Visible Light Photochemical Reactions for Nucleic Acid-Based Technologies
Abstract
1. Introduction
2. Nucleic Acid Strand Photocleavage
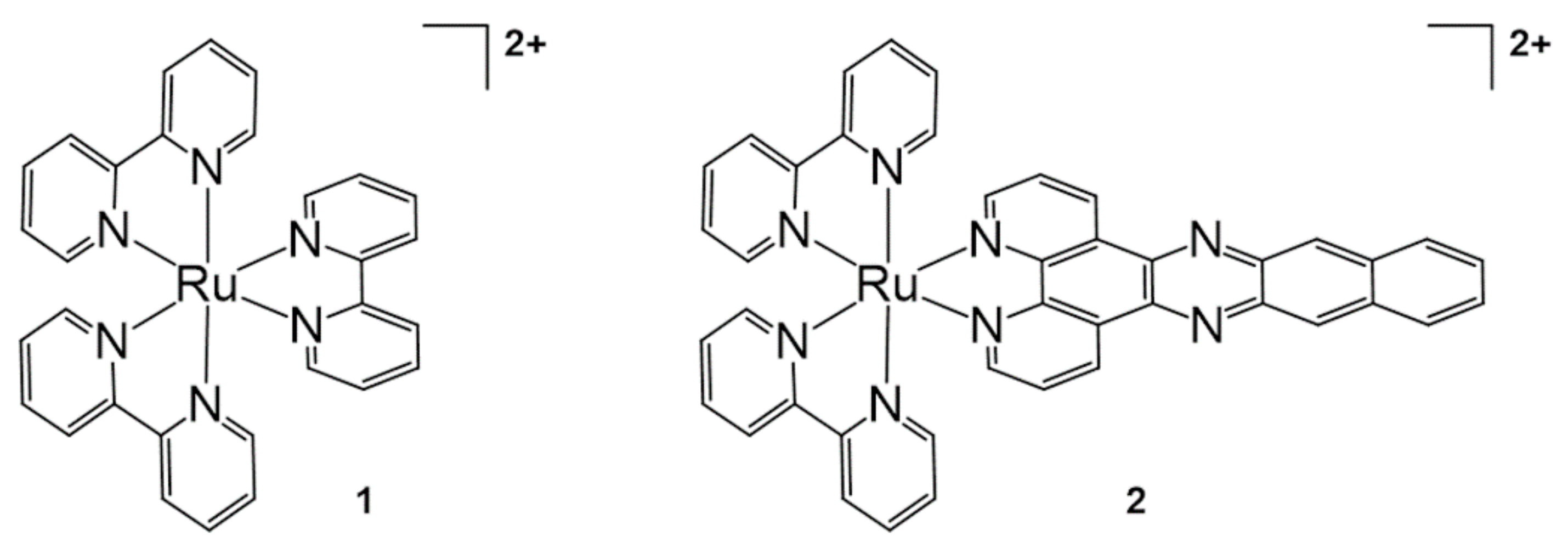
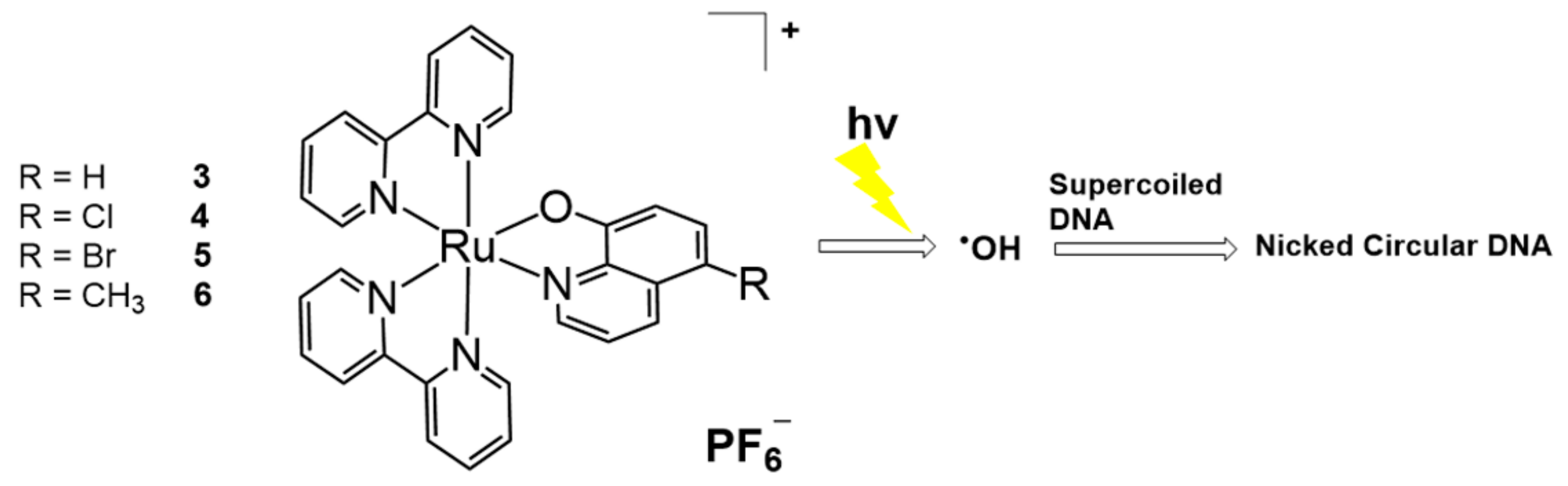
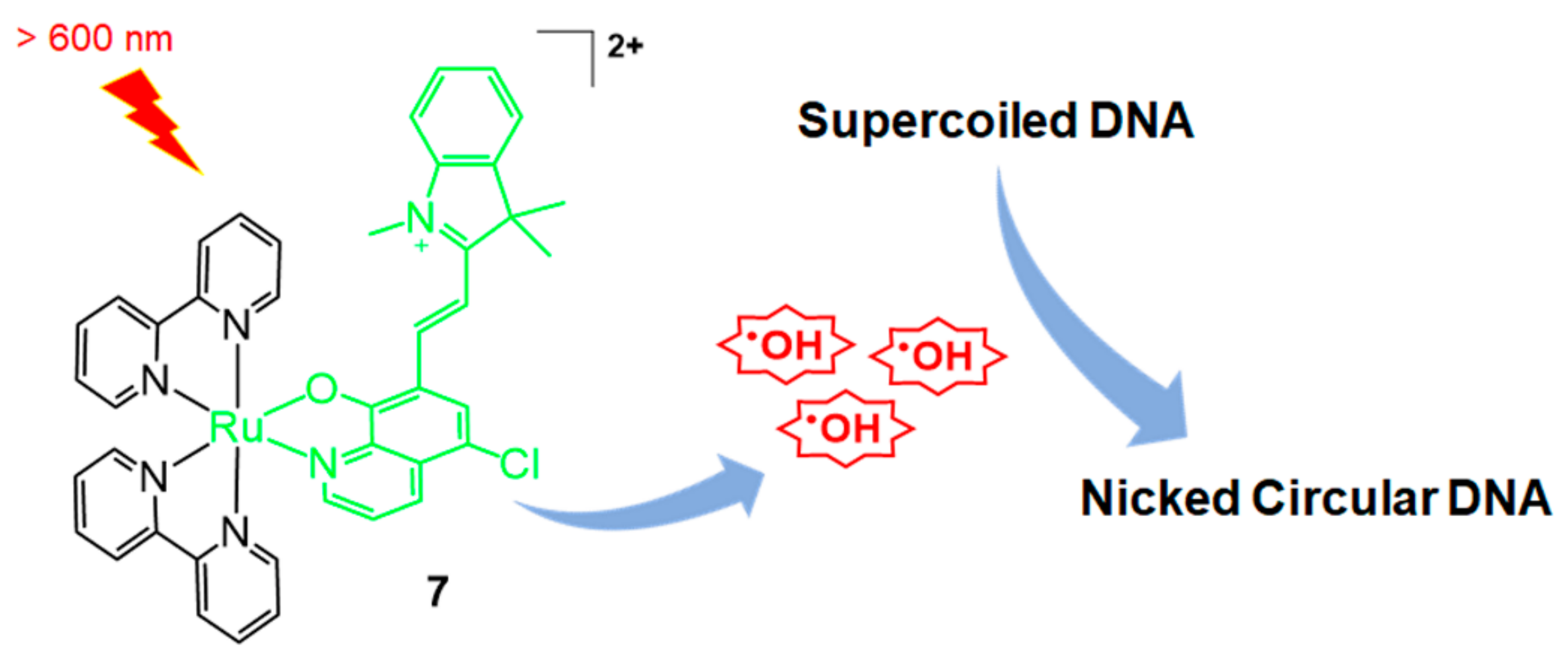
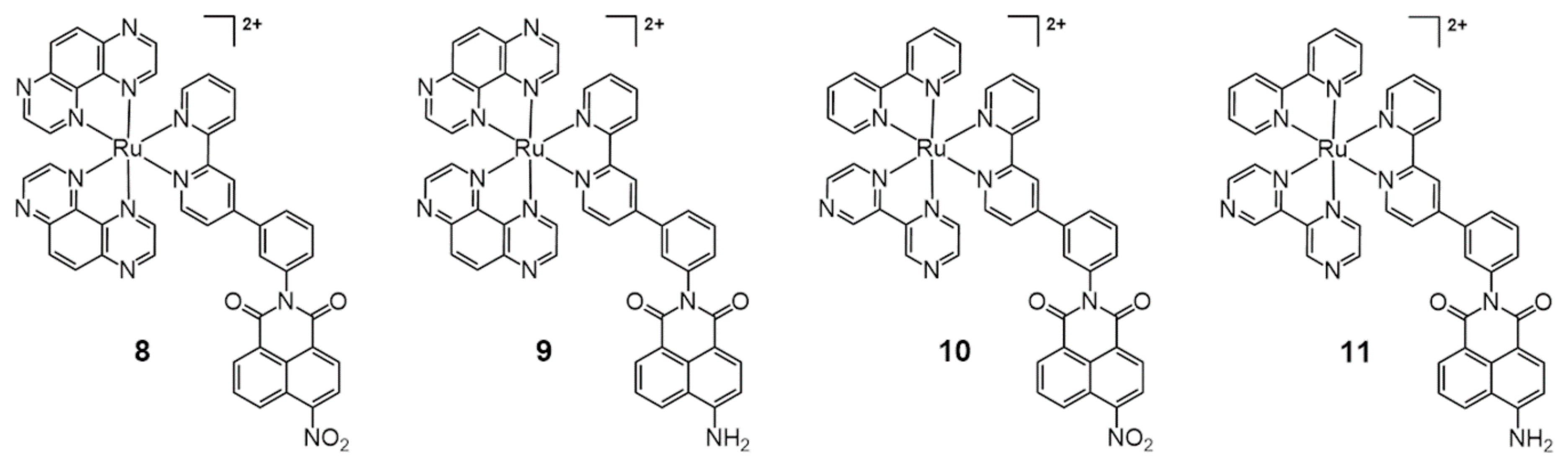
2.1. Ru Complex-Based Methods
2.2. Methods Based on Other Photosensitizers
3. Nucleic Acid Strand Photoligation
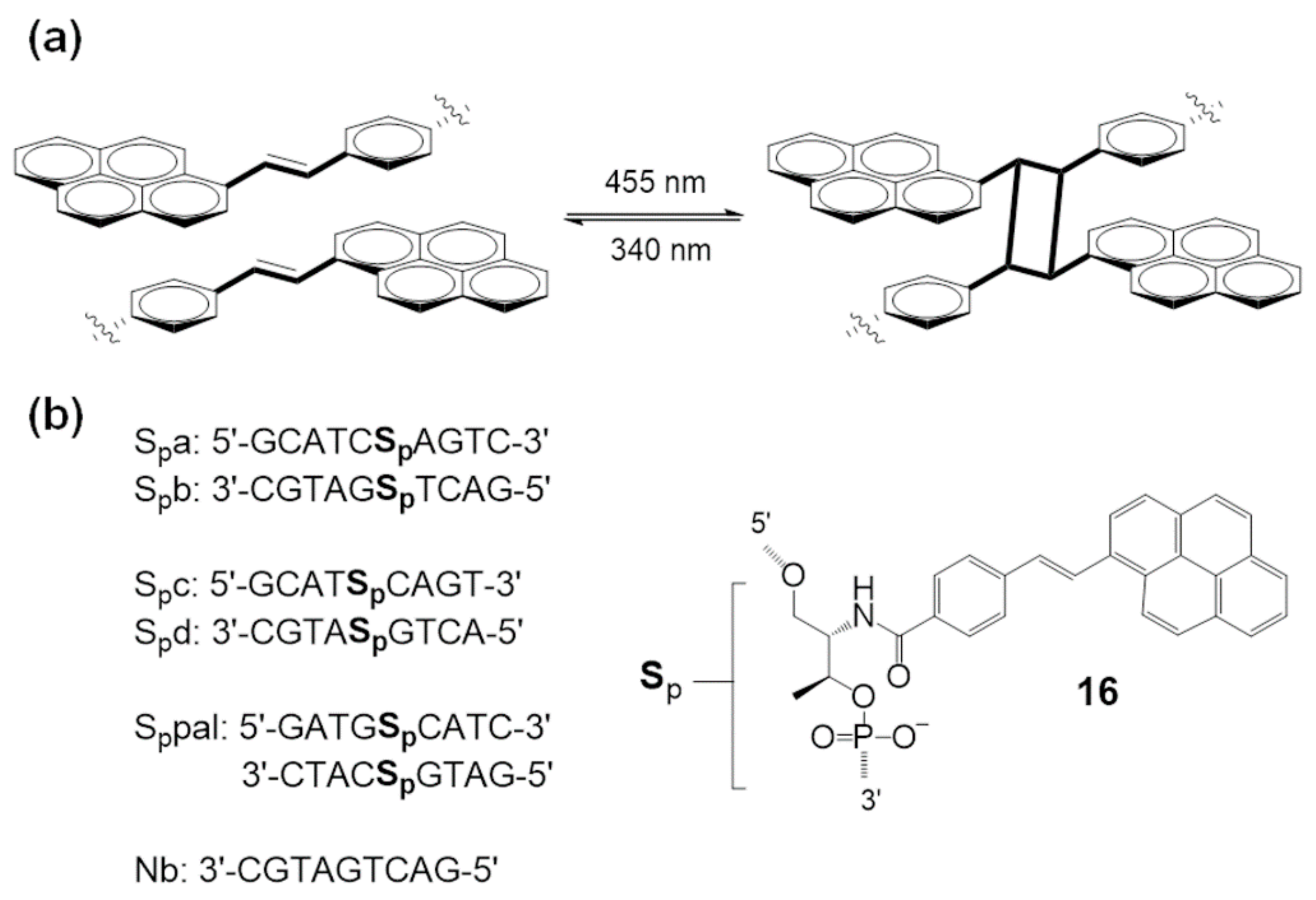
3.1. Photo-Cross-Linking Based on [2 + 2] Photocycloaddition Reaction
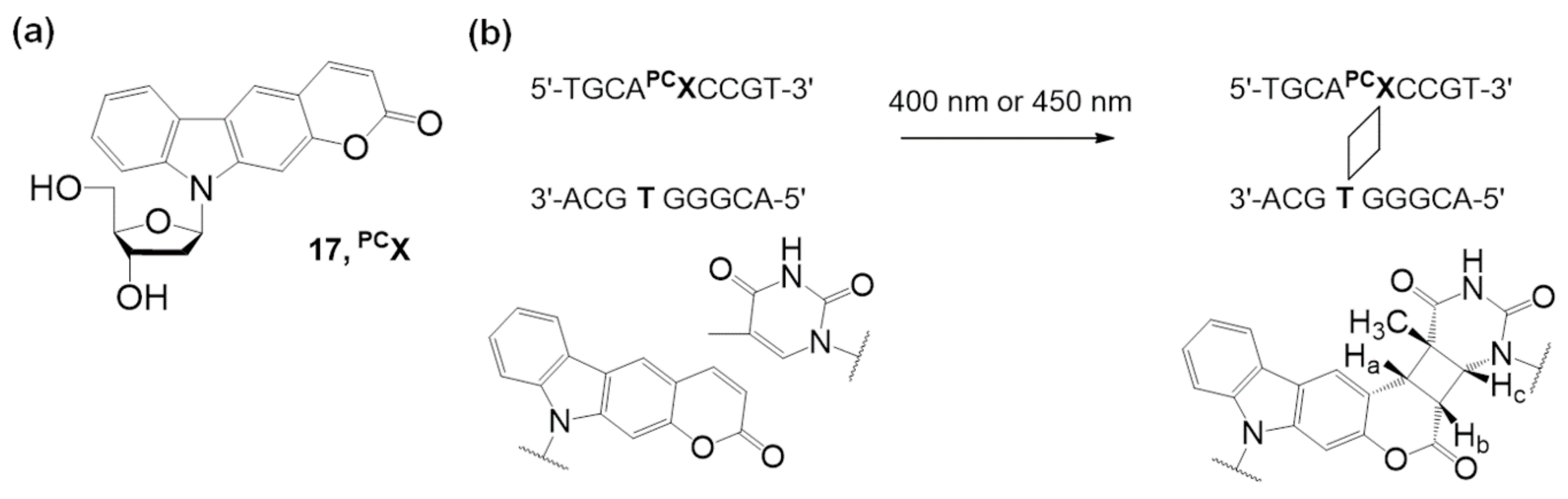
3.2. Other Photo-Cross-Linking Methods
4. Chemical Bond Photocleavage of Nucleic Acids
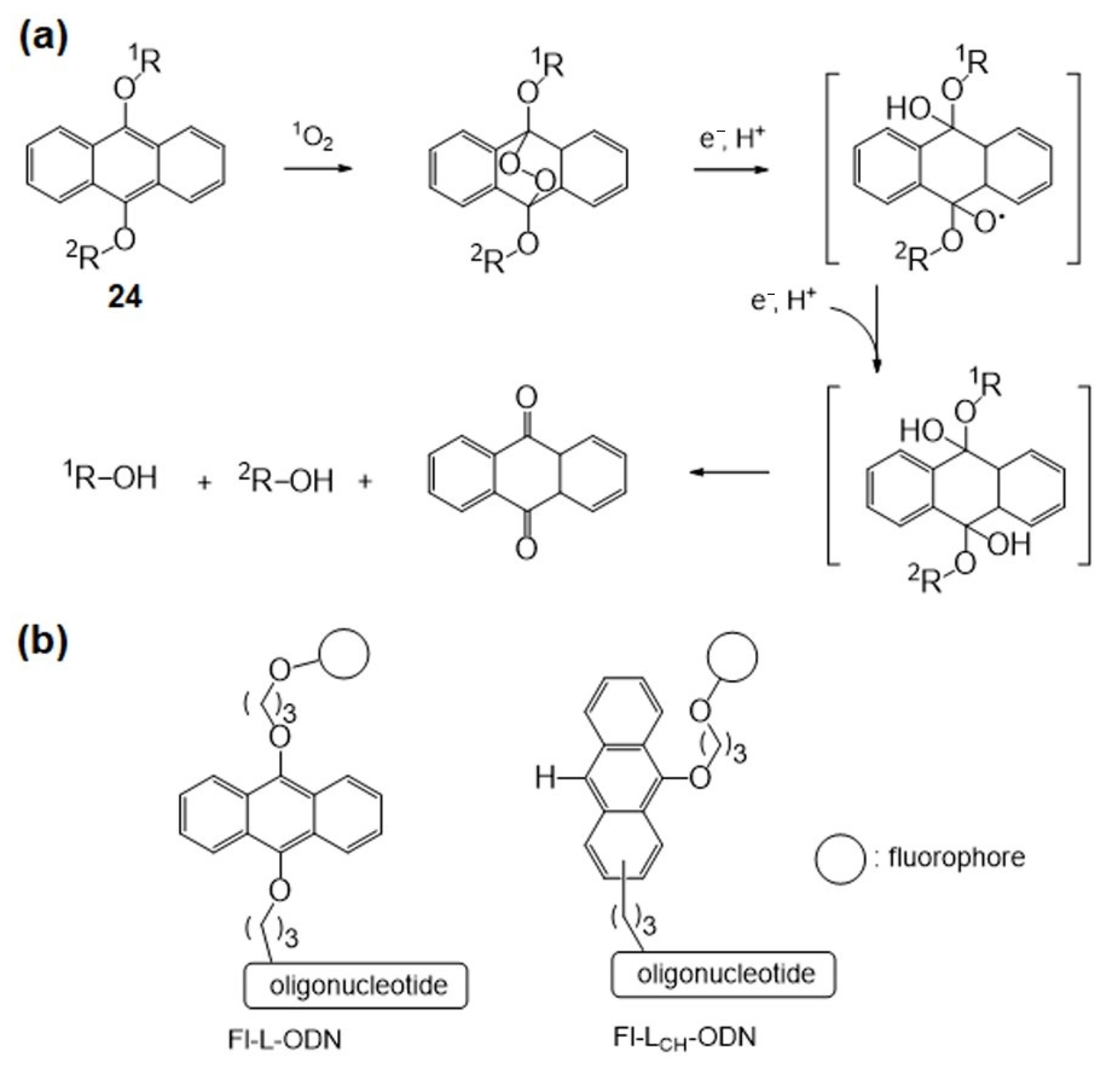
4.1. Singlet Oxygen-Mediated Fluorogenic Reaction
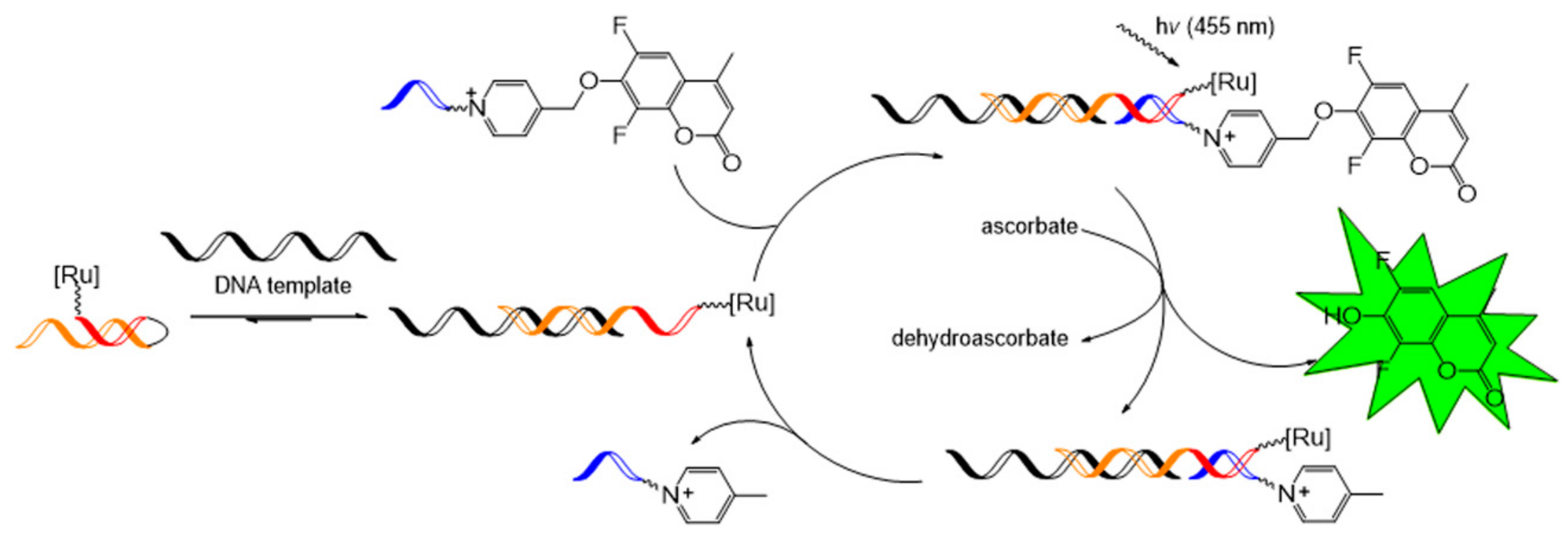
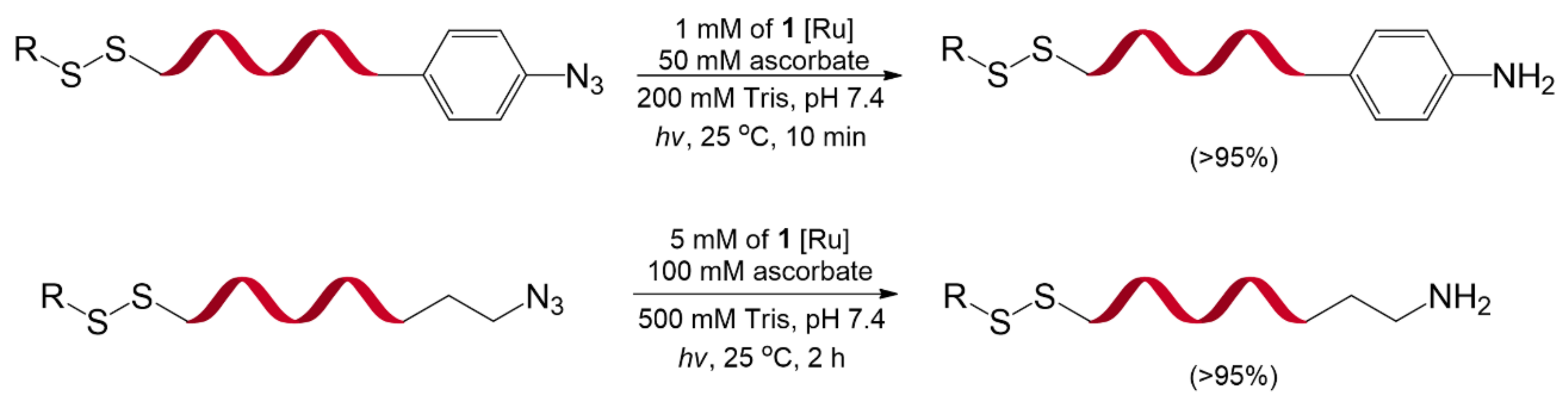
4.2. Ru-Catalyzed Photoreduction
5. Chemical Bond Formation in Nucleic Acids
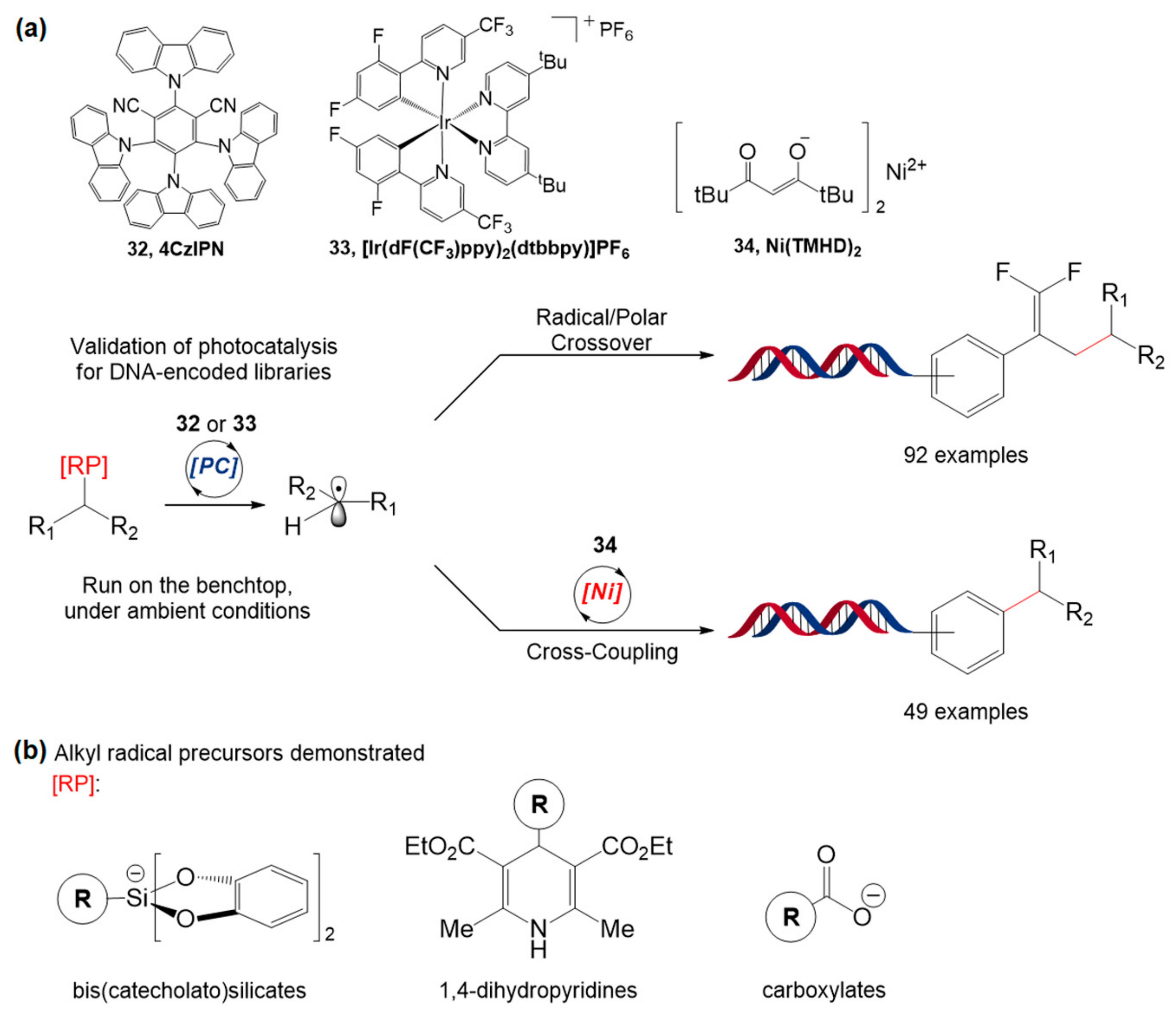
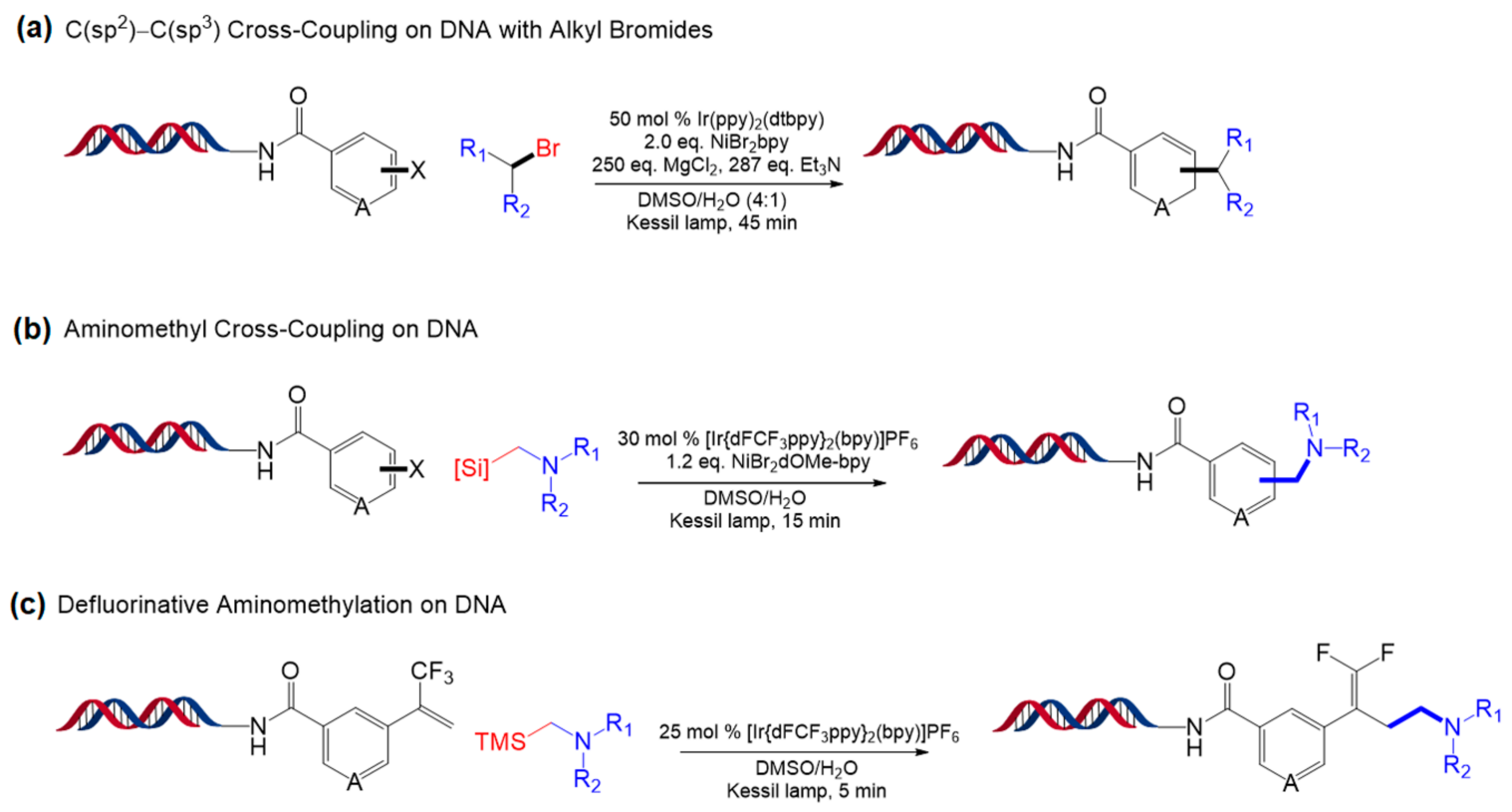
5.1. Ir-Based Photocatalytic Reactions
5.2. Other Photocatalytic Reactions
6. Conclusions and Outlook
Author Contributions
Funding
Conflicts of Interest
Data Availability
References
- Hershey, A.D.; Chase, M. Independent Functions of Viral Protein and Nucleic Acid in Growth of Bacteriophage. J. Gen. Physiol. 1952, 36, 39–56. [Google Scholar] [CrossRef] [PubMed]
- S Khan, A. Rapid Advances in Nucleic Acid Technologies for Detection and Diagnostics of Pathogens. J. Microbiol. Exp. 2014, 1, 56–61. [Google Scholar] [CrossRef][Green Version]
- Levsky, J.M. Fluorescence in situ hybridization: Past, present and future. J. Cell Sci. 2003, 116, 2833–2838. [Google Scholar] [CrossRef] [PubMed]
- Saiki, R.K.; Gelfand, D.H.; Stoffel, S.; Scharf, S.J.; Higuchi, R.; Horn, G.T.; Mullis, K.B.; Erlich, H.A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 1988, 239, 487–491. [Google Scholar] [CrossRef]
- Zhao, Y.X.; Chen, F.; Li, Q.; Wang, L.H.; Fan, C.H. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef]
- Shendure, J.; Balasubramanian, S.; Church, G.M.; Gilbert, W.; Rogers, J.; Schloss, J.A.; Waterston, R.H. DNA sequencing at 40: Past, present and future. Nature 2017, 550, 345–353. [Google Scholar] [CrossRef]
- Tomkinson, A.E.; Vijayakumar, S.; Pascal, J.M.; Ellenberger, T. DNA Ligases: Structure, Reaction Mechanism, and Function. Chem. Rev. 2006, 106, 687–699. [Google Scholar] [CrossRef]
- Li, X.Y.; Liu, D.R. DNA-Templated organic synthesis: Nature’s strategy for controlling chemical reactivity applied to synthetic molecules. Angew. Chem. Int. Ed. 2004, 43, 4848–4870. [Google Scholar] [CrossRef]
- Di Pisa, M.; Seitz, O. Nucleic Acid Templated Reactions for Chemical Biology. ChemMedChem 2017, 12, 872–882. [Google Scholar] [CrossRef]
- Silverman, A.P.; Kool, E.T. Detecting RNA and DNA with templated chemical reactions. Chem. Rev. 2006, 106, 3775–3789. [Google Scholar] [CrossRef]
- Seitz, O. Templated chemistry for bioorganic synthesis and chemical biology. J. Pep. Sci. 2019, 25, e3198. [Google Scholar] [CrossRef] [PubMed]
- Michaelis, J.; Roloff, A.; Seitz, O. Amplification by nucleic acid-templated reactions. Org. Biomol. Chem. 2014, 12, 2821–2833. [Google Scholar] [CrossRef] [PubMed]
- Madsen, M.; Gothelf, K.V. Chemistries for DNA Nanotechnology. Chem. Rev. 2019, 119, 6384–6458. [Google Scholar] [CrossRef] [PubMed]
- Wilner, O.I.; Willner, I. Functionalized DNA Nanostructures. Chem. Rev. 2012, 112, 2528–2556. [Google Scholar] [CrossRef]
- Pavagada, S.; Ladame, S. Platforms for Bioorthogonal Oligonucleotide-templated Reactions: Translating Concepts into Devices. Chim. Int. J. Chem. 2018, 72, 809–814. [Google Scholar] [CrossRef] [PubMed]
- Percivalle, C.; Bartolo, J.F.; Ladame, S. Oligonucleotide-templated chemical reactions: Pushing the boundaries of a nature-inspired process. Org. Biomol. Chem. 2013, 11, 16–26. [Google Scholar] [CrossRef]
- Gorska, K.; Winssinger, N. Reactions Templated by Nucleic Acids: More Ways to Translate Oligonucleotide-Based Instructions into Emerging Function. Angew. Chem. Int. Ed. 2013, 52, 6820–6843. [Google Scholar] [CrossRef]
- Ihara, T.; Fujii, T.; Mukae, M.; Kitamura, Y.; Jyo, A. Photochemical ligation of DNA conjugates through anthracene cyclodimer formation and its fidelity to the template sequences. J. Am. Chem. Soc. 2004, 126, 8880–8881. [Google Scholar] [CrossRef]
- Dall‘Acqua, F.; Vedaldi, D.; Recher, M. The Photoreaction between Furocoumarins and Various DNA with Different Base Compositions. Photochem. Photobiol. 1978, 27, 33–36. [Google Scholar] [CrossRef]
- Dallacqua, F.; Magno, S.M.; Zambon, F.; Rodighiero, G. Kinetic-Analysis of the Photoreaction (365nm) between Psoralen and DNA. Photochem. Photobiol. 1979, 29, 489–495. [Google Scholar] [CrossRef]
- Lewis, R.J.; Hanawalt, P.C. Ligation of oligonucleotides by pyrimidine dimers—a missing ‘link’ in the origin of life? Nature 1982, 298, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Schultz, D.M.; Yoon, T.P. Solar Synthesis: Prospects in Visible Light Photocatalysis. Science 2014, 343, 1239176. [Google Scholar] [CrossRef] [PubMed]
- Angerani, S.; Winssinger, N. Visible Light Photoredox Catalysis Using Ruthenium Complexes in Chemical Biology. Chem. Eur. J. 2019, 25, 6661–6672. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wu, W.; Sun, J.; Guo, S. Triplet photosensitizers: From molecular design to applications. Chem. Soc. Rev. 2013, 42, 5323–5351. [Google Scholar] [CrossRef] [PubMed]
- Benov, L. Photodynamic therapy: Current status and future directions. Med. Princ. Pract. 2015, 24, 14–28. [Google Scholar] [CrossRef] [PubMed]
- Armitage, B. Photocleavage of Nucleic Acids. Chem. Rev. 1998, 98, 1171–1200. [Google Scholar] [CrossRef]
- Detty, M.R.; Gibson, S.L.; Wagner, S.J. Current Clinical and Preclinical Photosensitizers for Use in Photodynamic Therapy. J. Med. Chem. 2004, 47, 3897–3915. [Google Scholar] [CrossRef]
- Liu, Y.; Hammitt, R.; Lutterman, D.A.; Joyce, L.E.; Thummel, R.P.; Turro, C. Ru(II) complexes of new tridentate ligands: Unexpected high yield of sensitized 1O2. Inorg. Chem. 2009, 48, 375–385. [Google Scholar] [CrossRef]
- Mari, C.; Pierroz, V.; Ferrari, S.; Gasser, G. Combination of Ru(II) complexes and light: New frontiers in cancer therapy. Chem. Sci. 2015, 6, 2660–2686. [Google Scholar] [CrossRef]
- Sun, Y.; Joyce, L.E.; Dickson, N.M.; Turro, C. Efficient DNA photocleavage by [Ru(bpy)2(dppn)]2+ with visible light. Chem. Commun. (Camb.) 2010, 46, 2426–2428. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, Q.; Zheng, Y.; Li, K.; Jiang, G.; Hou, Y.; Zhang, B.; Wang, X. DNA Photocleavage by Non-innocent Ligand-Based Ru(II) Complexes. Inorg. Chem. 2016, 55, 4296–4300. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, Q.; Tian, N.; Li, C.; Wang, X. Ru(II)-Complex-Based DNA Photocleaver Having Intense Absorption in the Phototherapeutic Window. Inorg. Chem. 2017, 56, 1865–1873. [Google Scholar] [CrossRef] [PubMed]
- Elmes, R.B.P.; Ryan, G.J.; Erby, M.L.; Frimannsson, D.O.; Kitchen, J.A.; Lawler, M.; Williams, D.C.; Quinn, S.J.; Gunnlaugsson, T. Synthesis, Characterization, and Biological Profiling of Ruthenium(II)-Based 4-Nitro- and 4-Amino-1,8-naphthalimide Conjugates. Inorg. Chem. 2020, 59, 10874–10893. [Google Scholar] [CrossRef] [PubMed]
- Ryan, G.J.; Quinn, S.; Gunnlaugsson, T. Highly effective DNA photocleavage by novel “rigid” Ru(bpy)3-4-nitro- and -4-amino-1,8-naphthalimide conjugates. Inorg. Chem. 2008, 47, 401–403. [Google Scholar] [CrossRef]
- Cheng, N.; Chen, Y.; Yu, J.; Li, J.J.; Liu, Y. Enhanced DNA Binding and Photocleavage Abilities of beta-Cyclodextrin Appended Ru(II) Complex through Supramolecular Strategy. Bioconj. Chem. 2018, 29, 1829–1833. [Google Scholar] [CrossRef]
- Zeng, L.; Gupta, P.; Chen, Y.; Wang, E.; Ji, L.; Chao, H.; Chen, Z.S. The development of anticancer ruthenium(ii) complexes: From single molecule compounds to nanomaterials. Chem. Soc. Rev. 2017, 46, 5771–5804. [Google Scholar] [CrossRef]
- Weynand, J.; Moreno-Betancourt, A.; Loiseau, F.; Berthet, N.; Defrancq, E.; Elias, B. Redox-Active Bis-Cyclometalated Iridium(III) Complex as a DNA Photo-Cleaving Agent. Inorg. Chem. 2020, 59, 2426–2433. [Google Scholar] [CrossRef]
- Weynand, J.; Bonnet, H.; Loiseau, F.; Ravanat, J.L.; Dejeu, J.; Defrancq, E.; Elias, B. Targeting G-Rich DNA Structures with Photoreactive Bis-Cyclometallated Iridium(III) Complexes. Chemistry 2019, 25, 12730–12739. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhao, Y.; Chen, L.; Wu, J.; Chen, G.; Li, S.; Zou, J.; Chen, R.; Wang, J.; Jiang, F.; et al. Selective tumor cell death induced by irradiated riboflavin through recognizing DNA G-T mismatch. Nucleic Acids Res. 2017, 45, 8676–8683. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, G.; Yuan, Y.; Li, N.; Dong, J.; Huang, X.; Cui, X.; Tang, Z. Sequence-specific RNA Photocleavage by Single-stranded DNA in Presence of Riboflavin. Sci. Rep. 2015, 5, 15039. [Google Scholar] [CrossRef]
- Zhou, J.; Fang, C.; Liu, Y.; Zhao, Y.; Zhang, N.; Liu, X.; Wang, F.; Shangguan, D. Visible-light-induced cleavage of 4-alpha-amino acid substituted naphthalimides and its application in DNA photocleavage. Org. Biomol. Chem. 2015, 13, 3931–3935. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhang, Y.-M.; Li, P.-Y.; Liu, Y. Efficient energy transfer between coronene-modified permethyl-β-cyclodextrins and porphyrin for light induced DNA cleavage. Chem. Commun. (Camb.) 2017, 53, 3717–3720. [Google Scholar] [CrossRef] [PubMed]
- Ahoulou, E.O.; Drinkard, K.K.; Basnet, K.; St Lorenz, A.; Taratula, O.; Henary, M.; Grant, K.B. DNA Photocleavage in the Near-Infrared Wavelength Range by 2-Quinolinium Dicarbocyanine Dyes. Molecules 2020, 25, 2926. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Browne, W.R.; Roelfes, G. DNA Cleavage Activity of Fe(II)N4Py under Photo Irradiation in the Presence of 1,8-Naphthalimide and 9-Aminoacridine: Unexpected Effects of Reactive Oxygen Species Scavengers. Inorg. Chem. 2011, 50, 8318–8325. [Google Scholar] [CrossRef]
- Dąbrowski, J.M.; Pucelik, B.; Regiel-Futyra, A.; Brindell, M.; Mazuryk, O.; Kyzioł, A.; Stochel, G.; Macyk, W.; Arnaut, L.G. Engineering of relevant photodynamic processes through structural modifications of metallotetrapyrrolic photosensitizers. Coord. Chem. Rev. 2016, 325, 67–101. [Google Scholar] [CrossRef]
- Kuncewicz, J.; Dąbrowski, J.M.; Kyzioł, A.; Brindell, M.; Łabuz, P.; Mazuryk, O.; Macyk, W.; Stochel, G. Perspectives of molecular and nanostructured systems with d- and f-block metals in photogeneration of reactive oxygen species for medical strategies. Coord. Chem. Rev. 2019, 398, 113012. [Google Scholar] [CrossRef]
- Meisenheimer, K.M.; Koch, T.H. Photocross-linking of nucleic acids to associated proteins. Crit. Rev. Biochem. Mol. Biol. 1997, 32, 101–140. [Google Scholar] [CrossRef]
- Noll, D.M.; Mason, T.M.; Miller, P.S. Formation and repair of interstrand cross-links in DNA. Chem. Rev. 2006, 106, 277–301. [Google Scholar] [CrossRef]
- Gasparro, F.P.; Havre, P.A.; Olack, G.A.; Gunther, E.J.; Glazer, P.M. Site-specific targeting of psoralen photoadducts with a triple helix-forming oligonucleotide: Characterization of psoralen monoadduct and crosslink formation. Nucleic Acids Res. 1994, 22, 2845–2852. [Google Scholar] [CrossRef][Green Version]
- Fujimoto, K.; Matsuda, S.; Takahashi, N.; Saito, I. Template-directed photoreversible ligation of deoxyoligonucleotides via 5-vinyldeoxyuridine. J. Am. Chem. Soc. 2000, 122, 5646–5647. [Google Scholar] [CrossRef]
- Sakamoto, T.; Ooe, M.; Fujimoto, K. Critical Effect of Base Pairing of Target Pyrimidine on the Interstrand Photo-Cross-Linking of DNA via 3-Cyanovinylcarbazole Nucleoside. Bioconj. Chem. 2015, 26, 1475–1478. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Tanaka, Y.; Fujimoto, K. DNA photo-cross-linking using 3-cyanovinylcarbazole modified oligonucleotide with threoninol linker. Org. Lett. 2015, 17, 936–939. [Google Scholar] [CrossRef] [PubMed]
- Harimech, P.K.; Gerrard, S.R.; El-Sagheer, A.H.; Brown, T.; Kanaras, A.G. Reversible Ligation of Programmed DNA-Gold Nanoparticle Assemblies. J. Am. Chem. Soc. 2015, 137, 9242–9245. [Google Scholar] [CrossRef] [PubMed]
- Ogino, M.; Taya, Y.; Fujimoto, K. Highly selective detection of 5-methylcytosine using photochemical ligation. Chem. Commun. (Camb.) 2008, 45, 5996–5998. [Google Scholar] [CrossRef]
- Sun, H.; Fan, H.; Peng, X. Quantitative DNA interstrand cross-link formation by coumarin and thymine: Structure determination, sequence effect, and fluorescence detection. J. Org. Chem. 2014, 79, 11359–11369. [Google Scholar] [CrossRef]
- Albagli, D.; Van Atta, R.; Cheng, P.; Huan, B.F.; Wood, M.L. Chemical amplification (CHAMP) by a continuous, self-replicating oligonucleotide-based system. J. Am. Chem. Soc. 1999, 121, 6954–6955. [Google Scholar] [CrossRef]
- Kashida, H.; Doi, T.; Sakakibara, T.; Hayashi, T.; Asanuma, H. p-Stilbazole moieties as artificial base pairs for photo-cross-linking of DNA duplex. J. Am. Chem. Soc. 2013, 135, 7960–7966. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, S.; Lin, Z.; Fan, Y.; Wang, Y.; Peng, X. Photochemical Generation of Benzyl Cations That Selectively Cross-Link Guanine and Cytosine in DNA. Org. Lett. 2016, 18, 2544–2547. [Google Scholar] [CrossRef]
- Lin, Z.; Fan, H.; Zhang, Q.; Peng, X. Design, Synthesis, and Characterization of Binaphthalene Precursors as Photoactivated DNA Interstrand Cross-Linkers. J. Org. Chem. 2018, 83, 8815–8826. [Google Scholar] [CrossRef]
- Kamiya, Y.; Takagi, T.; Ooi, H.; Ito, H.; Liang, X.; Asanuma, H. Synthetic gene involving azobenzene-tethered T7 promoter for the photocontrol of gene expression by visible light. ACS Synth. Biol. 2015, 4, 365–370. [Google Scholar] [CrossRef]
- Doi, T.; Kawai, H.; Murayama, K.; Kashida, H.; Asanuma, H. Visible-Light-Triggered Cross-Linking of DNA Duplexes by Reversible [2 + 2] Photocycloaddition of Styrylpyrene. Chemistry 2016, 22, 10533–10538. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, K.; Sasago, S.; Mihara, J.; Nakamura, S. DNA Photo-cross-linking Using Pyranocarbazole and Visible Light. Org. Lett. 2018, 20, 2802–2805. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, K.; Yamaguchi, T.; Inatsugi, T.; Takamura, M.; Ishimaru, I.; Koto, A.; Nakamura, S. DNA photo-cross-linking using a pyranocarbazole-modified oligodeoxynucleotide with a D-threoninol linker. RSC Adv. 2019, 9, 30693–30697. [Google Scholar] [CrossRef]
- Onizuka, K.; Ishida, K.; Mano, E.; Nagatsugi, F. Alkyne-Alkyne Photo-cross-linking on the Flipping-out Field. Org. Lett. 2019, 21, 2833–2837. [Google Scholar] [CrossRef] [PubMed]
- Manicardi, A.; Cadoni, E.; Madder, A. Visible-light triggered templated ligation on surface using furan-modified PNAs. Chem. Sci. 2020, 11, 11729–11739. [Google Scholar] [CrossRef]
- Op de Beeck, M.; Madder, A. Unprecedented C-selective interstrand cross-linking through in situ oxidation of furan-modified oligodeoxynucleotides. J. Am. Chem. Soc. 2011, 133, 796–807. [Google Scholar] [CrossRef]
- Manicardi, A.; Gyssels, E.; Corradini, R.; Madder, A. Furan-PNA: A mildly inducible irreversible interstrand crosslinking system targeting single and double stranded DNA. Chem. Commun. (Camb.) 2016, 52, 6930–6933. [Google Scholar] [CrossRef]
- Antonatou, E.; Hoogewijs, K.; Kalaitzakis, D.; Baudot, A.; Vassilikogiannakis, G.; Madder, A. Singlet Oxygen-Induced Furan Oxidation for Site-Specific and Chemoselective Peptide Ligation. Chemistry 2016, 22, 8457–8461. [Google Scholar] [CrossRef]
- Antonatou, E.; Verleysen, Y.; Madder, A. Singlet oxygen-mediated one-pot chemoselective peptide-peptide ligation. Org. Biomol. Chem. 2017, 15, 8140–8144. [Google Scholar] [CrossRef]
- Saarbach, J.; Lindberg, E.; Winssinger, N. Ruthenium-based Photocatalysis in Templated Reactions. CHIMIA Int. J. Chem. (Aarau) 2018, 72, 207–211. [Google Scholar] [CrossRef]
- Tanabe, K.; Nakata, H.; Mukai, S.; Nishimoto, S. Modulated drug release from the stem-and-loop structured oligodeoxynucleotide upon UV-A irradiation in the presence of target DNA. Org. Biomol. Chem. 2005, 3, 3893–3897. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Mokhir, A. An autocatalytic chromogenic and fluorogenic photochemical reaction controlled by nucleic acids. Chem. Commun. (Camb.) 2011, 47, 1243–1245. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Flottmann, B.; Heilemann, M.; Mokhir, A. Hybridization and reaction-based fluorogenic nucleic acid probes. Chem. Commun. (Camb.) 2012, 48, 9664–9666. [Google Scholar] [CrossRef] [PubMed]
- Zozulia, O.; Bachmann, T.; Mokhir, A. Red Light Triggered Fluorogenic Reaction with Picomolar Sensitivity Toward Nucleic Acids. Bioconj. Chem. 2019, 30, 2023–2031. [Google Scholar] [CrossRef]
- Dutta, S.; Fülöp, A.; Mokhir, A. Fluorogenic, Catalytic, Photochemical Reaction for Amplified Detection of Nucleic Acids. Bioconj. Chem. 2013, 24, 1533–1542. [Google Scholar] [CrossRef]
- Rothlingshofer, M.; Gorska, K.; Winssinger, N. Nucleic acid templated uncaging of fluorophores using Ru-catalyzed photoreduction with visible light. Org. Lett. 2012, 14, 482–485. [Google Scholar] [CrossRef]
- Sadhu, K.K.; Winssinger, N. Detection of miRNA in Live Cells by Using Templated RuII-Catalyzed Unmasking of a Fluorophore. Chem. Eur. J. 2013, 19, 8182–8189. [Google Scholar] [CrossRef]
- Gorska, K.; Keklikoglou, I.; Tschulena, U.; Winssinger, N. Rapid fluorescence imaging of miRNAs in human cells using templated Staudinger reaction. Chem. Sci. 2011, 2, 1969–1975. [Google Scholar] [CrossRef]
- Sletten, E.M.; Bertozzi, C.R. Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angew. Chem. Int. Ed. 2009, 48, 6974–6998. [Google Scholar] [CrossRef]
- Chen, Y.; Kamlet, A.S.; Steinman, J.B.; Liu, D.R. A biomolecule-compatible visible-light-induced azide reduction from a DNA-encoded reaction-discovery system. Nat. Chem. 2011, 3, 146–153. [Google Scholar] [CrossRef]
- Chang, D.; Lindberg, E.; Winssinger, N. Critical Analysis of Rate Constants and Turnover Frequency in Nucleic Acid-Templated Reactions: Reaching Terminal Velocity. J. Am. Chem. Soc. 2017, 139, 1444–1447. [Google Scholar] [CrossRef] [PubMed]
- Anzola, M.; Winssinger, N. Turn On of a Ruthenium(II) Photocatalyst by DNA-Templated Ligation. Chem. Eur. J. 2019, 25, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.T.; Angerani, S.; Chang, D.L.; Winssinger, N. Coupling of DNA Circuit and Templated Reactions for Quadratic Amplification and Release of Functional Molecules. J. Am. Chem. Soc. 2019, 141, 16288–16295. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.T.; Chang, D.L.; Winssinger, N. Double-Stranded RNA-Specific Templated Reaction with Triplex Forming PNA. Helv. Chim. Acta 2018, 101, e1700295. [Google Scholar] [CrossRef]
- Kim, K.T.; Winssinger, N. Enhanced SNP-sensing using DNA-templated reactions through confined hybridization of minimal substrates (CHOMS). Chem. Sci. 2020, 11, 4150–4157. [Google Scholar] [CrossRef]
- Brenner, S.; Lerner, R.A. Encoded Combinatorial Chemistry. Proc. Natl. Acad. Sci. USA 1992, 89, 5381–5383. [Google Scholar] [CrossRef]
- Song, M.; Hwang, G.T. DNA-Encoded Library Screening as Core Platform Technology in Drug Discovery: Its Synthetic Method Development and Applications in DEL Synthesis. J. Med. Chem. 2020, 63, 6578–6599. [Google Scholar] [CrossRef]
- Fitzgerald, P.R.; Paegel, B.M. DNA-Encoded Chemistry: Drug Discovery from a Few Good Reactions. Chem. Rev. 2020, in press. [Google Scholar] [CrossRef]
- Decurtins, W.; Wichert, M.; Franzini, R.M.; Buller, F.; Stravs, M.A.; Zhang, Y.; Neri, D.; Scheuermann, J. Automated screening for small organic ligands using DNA-encoded chemical libraries. Nat. Protoc. 2016, 11, 764–780. [Google Scholar] [CrossRef]
- Zhu, Z.; Shaginian, A.; Grady, L.C.; O’Keeffe, T.; Shi, X.E.; Davie, C.P.; Simpson, G.L.; Messer, J.A.; Evindar, G.; Bream, R.N.; et al. Design and Application of a DNA-Encoded Macrocyclic Peptide Library. ACS Chem. Biol. 2017, 13, 53–59. [Google Scholar] [CrossRef]
- Ottl, J.; Leder, L.; Schaefer, J.V.; Dumelin, C.E. Encoded Library Technologies as Integrated Lead Finding Platforms for Drug Discovery. Molecules 2019, 24, 1629. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.L.; Ohta, C.; Zuo, Z.W.; MacMillan, D.W.C. Carboxylic Acids as A Traceless Activation Group for Conjugate Additions: A Three-Step Synthesis of (+/−)-Pregabalin. J. Am. Chem. Soc. 2014, 136, 10886–10889. [Google Scholar] [CrossRef] [PubMed]
- Kolmel, D.K.; Loach, R.P.; Knauber, T.; Flanagan, M.E. Employing Photoredox Catalysis for DNA-Encoded Chemistry: Decarboxylative Alkylation of alpha-Amino Acids. ChemMedChem 2018, 13, 2159–2165. [Google Scholar] [CrossRef] [PubMed]
- Kolmel, D.K.; Meng, J.; Tsai, M.H.; Que, J.; Loach, R.P.; Knauber, T.; Wan, J.; Flanagan, M.E. On-DNA Decarboxylative Arylation: Merging Photoredox with Nickel Catalysis in Water. ACS Comb. Sci. 2019, 21, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Z.; Ahneman, D.T.; Chu, L.; Terrett, J.A.; Doyle, A.G.; MacMillan, D.W. Dual catalysis. Merging photoredox with nickel catalysis: Coupling of alpha-carboxyl sp(3)-carbons with aryl halides. Science 2014, 345, 437–440. [Google Scholar] [CrossRef]
- Ruff, Y.; Martinez, R.; Pelle, X.; Nimsgern, P.; Fille, P.; Ratnikov, M.; Berst, F. An Amphiphilic Polymer-Supported Strategy Enables Chemical Transformations under Anhydrous Conditions for DNA-Encoded Library Synthesis. ACS Comb. Sci. 2020, 22, 120–128. [Google Scholar] [CrossRef]
- Phelan, J.P.; Lang, S.B.; Sim, J.; Berritt, S.; Peat, A.J.; Billings, K.; Fan, L.; Molander, G.A. Open-Air Alkylation Reactions in Photoredox-Catalyzed DNA-Encoded Library Synthesis. J. Am. Chem. Soc. 2019, 141, 3723–3732. [Google Scholar] [CrossRef]
- Badir, S.O.; Sim, J.; Billings, K.; Csakai, A.; Zhang, X.; Dong, W.; Molander, G.A. Multifunctional Building Blocks Compatible with Photoredox-Mediated Alkylation for DNA-Encoded Library Synthesis. Org. Lett. 2020, 22, 1046–1051. [Google Scholar] [CrossRef]
- Lei, T.; Zhou, C.; Wei, X.Z.; Yang, B.; Chen, B.; Tung, C.H.; Wu, L.Z. Construction of Cyclobutanes by Multicomponent Cascade Reactions in Homogeneous Solution through Visible-Light Catalysis. Chem. Eur. J. 2019, 25, 879–884. [Google Scholar] [CrossRef]
- Poplata, S.; Troster, A.; Zou, Y.Q.; Bach, T. Recent Advances in the Synthesis of Cyclobutanes by Olefin [2 + 2] Photocycloaddition Reactions. Chem. Rev. 2016, 116, 9748–9815. [Google Scholar] [CrossRef]
- Kolmel, D.K.; Ratnayake, A.S.; Flanagan, M.E.; Tsai, M.H.; Duan, C.; Song, C. Photocatalytic [2 + 2] Cycloaddition in DNA-Encoded Chemistry. Org. Lett. 2020, 22, 2908–2913. [Google Scholar] [CrossRef] [PubMed]
- Lueckerath, T.; Strauch, T.; Koynov, K.; Barner-Kowollik, C.; Ng, D.Y.W.; Weil, T. DNA-Polymer Conjugates by Photoinduced RAFT Polymerization. Biomacromolecules 2019, 20, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Lunn, D.J.; Pusuluri, A.; Yoo, J.I.; O’Malley, M.A.; Mitragotri, S.; Soh, H.T.; Hawker, C.J. Engineering live cell surfaces with functional polymers via cytocompatible controlled radical polymerization. Nat. Chem. 2017, 9, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.Q.; Zou, Y.Q.; Lu, L.Q.; Xiao, W.J. Visible-Light-Induced Organic Photochemical Reactions through Energy-Transfer Pathways. Angew. Chem. Int. Ed. 2019, 58, 1586–1604. [Google Scholar] [CrossRef] [PubMed]
- Prier, C.K.; Rankic, D.A.; MacMillan, D.W. Visible light photoredox catalysis with transition metal complexes: Applications in organic synthesis. Chem. Rev. 2013, 113, 5322–5363. [Google Scholar] [CrossRef]
- Koike, T.; Akita, M. Visible-light radical reaction designed by Ru- and Ir-based photoredox catalysis. Inorg. Chem. Front. 2014, 1, 562–576. [Google Scholar] [CrossRef]
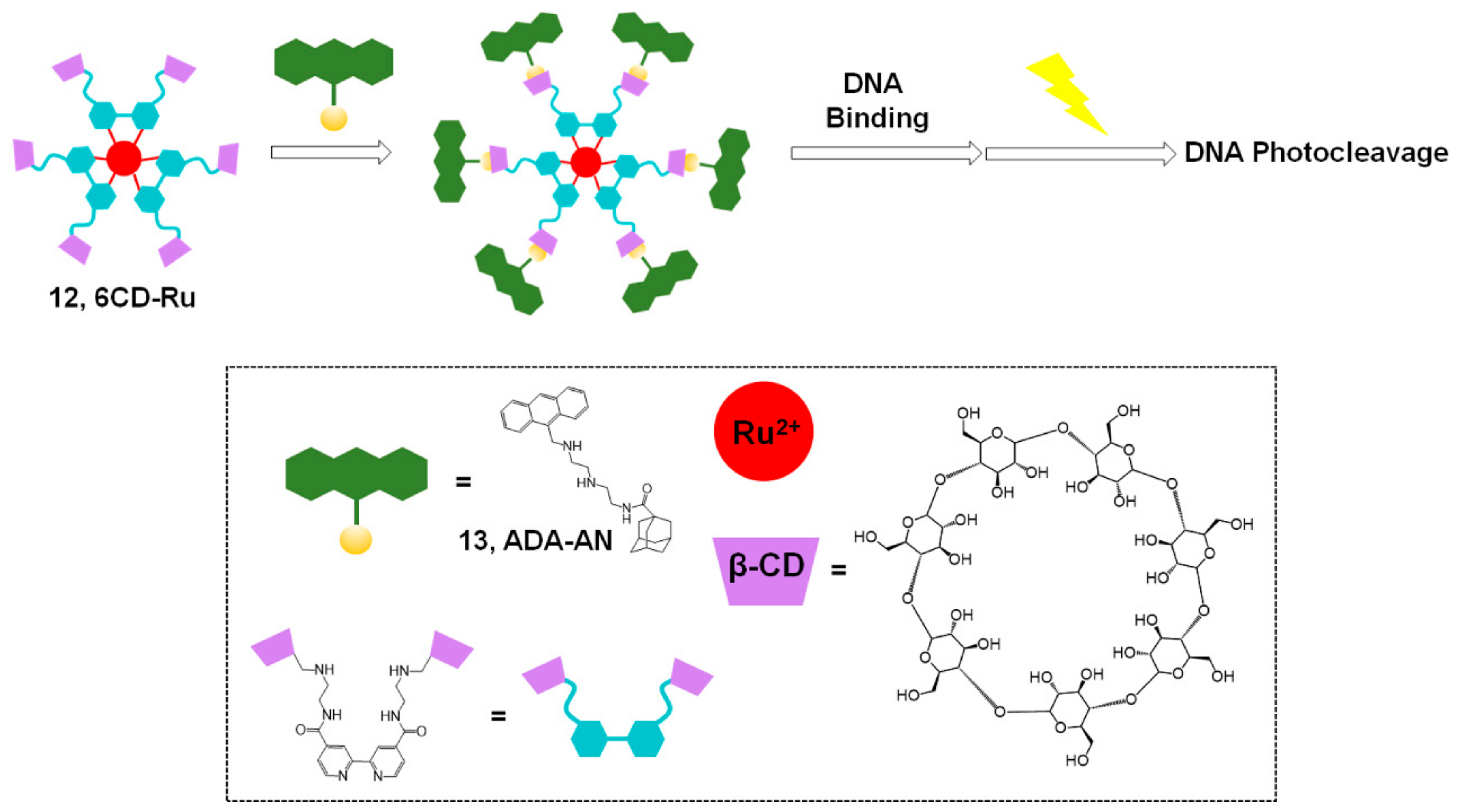
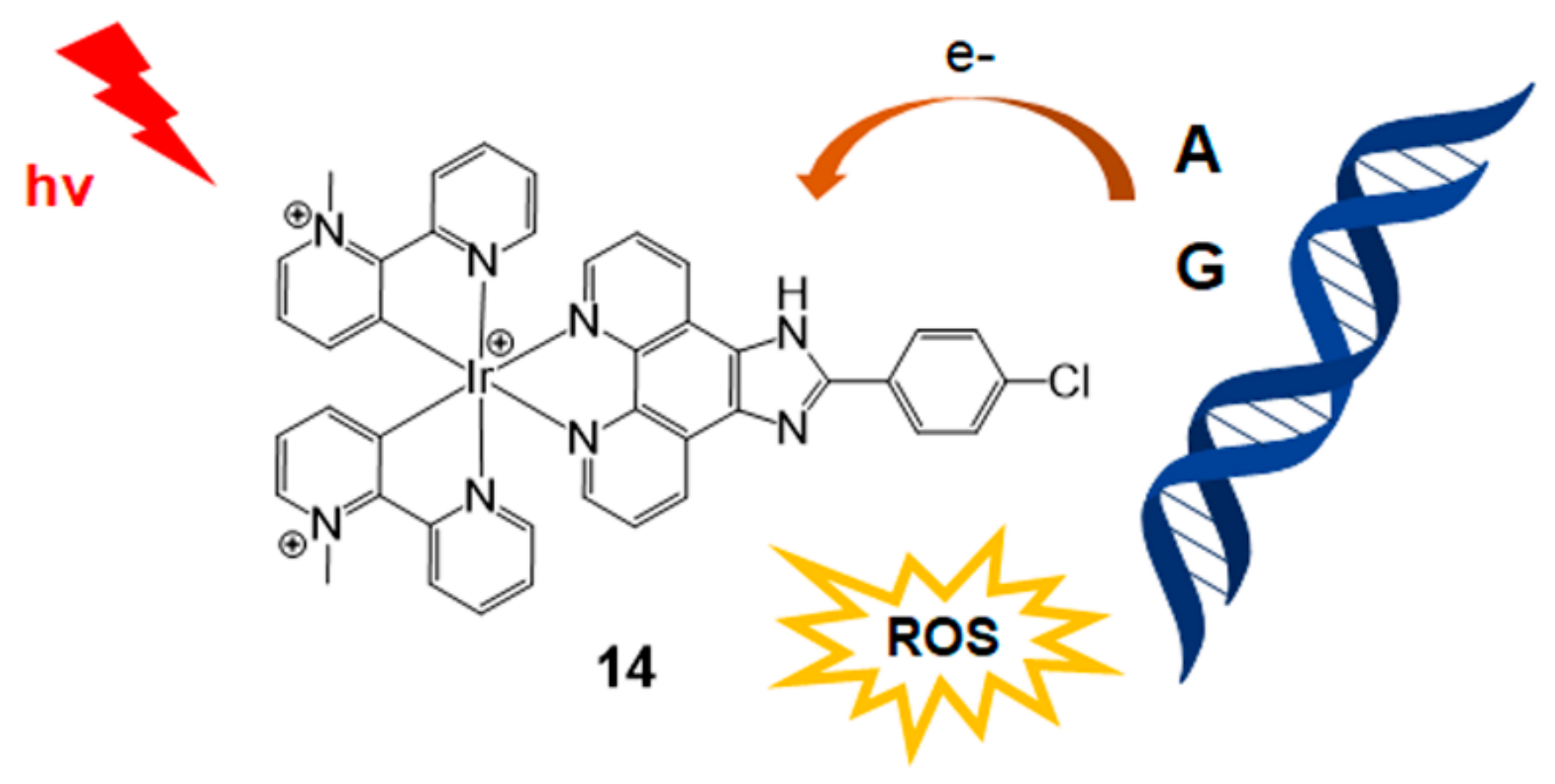

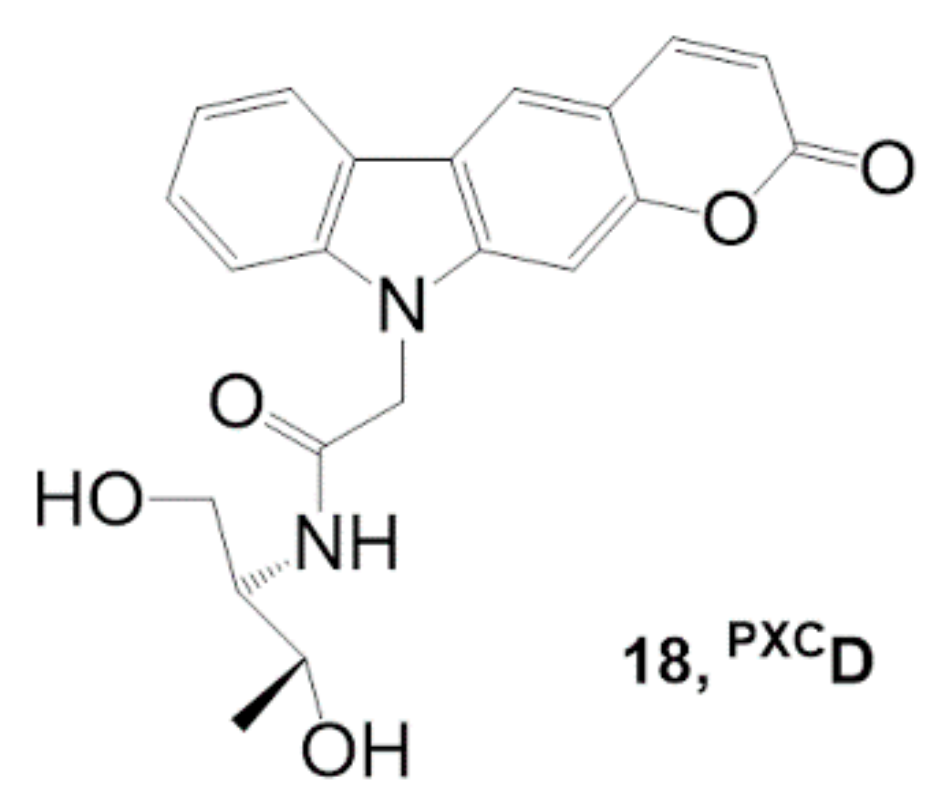
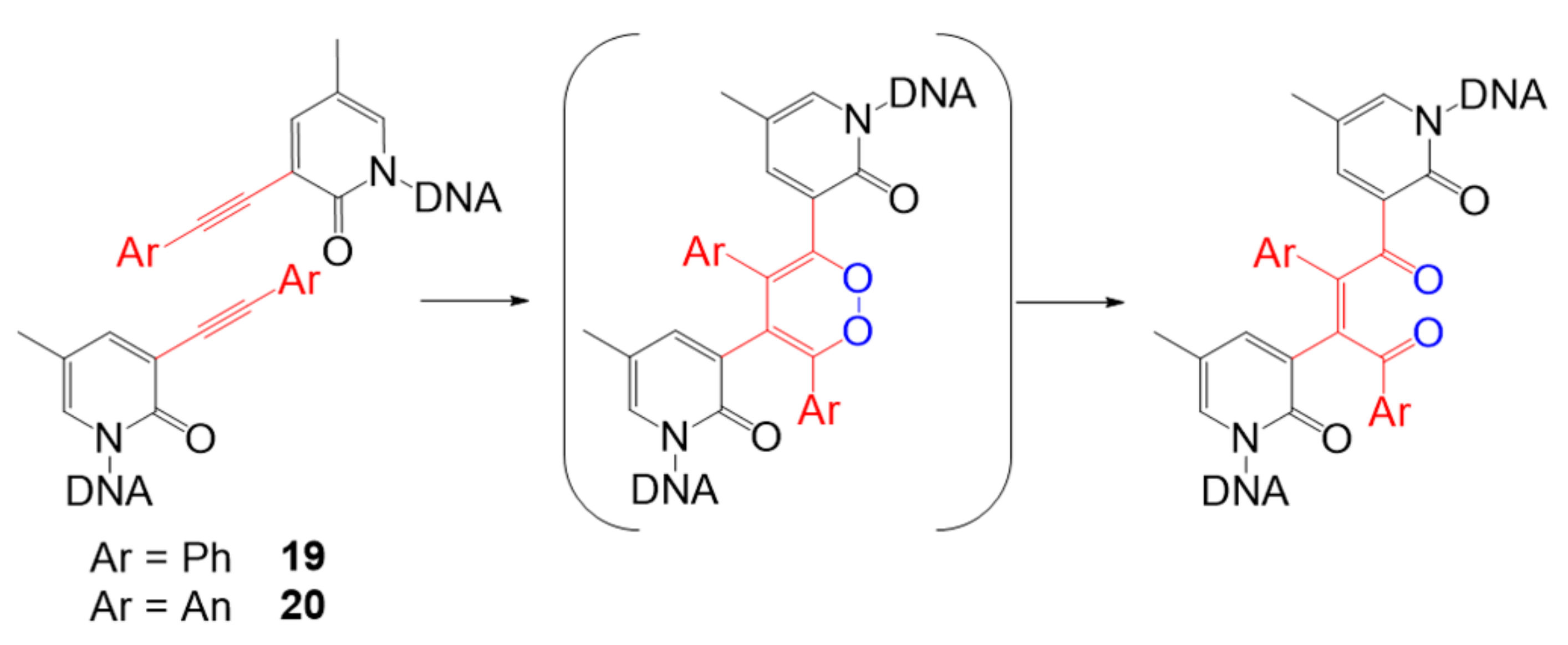
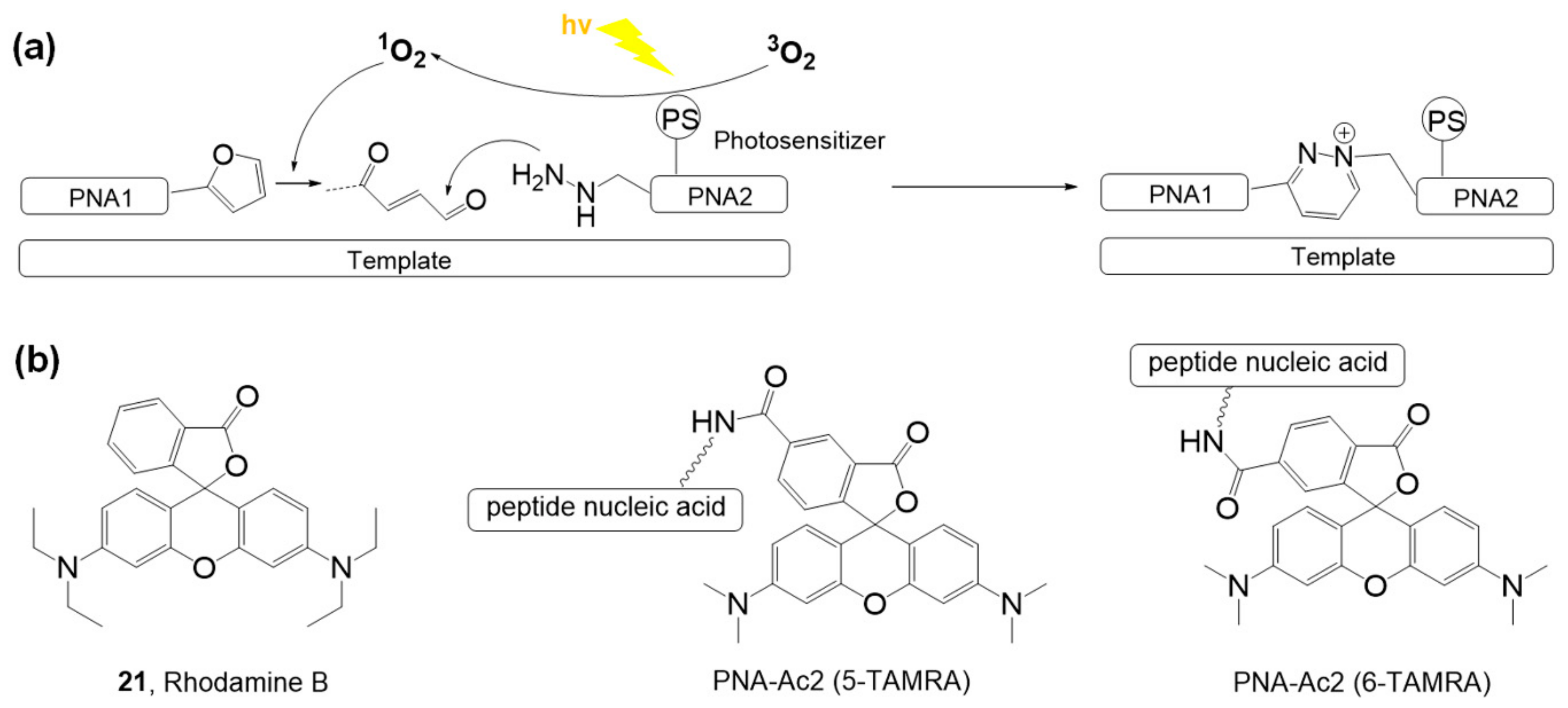
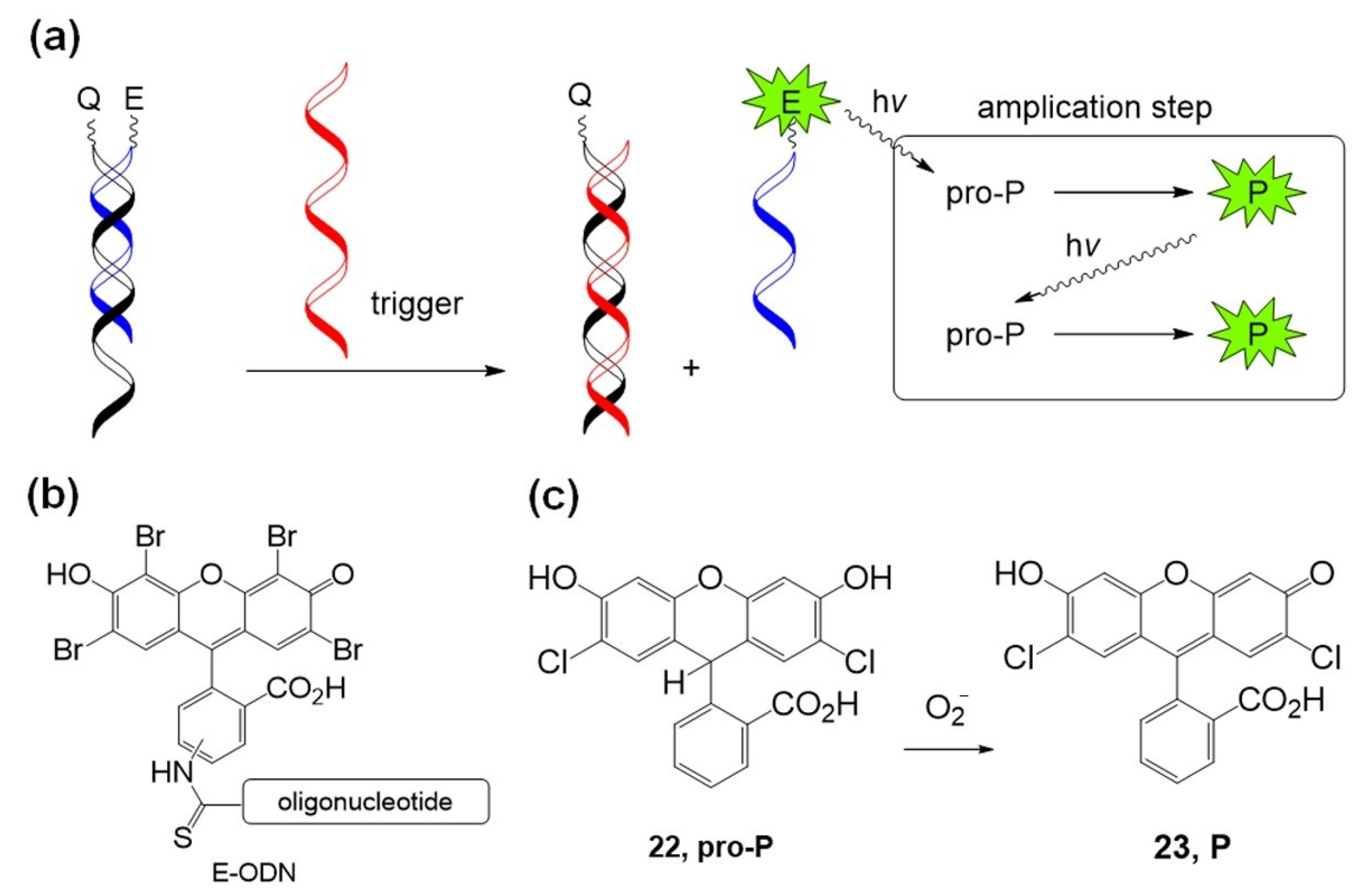






| Photosensitizer | Ligands | Properties | References |
|---|---|---|---|
| Ru complex (Figure 5) | ADA-AN | - PDT with visible light - Water-soluble | [35] |
| Ir complex (Figure 6) | CPIP | - G–A DNA base pair triggered PET | [37] |
| Riboflavin (Figure 7) | - Selective DNA photocleavage of the G–T mismatch | [39] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koo, B.; Yoo, H.; Choi, H.J.; Kim, M.; Kim, C.; Kim, K.T. Visible Light Photochemical Reactions for Nucleic Acid-Based Technologies. Molecules 2021, 26, 556. https://doi.org/10.3390/molecules26030556
Koo B, Yoo H, Choi HJ, Kim M, Kim C, Kim KT. Visible Light Photochemical Reactions for Nucleic Acid-Based Technologies. Molecules. 2021; 26(3):556. https://doi.org/10.3390/molecules26030556
Chicago/Turabian StyleKoo, Bonwoo, Haneul Yoo, Ho Jeong Choi, Min Kim, Cheoljae Kim, and Ki Tae Kim. 2021. "Visible Light Photochemical Reactions for Nucleic Acid-Based Technologies" Molecules 26, no. 3: 556. https://doi.org/10.3390/molecules26030556
APA StyleKoo, B., Yoo, H., Choi, H. J., Kim, M., Kim, C., & Kim, K. T. (2021). Visible Light Photochemical Reactions for Nucleic Acid-Based Technologies. Molecules, 26(3), 556. https://doi.org/10.3390/molecules26030556








