NMR Structure Determinations of Small Proteins Using only One Fractionally 20% 13C- and Uniformly 100% 15N-Labeled Sample
Abstract
1. Introduction
2. Results
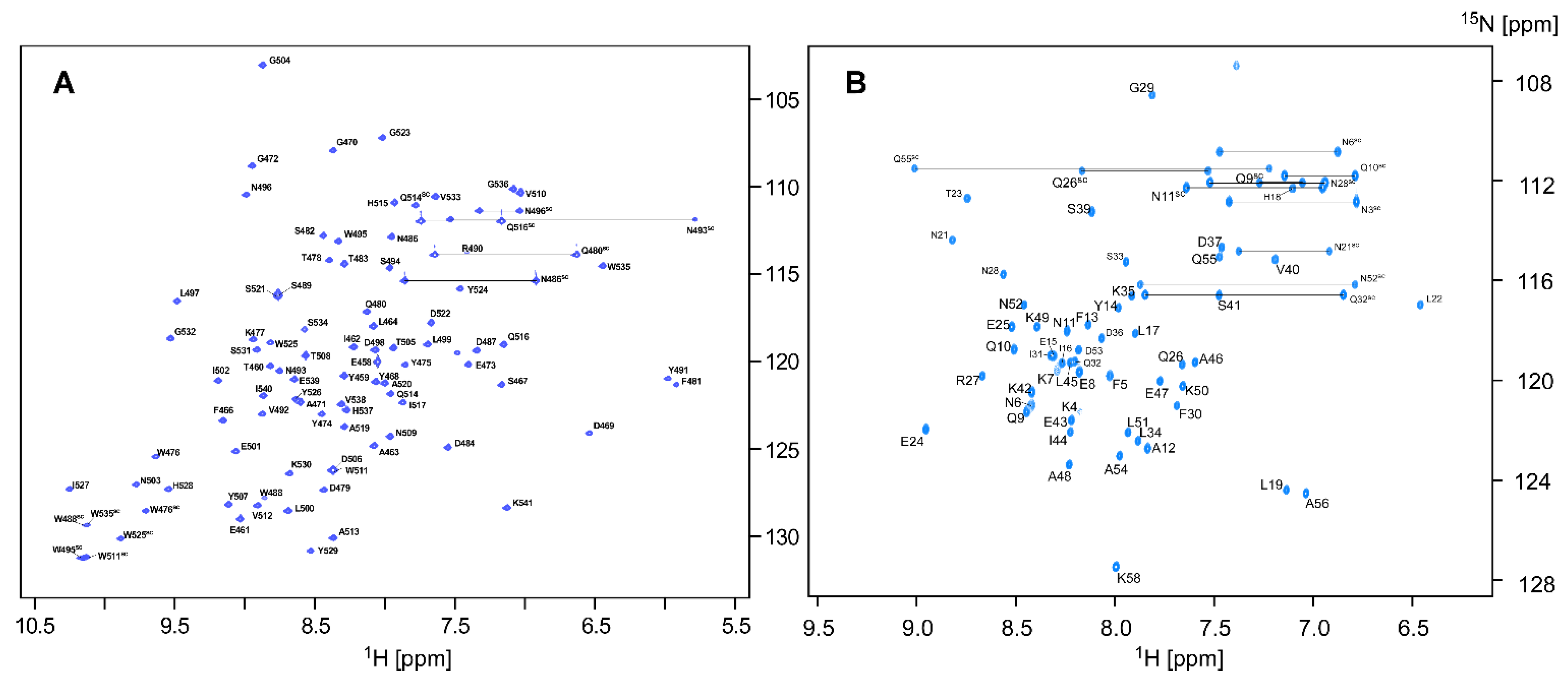
2.1. NMR Assignments
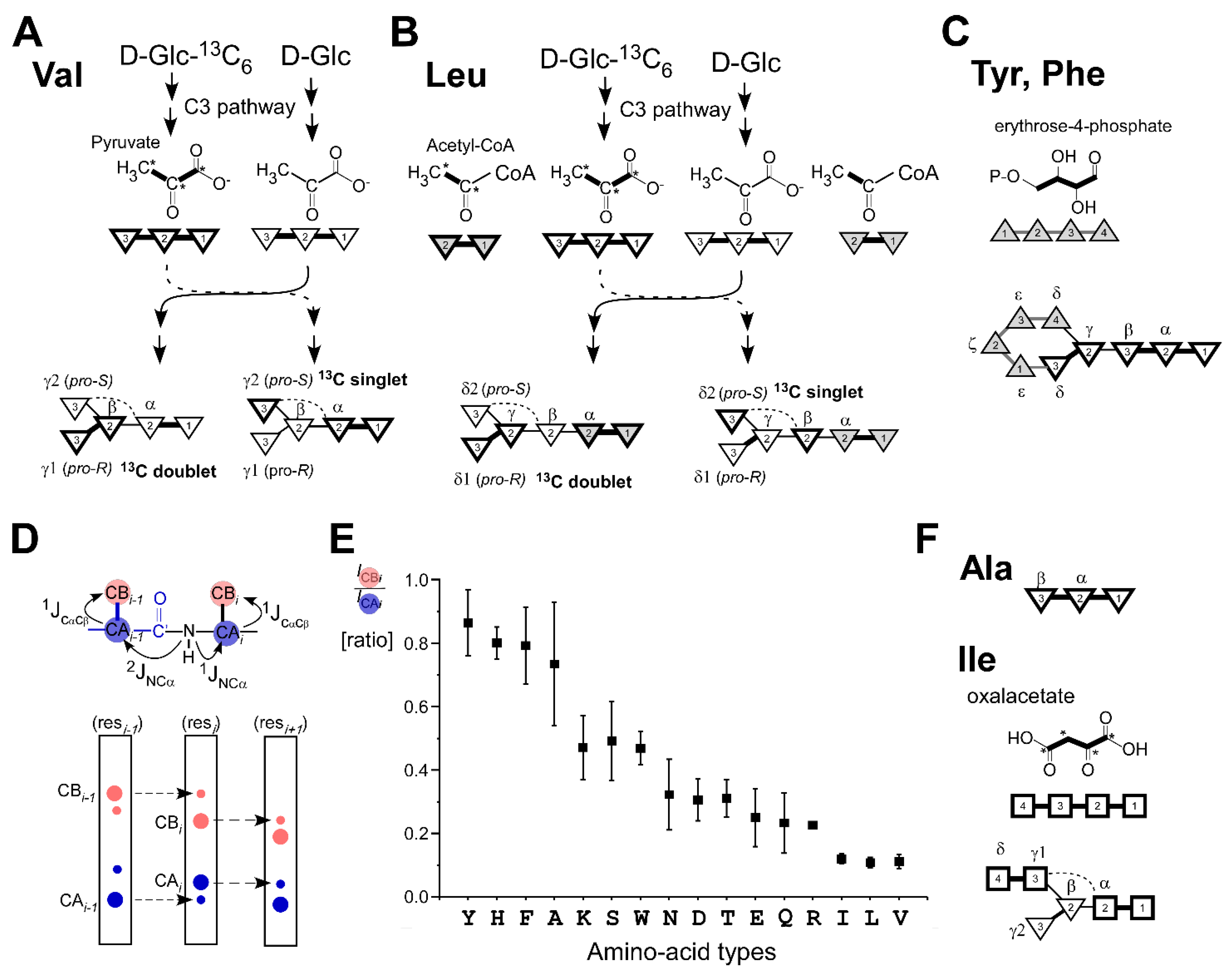
2.2. Effects of Fractional 13C-Labeling on NMR Spectra from the Different Biosynthetic Pathways
2.3. NMR Structure Determination of CBM64
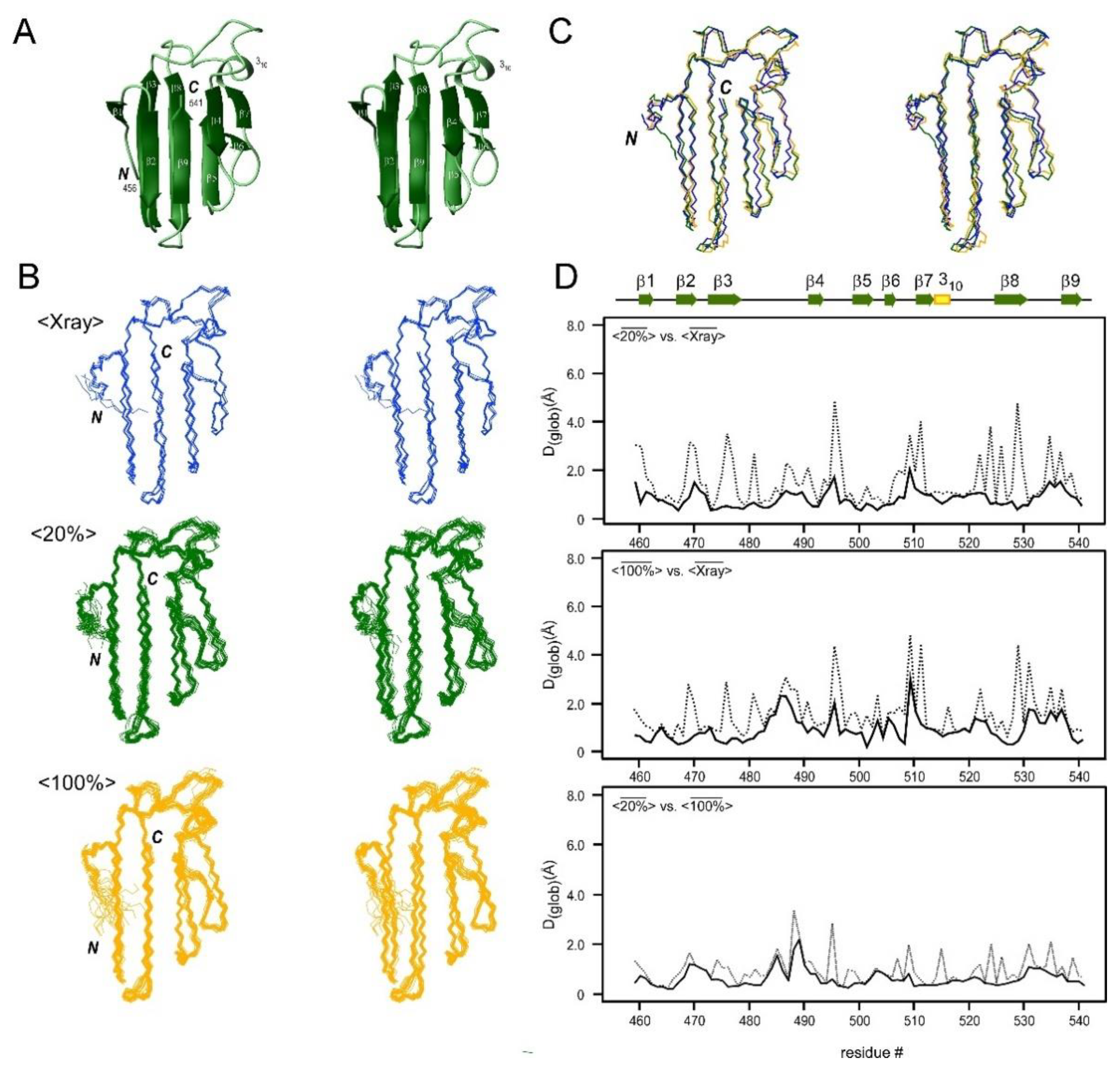
2.4. Comparison of the Structures Determined by Differently 13C-Labeled Samples of CBM64
2.5. Comparison of the NMR Structures with the Crystal Structures
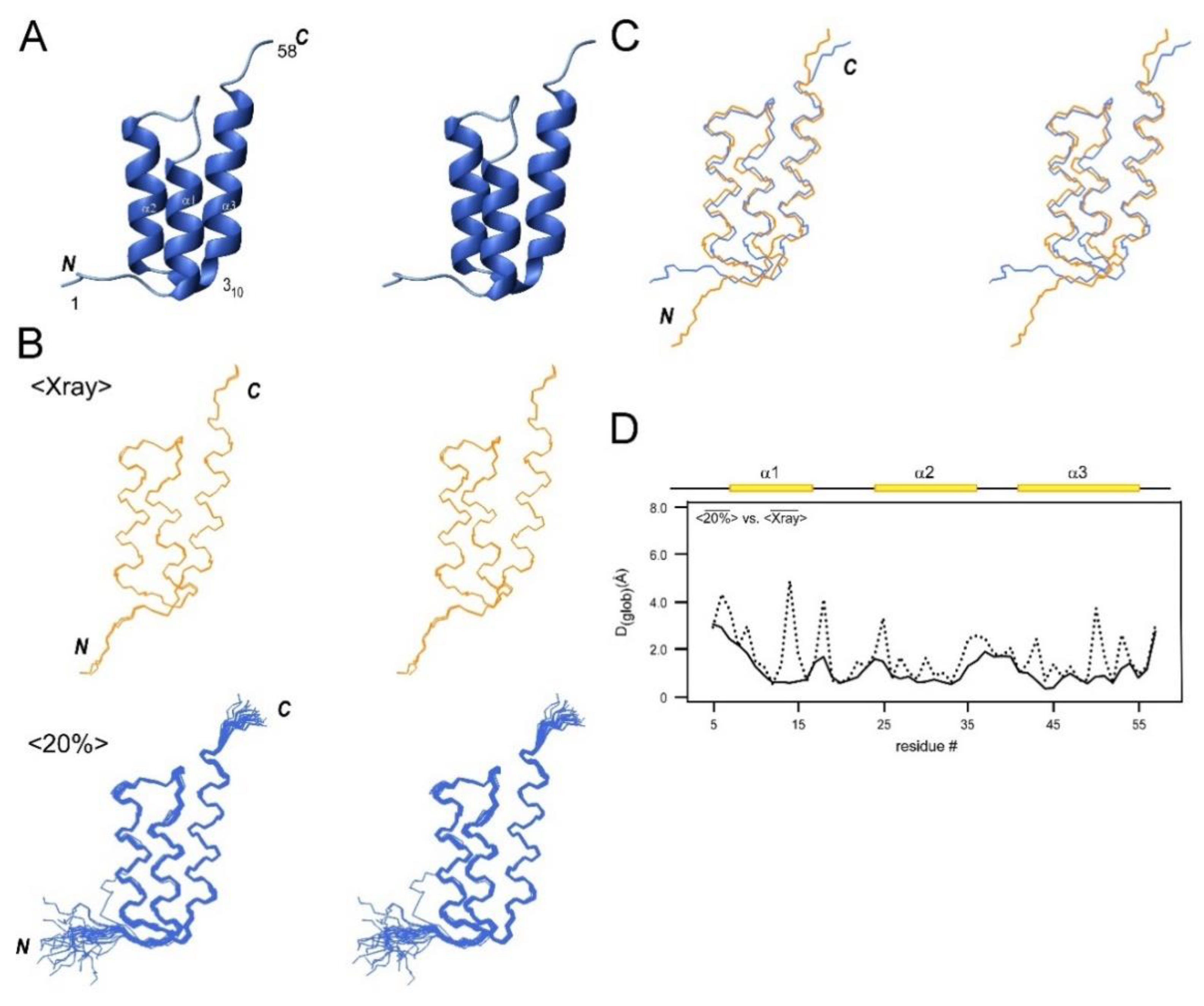
2.6. NMR Structures of a Helical Protein, SpaC, and Comparison with the Crystal Structures
2.7. Interaction Analysis Using Fractional 20% 13C-Labeled Sample by NMR
2.8. NMR Relaxation Analysis of the CBM64
3. Discussion
4. Materials and Methods
4.1. Cloning, Protein Production, and Purification for NMR Studies
4.2. Multidimensional NMR Spectroscopy
4.3. NMR Data Acquisition and Processing
4.4. NMR Structure Calculations
4.5. 15N-Nuclear Relaxation Analysis of CBM64
4.6. NMR Titrations of D-Cellobiose with CBM64
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Clore, G.M.; Gronenborn, A.M. Multidimensional heteronuclear-magnetic-resonance of proteins. Methods Enzymol. 1994, 239, 349–363. [Google Scholar] [PubMed]
- Wütrich, K. NMR of Proteins and Nucleic Acids, 1st ed.; Wiley: New York, NY, USA, 1986. [Google Scholar]
- Güntert, P. Automated NMR protein structure calculation. Prog. Nucl. Magn. Reson. Spectrosc. 2003, 43, 105–125. [Google Scholar] [CrossRef]
- Sakakibara, D.; Sasaki, A.; Ikeya, T.; Hamatsu, J.; Hanashima, T.; Mishima, M.; Yoshimasu, M.; Hayashi, N.; Mikawa, T.; Waelchli, M.; et al. Protein structure determination in living cells by in-cell NMR spectroscopy. Nat. Cell Biol. 2009, 458, 102–105. [Google Scholar] [CrossRef] [PubMed]
- Ikeya, T.; Sasaki, A.; Sakakibara, D.; Shigemitsu, Y.; Hamatsu, J.; Hanashima, T.; Mishima, M.; Yoshimasu, M.; Hayashi, N.; Mikawa, T.; et al. NMR protein structure determination in living E. coli cells using nonlinear sampling. Nat. Protoc. 2010, 5, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Wlodawer, A.; Minor, W.; Dauter, Z.; Jaskolski, M. Protein crystallography for non-crystallographers, or how to get the best (but not more) from published macromolecular structures. FEBS J. 2008, 275, 1–21. [Google Scholar] [CrossRef]
- Bartels, C.; Billeter, M. Automated sequence-specific NMR assignment of homologous proteins using the program GARANT. J. Biomol. NMR 1996, 7, 207–213. [Google Scholar] [CrossRef]
- Stratmann, D.; Guittet, E.; Van Heijenoort, C. Robust structure-based resonance assignment for functional protein studies by NMR. J. Biomol. NMR 2009, 46, 157–173. [Google Scholar] [CrossRef]
- Kovacs, H.; Moskau, D.; Spraul, M. Cryogenically cooled probes—a leap in NMR technology. Prog. Nucl. Magn. Reson. Spectrosc. 2005, 46, 131–155. [Google Scholar] [CrossRef]
- Iwai, H.; Fiaux, J. Use of biosynthetic fractional 13C-labeling for backbone NMR assignment of proteins. J. Biomol. NMR 2007, 37, 187–193. [Google Scholar] [CrossRef]
- Wenrich, B.R.; Sonstrom, R.E.; Gupta, R.A.; Rovnyak, D. Enhanced biosynthetically directed fractional carbon-13 enrichment of proteins for backbone NMR assignments. Protein Expr. Purif. 2015, 115, 1–10. [Google Scholar] [CrossRef][Green Version]
- Szyperski, T. Biosynthetically Directed Fractional 13C-labelling of Proteinogenic Amino Acids. Eur. J. Biochem. 1995, 232, 433–448. [Google Scholar] [CrossRef] [PubMed]
- Neri, D.; Szyperski, T.; Otting, G.; Senn, H.; Wüthrich, K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional carbon-13 labeling. Biochemistry 1989, 28, 7510–7516. [Google Scholar] [CrossRef]
- Hiroaki, H.; Klaus, W.; Senn, H. Determination of the solution structure of the SH3 domain of human p56 Lck tyrosine kinase. J. Biomol. NMR 1996, 8, 105–122. [Google Scholar] [CrossRef] [PubMed]
- Jacob, J.; Louis, J.M.; Nesheiwat, I.; Torchia, D.A. Biosynthetically directed fractional 13C labeling facilitates identification of Phe and Tyr aromatic signals in proteins. J. Biomol. NMR 2002, 24, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Tejero, R.; Montelione, G.T. Evaluating protein structures determined by structural genomics consortia. Proteins: Struct. Funct. Bioinform. 2006, 66, 778–795. [Google Scholar] [CrossRef]
- Schiefner, A.; Angelov, A.; Liebl, W.; Skerra, A. Structural basis for cellulose binding by the type A carbohydrate-binding module 64 ofSpirochaeta thermophila. Proteins Struct. Funct. Bioinform. 2016, 84, 855–858. [Google Scholar] [CrossRef] [PubMed]
- Pires, V.M.R.; Pereira, P.M.M.; Brás, J.L.A.; Correia, M.; Cardoso, V.; Bule, P.; Alves, V.D.; Najmudin, S.; Venditto, I.; Ferreira, L.M.A.; et al. Stability and Ligand Promiscuity of Type A Carbohydrate-binding Modules Are Illustrated by the Structure of Spirochaeta thermophila StCBM64C. J. Biol. Chem. 2017, 292, 4847–4860. [Google Scholar] [CrossRef]
- Clore, G.M.; Polenova, T. Structures of larger proteins in solution: Three- and four-dimensional heteronuclear NMR spectroscopy. Science 1991, 252, 1390–1399. [Google Scholar] [CrossRef]
- Koradi, R.; Billeter, M.; Wüthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996, 14, 51–55. [Google Scholar] [CrossRef]
- Deis, L.N.; Pemble, C.W.; Qi, Y.; Hagarman, A.; Richardson, D.C.; Richardson, J.S.; Oas, T.G. Multiscale Conformational Heterogeneity in Staphylococcal Protein A: Possible Determinant of Functional Plasticity. Structure 2014, 22, 1467–1477. [Google Scholar] [CrossRef]
- Gouda, H.; Torigoe, H.; Saito, A.; Sato, M.; Arata, Y.; Shimada, I. Three-dimensional solution structure of the B domain of staphylococcal protein A: Comparisons of the solution and crystal structures. Biochemistry 1992, 31, 9665–9672. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2008, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Stevens, R.C.; Yokoyama, S.; Wilson, I.A. Global Efforts in Structural Genomics. Science 2001, 294, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Khafizov, K.; Madrid-Aliste, C.; Almo, S.C.; Fiser, A. Trends in structural coverage of the protein universe and the impact of the Protein Structure Initiative. Proc. Natl. Acad. Sci. USA 2014, 111, 3733–3738. [Google Scholar] [CrossRef] [PubMed]
- Armenta, S.; Moreno-Mendieta, S.; Sánchez-Cuapio, Z.; Sanchez, S.; Rodríguez-Sanoja, R. Advances in molecular engineering of carbohydrate-binding modules. Proteins Struct. Funct. Bioinform. 2017, 85, 1602–1617. [Google Scholar] [CrossRef]
- Orekhov, V.Y.; Nolde, D.E.; Golovanov, A.P.; Korzhnev, D.M.; Arseniev, A.S. Processing of heteronuclear NMR relaxation data with the new software DASHA. Appl. Magn. Reson. 1995, 9, 581–588. [Google Scholar] [CrossRef]
- Cavanagh, J.; Fairbrother, W.; Palmer, A., III; Rance, M.; Skelton, N. Protein NMR Spectroscopy: Principles and Practice, 2nd ed.; Academic Press: Cambridge, MA, USA, 2007. [Google Scholar]
- NESG Wiki: NMR Determined Rotational Correlation Time. Available online: http://www.nmr2.buffalo.edu/nesg.wiki/NMR_determined_Rotational_correlation_time (accessed on 7 January 2021).
- Banci, L.; Barbieri, L.; Calderone, V.; Cantini, F.; Cerofolini, L.; Ciofi-Baffoni, S.; Felli, I.C.; Fragai, M.; Lelli, M.; Luchinat, C.; et al. Biomolecular NMR at 1.2 GHz. arXiv 2019, arXiv:1910.07462. (Preprint). [Google Scholar]
- Coughlin, P.E.; Anderson, F.E.; Oliver, E.J.; Brown, J.M.; Homans, S.W.; Pollak, S.; Lustbader, J.W. Improved resolution and sensitivity of triple-resonance NMR methods for the structural analysis of proteins by use of a backbone-labelling strategy. J. Am. Chem. Soc. 1999, 121, 11871–11874. [Google Scholar] [CrossRef]
- Busche, A.E.L.; Aranko, A.S.; Talebzadeh-Farooji, M.; Bernhard, F.; Dötsch, V.; Iwaï, H. Segmental Isotopic Labeling of a Central Domain in a Multidomain Protein by ProteinTrans-Splicing Using Only One Robust DnaE Intein. Angew. Chem. Int. Ed. 2009, 48, 6128–6131. [Google Scholar] [CrossRef]
- Guerrero, F.; Ciragan, A.; Iwaï, H. Tandem SUMO fusion vectors for improving soluble protein expression and purification. Protein Expr. Purif. 2015, 116, 42–49. [Google Scholar] [CrossRef]
- Sattler, M.; Schleucher, J.; Griesinger, C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrosc. 1999, 34, 93–158. [Google Scholar] [CrossRef]
- Lescop, E.; Schanda, P.; Brutscher, B. A set of BEST triple-resonance experiments for time-optimized protein resonance assignment. J. Magn. Reson. 2007, 187, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Bermel, W.; Bertini, I.; Duma, L.; Felli, I.C.; Emsley, L.; Pierattelli, R.; Vasos, P.R. Complete assignment of heteronuclear protein reso-nances by protonless NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2005, 44, 3089–3092. [Google Scholar] [CrossRef] [PubMed]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins Struct. Funct. Bioinform. 2005, 59, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Güntert, P.; Mumenthaler, C.; Wüthrich, K. Torsion angle dynamics for NMR structure calculation with the new program Dyana. J. Mol. Biol. 1997, 273, 283–298. [Google Scholar] [CrossRef]
- Güntert, P. Automated NMR protein structure calculation with CYANA. Methods Mol. Biol. 2004, 278, 353–378. [Google Scholar]
- Güntert, P. Automated structure determination from NMR spectra. Eur. Biophys. J. 2009, 38, 129–143. [Google Scholar] [CrossRef]
- Schmidt, E.; Güntert, P. A New Algorithm for Reliable and General NMR Resonance Assignment. J. Am. Chem. Soc. 2012, 134, 12817–12829. [Google Scholar] [CrossRef]
- Shen, Y.; Bax, A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 2013, 56, 227–241. [Google Scholar] [CrossRef]
- Cornilescu, G.; Delaglio, F.; Bax, A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 1999, 13, 289–302. [Google Scholar] [CrossRef]
- Cornell, W.D.; Cieplak, P.; Bayly, C.I.; Gould, I.R.; Merz, K.M.; Ferguson, D.M.; Spellmeyer, D.C.; Fox, T.; Caldwell, J.W.; Kollman, P.A. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc. 1995, 117, 5179–5197. [Google Scholar] [CrossRef]
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the worldwide Protein Data Bank. Nat. Struct. Mol. Biol. 2003, 10, 980. [Google Scholar] [CrossRef] [PubMed]
- Biological Magnetic Resonance Data Bank. Available online: http://bmrb.io/ (accessed on 28 January 2021).
- Farrow, N.A.; Muhandiram, R.; Singer, A.U.; Pascal, S.M.; Kay, C.M.; Gish, G.; Shoelson, S.E.; Pawson, T.; Forman-Kay, J.D.; Kay, L.E. Backbone Dynamics of a Free and a Phosphopeptide-Complexed Src Homology 2 Domain Studied by 15N NMR Relaxation. Biochemistry 1994, 33, 5984–6003. [Google Scholar] [CrossRef] [PubMed]
- Kay, L.E.; Torchia, D.A.; Bax, A. Backbone dynamics of proteins as studied by nitrogen-15 inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry 1989, 28, 8972–8979. [Google Scholar] [CrossRef] [PubMed]
- Fielding, L. NMR methods for the determination of protein–ligand dissociation constants. Prog. Nucl. Magn. Reson. Spectrosc. 2007, 51, 219–242. [Google Scholar] [CrossRef]
| Protein | CBM64 | CBM64 | SpaC |
|---|---|---|---|
| Completeness of resonance assignments (%) a | [20% 13C, 100% 15N] | 100 % [13C, 15N] | [20% 13C, 100% 15N] |
| Backbone | 96.9 | 96.9 | 99.1 |
| Side-chain Aromatic | 93.6 88.5 | 93.6 88.5 | 97.8 84.6 |
| Stereospecific methyl | 100 | 100 | 100 |
| Distance restraints | |||
| Total | 1396 | 1575 | 1163 |
| Sequential (|i − j| ≤ 1) | 680 | 752 | 544 |
| Medium range (1 < |i − j| < 5) | 178 | 196 | 357 |
| Long range (|i − j| ≥ 5) | 538 | 627 | 262 |
| No. of restraints per residue | 16.2 | 18.3 | 20.1 |
| No. of long-range restraints per residue | 6.3 | 7.3 | 4.5 |
| Residual restraint violations | |||
| Average no. of distance violation per structure | |||
| 0.1–0.2 Å | 5.5 | 6.7 | 8.3 |
| >0.2 Å | 0 (max 0.20) | 0.2 (max 0.26) | 0.3 (max 0.25) |
| No. of dihedral angle violations per structure >5° | 0 | 0 | 0 |
| Model quality b | |||
| Rmsd backbone atoms (Å) | 0.4 | 0.4 | 0.3 |
| Rmsd heavy atoms (Å) | 0.8 | 0.9 | 0.7 |
| Rmsd bond lengths (Å) | 0.015 | 0.015 | 0.013 |
| Rmsd bond angles (°) | 2.2 | 2.1 | 2.0 |
| MolProbity Ramachandran statistics b | |||
| Most favored regions (%) | 95.6 | 95.4 | 98.5 |
| Allowed regions (%) | 4.3 | 2.3 | 1.5 |
| Disallowed regions (%) | 0.1 | 2.3 | 0 |
| Global quality scores (raw/Z score) b | |||
| Verify3D | 0.41/−0.80 | 0.42/−0.64 | 0.35/−1.77 |
| ProsaII | 0.39/−1.08 | 0.43/−0.91 | 1.23/2.40 |
| PROCHECK (φ − ψ) | −0.51/−1.69 | −0.49/−1.61 | 0.18/1.02 |
| PROCHECK (all) | −0.59/−3.49 | −0.58/−3.43 | −0.00/−0.47 |
| MolProbity clash score | 1.20/1.32 | 2.95/1.02 | 1.51/1.27 |
| Model contents | |||
| Ordered residue ranges | 459−541 | 459−541 | 5−57 |
| Total no. of residues | 86 | 86 | 58 |
| BMRB accession number | 34229 | 34227 | 34430 |
| PDB ID code | 6FFU | 6FFQ | 6SOW |
| RMSD (Å) for Residues 459–541 of CBM64 a | |||
|---|---|---|---|
| Mean | <Xray> | <20%> | <100%> |
| 0.23 ± 0.05 (0.53 ± 0.21) | 0.97 ± 0.09 (2.12 ± 0.10) | 1.11 ± 0.13 (2.04 ± 0.12) | |
| 0.94 ± 0.05 (2.00 ± 0.07) | 0.37 ± 0.09 (0.77 ± 0.11) | 0.98 ± 0.15 (1.70 ± 0.12) | |
| 1.23 ± 0.07 (1.97 ± 0.09) | 0.93 ± 0.10 (1.65 ± 0.11) | 0.42 ± 0.13 (0.81 ± 0.21) | |
| RMSD (Å) for Residues 5–57 of SpaC a | ||
|---|---|---|
| Mean | <Xray> | <20%> |
| 0.09 ± 0.00 (0.30 ± 0.00) | 1.34 ± 0.13 (2.23 ± 0.14) | |
| 1.32 ± 0.02 (2.14 ± 0.04) | 0.30 ± 0.06 (0.68 ± 0.10) | |
Sample Availability: Plasmids are available from www.addgene.org/Hideo_Iwai with ID# 165159 and 165160. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heikkinen, H.A.; Backlund, S.M.; Iwaï, H. NMR Structure Determinations of Small Proteins Using only One Fractionally 20% 13C- and Uniformly 100% 15N-Labeled Sample. Molecules 2021, 26, 747. https://doi.org/10.3390/molecules26030747
Heikkinen HA, Backlund SM, Iwaï H. NMR Structure Determinations of Small Proteins Using only One Fractionally 20% 13C- and Uniformly 100% 15N-Labeled Sample. Molecules. 2021; 26(3):747. https://doi.org/10.3390/molecules26030747
Chicago/Turabian StyleHeikkinen, Harri A., Sofia M. Backlund, and Hideo Iwaï. 2021. "NMR Structure Determinations of Small Proteins Using only One Fractionally 20% 13C- and Uniformly 100% 15N-Labeled Sample" Molecules 26, no. 3: 747. https://doi.org/10.3390/molecules26030747
APA StyleHeikkinen, H. A., Backlund, S. M., & Iwaï, H. (2021). NMR Structure Determinations of Small Proteins Using only One Fractionally 20% 13C- and Uniformly 100% 15N-Labeled Sample. Molecules, 26(3), 747. https://doi.org/10.3390/molecules26030747






