | 164.20 | |
| FOL08@O | SER174@HG | SER174@OG | 10.11 | 2.80 | 155.58 | |
| Property | Model Name | Predicted Value | ||||
|---|---|---|---|---|---|---|
| FA | MTX | PTX | FOL03 | FOL08 | ||
| Absorption | Water solubility (log mol/L) | −2.88 | −2.859 | −2.842 | −2.892 | −2.905 |
| Caco2 permeability (log cm/s) | −0.877 | −0.77 | −0.954 | −0.92 | −0.667 | |
| Human intestinal absorption (% absorbed) | 17.745 | 35.716 | 37.981 | 7.719 | 76.253 | |
| Skin permeability (log Kp) | −2.735 | −2.735 | −2.735 | −2.735 | −2.735 | |
| P-glycoprotein substrate | Yes | Yes | Yes | Yes | Yes | |
| P-glycoprotein I inhibitor | No | No | No | No | No | |
| P-glycoprotein II inhibitor | No | No | No | No | No | |
| Distribution | Human volume of distribution (log L/kg) | 0.046 | −0.883 | −0.927 | −0.548 | −0.720 |
| Human fraction unbound (Fu) | 0.370 | 0.183 | 0.160 | 0.276 | 0.127 | |
| Blood Brain Barrier (BBB)permeability (log BB) | −1.615 | −1.865 | −1.442 | −3.458 | −2.372 | |
| CNS permeability (log PS) | −4.262 | −3.818 | −4.022 | −7.265 | −4.174 | |
| Metabolism | CYP2D6 substrate | No | No | No | No | No |
| CYP3A4 substrate | No | No | No | No | No | |
| CYP1A2 inhibitor | No | No | No | No | No | |
| CYP2C19 inhibitor | No | No | No | No | No | |
| CYP2C9 inhibitor | No | No | No | No | No | |
| CYP2D6 inhibitor | No | No | No | No | No | |
| CYP3A4 inhibitor | No | No | No | No | No | |
| Excretion | Total clearance (log ml/min/kg) | 0.527 | 0.378 | 0.285 | −0.196 | −0.111 |
| Renal OCT2 substrate | No | No | No | No | No | |
| Toxicity | Ames toxicity | No | No | No | No | No |
| Max. human tolerated dose (log mg/kg/day) | −0.586 | −0.827 | −0.292 | 0.366 | 0.489 | |
| hERG I inhibitor | No | No | No | No | No | |
| hERG II inhibitor | No | Yes | No | No | No | |
| Oral rat acute toxicity (LD50) (mol/kg) | 2.670 | 2.713 | 2.585 | 2.483 | 2.501 | |
| Oral rat chronic toxicity (LOAEL) (log mg/kg_bw/day) | 3.153 | 3.112 | 3.111 | 4.876 | 3.152 | |
| Skin sensitization | No | No | No | No | No | |
| T. pyriformis toxicity (log ug/L) | 0.285 | 0.285 | 0.285 | 0.285 | 0.285 | |
| Minnow toxicity (log mM) | 4.009 | 2.384 | 2.867 | 4.886 | 1.221 | |
| System | Total Numberof Atoms | Number of Heteroatoms | Water Atoms | Neutralizing Atoms |
|---|---|---|---|---|
| FRα + FA | 29,808 | 3634 | 9033 | 3 Cl− |
| FRα + MTX | 29,812 | 3639 | 9033 | 3 Cl− |
| FRα + PTX | 29,808 | 3643 | 9033 | 3 Cl− |
| FRα + FOL03 | 29,807 | 3635 | 9031 | 3 Cl− |
| FRα + FOL08 | 29,809 | 3643 | 9029 | 3 Cl− |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Thiabat, M.G.; Saqallah, F.G.; Gazzali, A.M.; Mohtar, N.; Yap, B.K.; Choong, Y.S.; Wahab, H.A. Heterocyclic Substitutions Greatly Improve Affinity and Stability of Folic Acid towards FRα. an In Silico Insight. Molecules 2021, 26, 1079. https://doi.org/10.3390/molecules26041079
Al-Thiabat MG, Saqallah FG, Gazzali AM, Mohtar N, Yap BK, Choong YS, Wahab HA. Heterocyclic Substitutions Greatly Improve Affinity and Stability of Folic Acid towards FRα. an In Silico Insight. Molecules. 2021; 26(4):1079. https://doi.org/10.3390/molecules26041079
Chicago/Turabian StyleAl-Thiabat, Mohammad G., Fadi G. Saqallah, Amirah Mohd Gazzali, Noratiqah Mohtar, Beow Keat Yap, Yee Siew Choong, and Habibah A Wahab. 2021. "Heterocyclic Substitutions Greatly Improve Affinity and Stability of Folic Acid towards FRα. an In Silico Insight" Molecules 26, no. 4: 1079. https://doi.org/10.3390/molecules26041079
APA StyleAl-Thiabat, M. G., Saqallah, F. G., Gazzali, A. M., Mohtar, N., Yap, B. K., Choong, Y. S., & Wahab, H. A. (2021). Heterocyclic Substitutions Greatly Improve Affinity and Stability of Folic Acid towards FRα. an In Silico Insight. Molecules, 26(4), 1079. https://doi.org/10.3390/molecules26041079







