Region-Specific Biomarkers and Their Mechanisms in the Treatment of Lung Adenocarcinoma: A Study of Panax quinquefolius from Wendeng, China
Abstract
:1. Introduction
2. Results
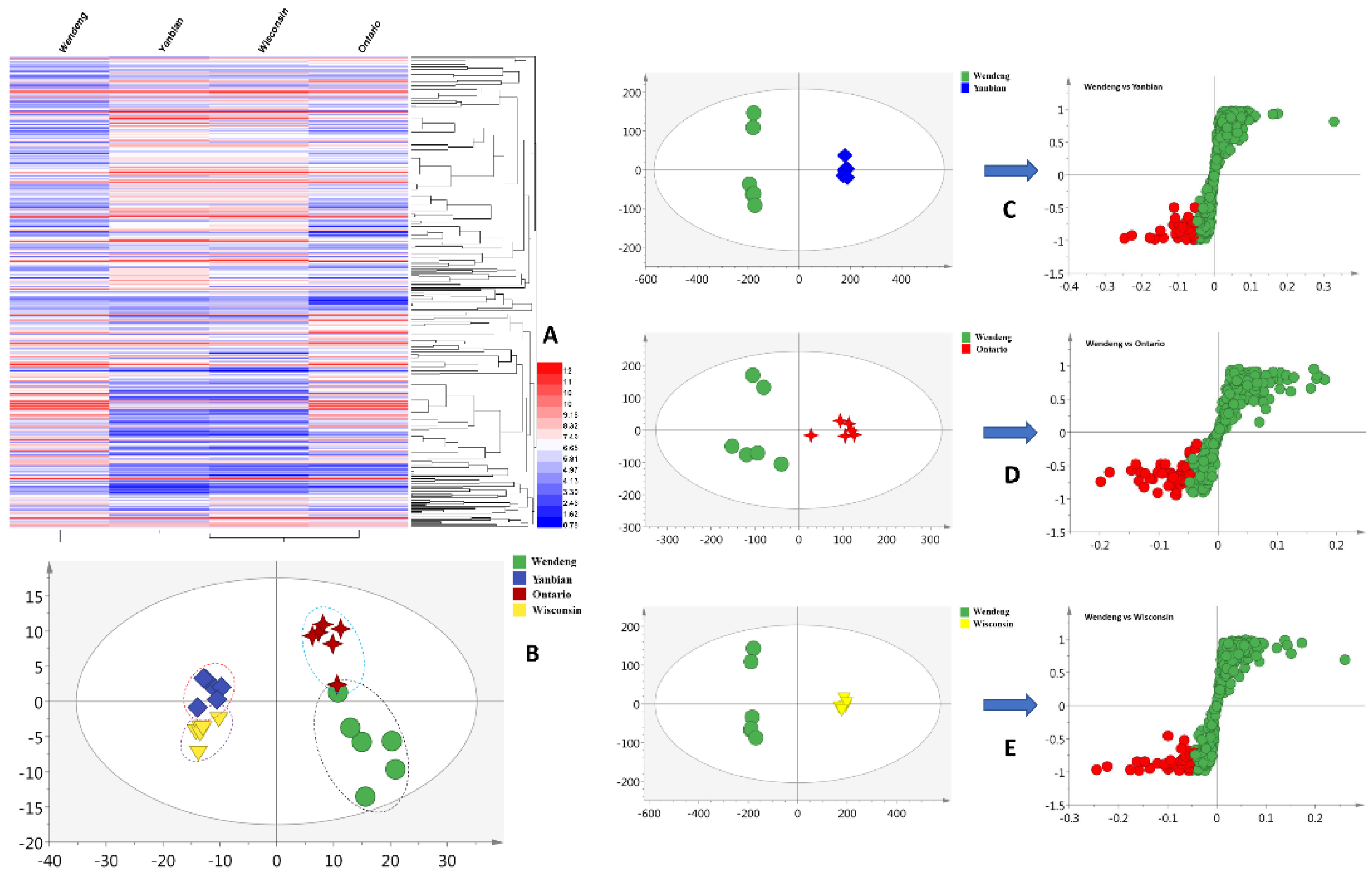
2.1. Statistical Analysis and Biomarker Discovery
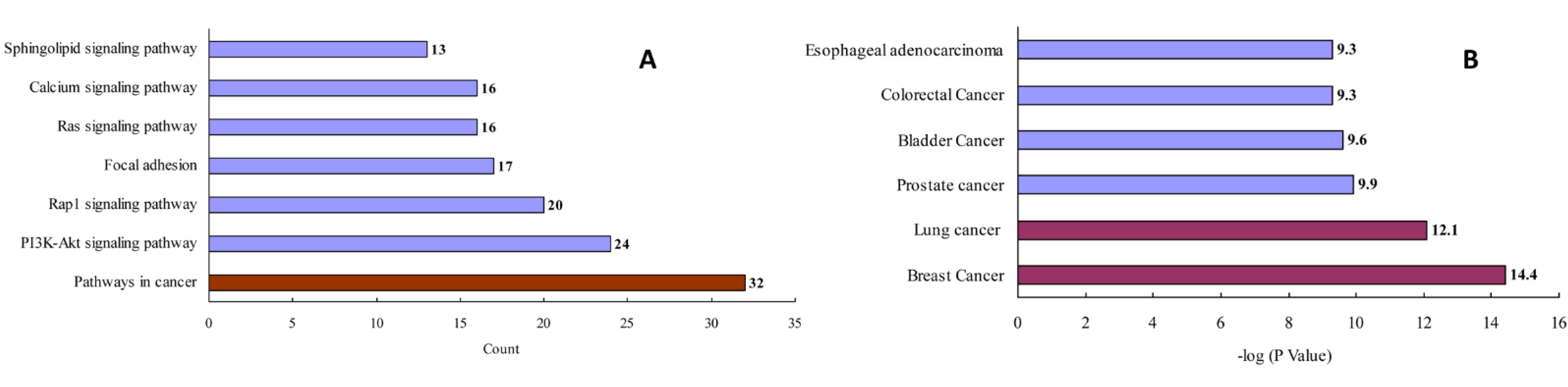
2.2. Construction and Analysis of Interaction Networks
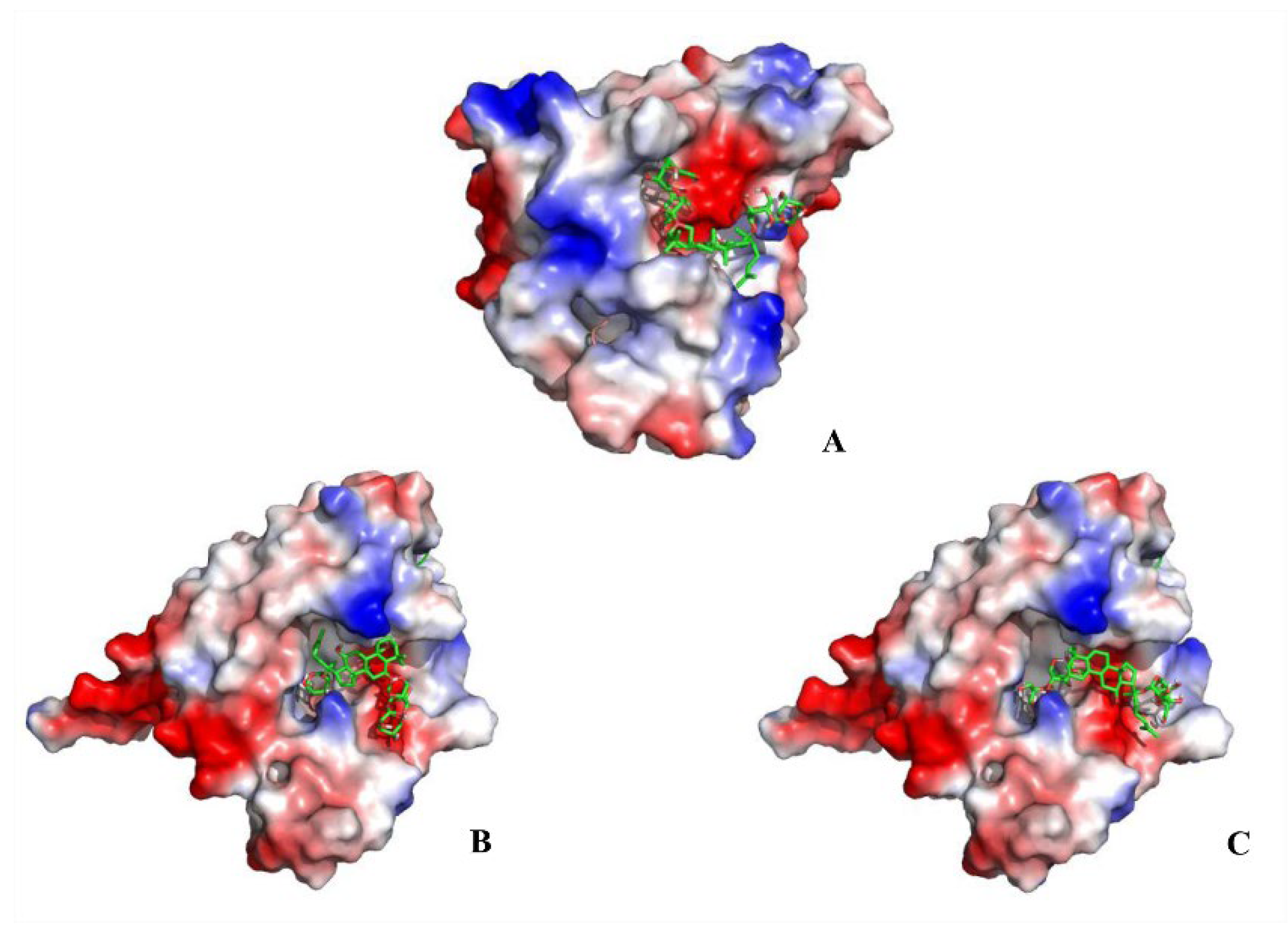
2.3. Docking Results Analysis
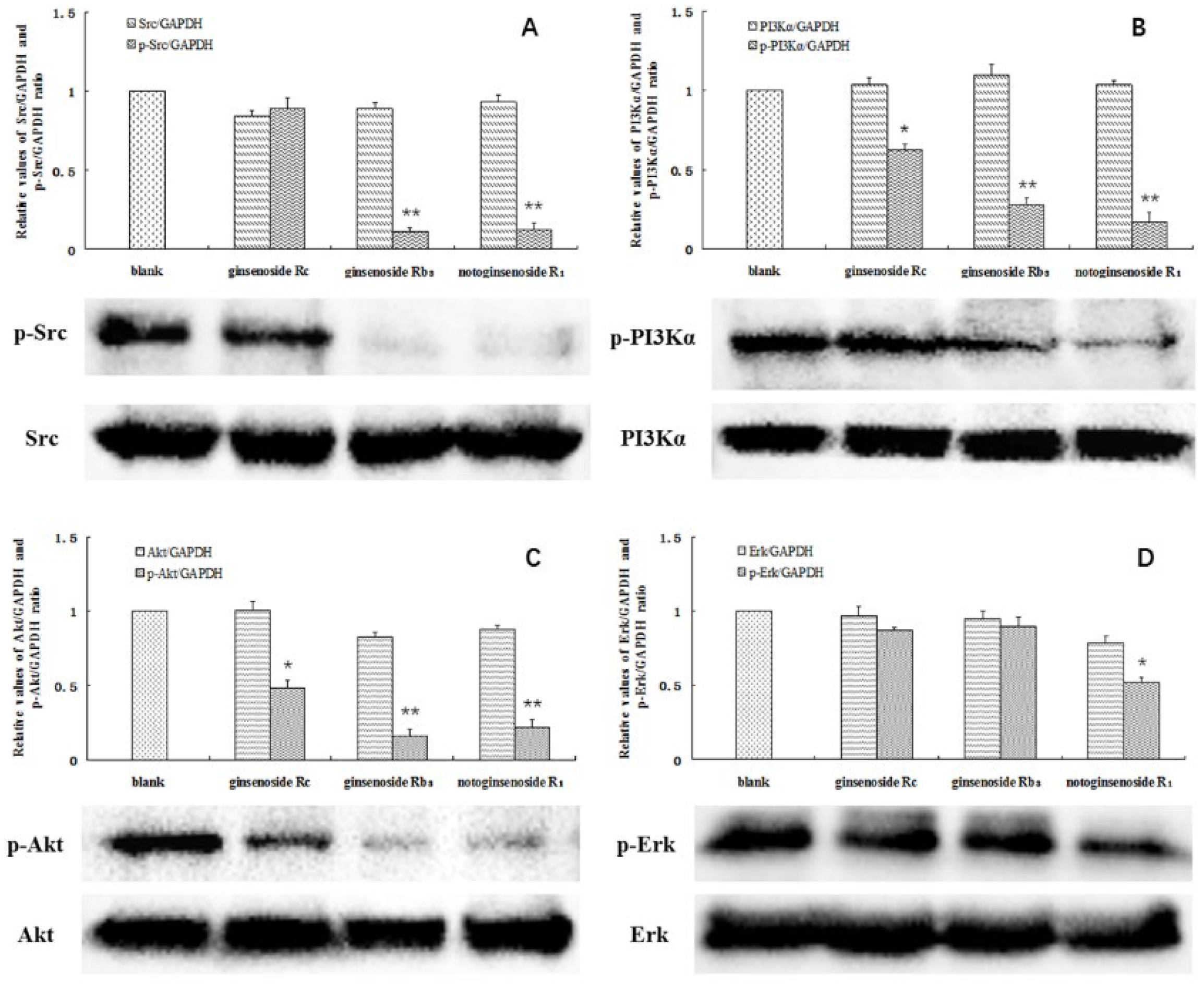
2.4. Phosphorylation of SRC, PI3Kα, Akt, and ERK
3. Discussion
4. Materials and Methods
4.1. Samples, Chemicals, and Reagents
4.2. LC–MS Assay and Metabolomic Analyses
4.3. Target Database Construction and Bioinformatics Analysis
4.4. Molecular Docking
4.5. Western Blot Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Chen, C.Y.O.; Ribaya-Mercado, J.D.; McKay, D.L.; Croom, E.; Blumberg, J.B. Differential antioxidant and quinone reductase inducing activity of American, Asian, and Siberian ginseng. Food Chem. 2010, 119, 445–451. [Google Scholar] [CrossRef]
- Ghosh, R.; Bryant, D.L.; Farone, A.L. Panax quinquefolius (North American Ginseng) polysaccharides as immunomodulators: Current research status and future directions. Molecules 2020, 25, 5854. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.Y.; Fan, Y.; Yu, Q.T.; Ge, Y.Z.; Yan, C.P.; Alolga, R.N.; Li, P.; Ma, Z.H.; Qi, L.W. Integrated evaluation of malonyl ginsenosides, amino acids and polysaccharides in fresh and processed ginseng. J. Pharmtl. Biomed. 2015, 107, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Schlag, E.M.; Mcintosh, M.S. Ginsenoside content and variation among and within American ginseng (Panax quinquefolius L.) populations. Phytochemistry 2006, 67, 1510–1519. [Google Scholar] [CrossRef] [PubMed]
- Li, L.L.; Wang, D.J.; Sun, C.L.; Li, Y.; Lu, H.; Wang, X. Comprehensive lipidome and metabolome profiling investigations of Panax quinquefolius and application in different growing regions using liquid chromatography coupled with mass spectrometry. J. Agric. Food Chem. 2021, 69, 6710–6719. [Google Scholar] [CrossRef]
- Yang, Y.G.; Ju, Z.C.; Yang, Y.B.; Zhang, Y.H.; Yang, L.; Wang, Z.T. Phytochemical analysis of Panax species: A review. J. Ginseng. Res. 2021, 1, 1–21. [Google Scholar] [CrossRef]
- Barton, D.L.; Liu, H.; Dakhil, S.R.; Linquist, B.; Sloan, J.A.; Nichols, C.R.; McGinn, T.W.; Stella, P.J.; Seeger, G.R.; Sood, A.; et al. Wisconsin ginseng (Panax quinquefolius) to improve cancer-related fatigue: A randomized, double-blind trial, N07C2. J. Natl. Cancer I 2013, 105, 1230–1238. [Google Scholar]
- Chen, C.F.; Chiou, W.F.; Zhang, J.T. Comparison of the pharmacological effects of Panax ginseng and Panax quinquefolium. Acta Pharm. Sin. 2008, 29, 1103–1108. [Google Scholar] [CrossRef] [Green Version]
- Kochan, E.; Nowak, A.; Zakłos-Szyda, M.; Szczuka, D.; Szymanska, G.; Motyl, I. Panax quinquefolium L. ginsenosides from hairy root cultures and their clones exert cytotoxic, genotoxic and pro-apoptotic activity towards human colon adenocarcinoma cell line caco-2. Molecules 2020, 25, 2262. [Google Scholar] [CrossRef]
- Qu, C.L.; Bai, Y.P.; Jin, X.Q.; Wang, Y.T.; Zhang, K.; You, J.Y.; Zhang, H.Q. Study on ginsenosides in different parts and ages of Panax quinquefolius L. Food Chem. 2009, 115, 340–346. [Google Scholar] [CrossRef]
- Sun, S.; Qi, L.W.; Du, G.J.; Mehendale, S.R.; Wang, C.Z.; Yuan, C.S. Red notoginseng: Higher ginsenoside content and stronger anticancer potential than Asian and American ginseng. Food Chem. 2011, 125, 1299–1305. [Google Scholar] [CrossRef] [Green Version]
- He, S.; Lyu, F.; Lou, L.X.; Liu, L.; Li, S.L.; Jakowitsch, J.; Ma, Y. Anti-tumor activities of Panax quinquefolius saponins and po-tential biomarkers in prostate cancer. J. Ginseng Res. 2021, 45, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.M.; Bi, Y.M.; Li, J.F.; Shao, H.H.; Jiao, X.L.; Gao, W.W. First report of root rot caused by Fusarium armeniacum on American ginseng in China. Plant. Dis 2021, 105, 1223. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Lv, C.N.; Li, Q.; Wang, J.; Song, D.; Liu, P.P.; Zhang, D.D.; Lu, J.C. Chemical and bioactive comparison of flowers of Panax ginseng Meyer, Panax quinquefolius L. and Panax notoginseng Burk. J. Gins Res. 2017, 41, 487–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ligor, T.; Ludwiczuk, A.; Wolski, T.; Buszewski, B. Isolation and determination of ginsenosides in American ginseng leaves and root extracts by LC–MS. Anal. Bioanal. Chem. 2005, 383, 1098–1105. [Google Scholar] [CrossRef]
- Wang, H.D.; Zhang, C.X.; Zuo, T.T.; Li, W.W.; Jia, L.; Wang, X.Y.; Qian, Y.X.; Guo, D.A.; Yang, W.Z. In-depth profiling, characterization, and comparison of the ginsenosides among three different parts (the root, stem leaf, and flower bud) of Panax quinquefolius L. by ultra-high performance liquid chromatography/quadrupole-Orbitrap mass spectrometry. Anal. Bioanal. Chem. 2019, 411, 7817–7829. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, H.; Gao, W.Y.; Zhang, L.M. Comparison of ginsenoside composition in native roots and cultured callus cells of Panax quinquefolium L. Acta Physiol. Plant. 2013, 35, 1363–1366. [Google Scholar] [CrossRef]
- Li, J.; Zuo, T.T.; Zhang, C.X.; Li, W.W.; Wang, H.D.; Hu, Y.; Wang, X.Y.; Qian, Y.X.; Yang, W.Z.; Yu, H.S. Simultaneous profiling and holistic comparison of the metabolomes among the flower buds of Panax ginseng, Panax quinquefolius, and Panax notoginseng by UHPLC/IM-QTOF-HDMSE-based metabolomics analysis. Molecules 2019, 24, 2188. [Google Scholar]
- Xing, J.J.; Hou, J.G.; Liu, Y.; Zhang, R.B.; Jiang, S.; Ren, S.; Wang, Y.P.; Shen, Q.; Li, W.; Li, X.D.; et al. Supplementation of saponins from leaves of Panax quinquefolius mitigates cisplatin-evoked cardiotoxicity via inhibiting oxidative stress-associated inflammation and apoptosis in mice. Antioxidants 2019, 8, 347. [Google Scholar] [CrossRef] [Green Version]
- Han, B.J.; Zhang, Z.; Xie, Y.X.; Hu, X.Q.; Wang, H.B.; Xia, W.; Wang, Y.L.; Li, H.Y.; Wang, Y.C.; Sun, H.Z. Multi-omics and temporal dynamics profiling reveal disruption of central metabolism in Helicobacter pylori on bismuth treatment. Chem. Sci. 2018, 9, 7488–7497. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Liu, Y.; Wang, Y.; Abozeid, A.; Zu, Y.G.; Tang, Z.H. The integration of GC–MS and LC–MS to assay the metabolomics profiling in Panax ginseng and Panax quinquefolius reveals a tissue- and species-specific connectivity of primary metabolites and ginsenosides accumulation. J. Pharm. Biomed. 2017, 135, 176–185. [Google Scholar] [CrossRef]
- Wang, H.Y.; Hua, H.Y.; Liu, X.Y.; Liu, J.H.; Yu, B.Y. In vitro biotransformation of red ginseng extract by human intestinal microflora: Metabolites identification and metabolic profile elucidation using LC-Q-TOF/MS. J Pharm. Biomed. 2014, 98, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Zhang, A.H.; Zhao, Q.Q.; Yan, G.L.; Sun, H.; Wang, X.J. Discovery of quality-marker ingredients of Panax quinquefolius driven by high-throughput chinmedomics approach. Phytomedicine 2020, 74, 152928. [Google Scholar] [CrossRef]
- Calvert, S.; Tacutu, R.; Sharifi, S.; Teixeira, R.; Ghosh, P.; Magalhaes, J.P. A network pharmacology approach reveals new candidate caloric restriction mimetics in C. elegans. Aging Cell 2016, 15, 256–266. [Google Scholar] [CrossRef]
- Gao, L.; Wang, X.D.; Niu, Y.Y.; Duan, D.D.; Yang, X.; Hao, J.; Zhu, C.H.; Chen, D.; Wang, K.X.; Qin, X.M.; et al. Molecular targets of Chinese herbs: A clinical study of hepatoma based on network pharmacology. Sci. Rep. 2016, 6, 24944. [Google Scholar] [CrossRef] [Green Version]
- Tan, Y.S.; Li, F.; Lv, Y.N.; Zhai, K.F.; Chai, C.Z.; Kou, J.P.; Yu, B.Y. Study on the multi-targets mechanism of YiQiFuMai powder injection on cardio-cerebral ischemic diseases based on network pharmacology. J. Proteom. Comput. Biol. 2014, 1, 9. [Google Scholar]
- Gazdar, A.F.; Bunn, P.A.; Minna, J.D. Small-cell lung cancer: What we know, what we need to know and the path forward. Nat. Rev. Cancer 2017, 17, 725–737. [Google Scholar] [CrossRef]
- Mclntyre, A.; Ganti, A.K. Lung cancer-A global perspective. J. Surg Oncol. 2017, 115, 550–554. [Google Scholar] [CrossRef]
- Triba, M.N.; Moyec, L.L.; Amathieu, R.; Goossens, C.; Bouchemal, N.; Nahon, P.; Rutledge, D.N.; Savarin, P. PLS/OPLS models in metabolomics: The impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. Biosyst. 2014, 11, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.Q.; Xiao, H.B.; Liang, X.M. Identification of ginsenosides in Panax quinquefolium by LC–MS. Chromatographia 2006, 64, 31–36. [Google Scholar] [CrossRef]
- Jega, J.; Jeong, E.J.; Yang, M.H. A review of the different methods applied in ginsenoside extraction from Panax ginseng and Panax quinquefolius roots. Nat. Prod. Commun. 2019, 14, 1934578X1986839. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.D. Hsp90 flexibility and development of its inhibitors for the treatment of cancer. Curr. Chem. Biol. 2018, 12, 53–64. [Google Scholar] [CrossRef]
- Solit, D.B.; Rosen, N. Hsp90: A novel target for cancer therapy. Curr. Top. Med. Chem. 2006, 6, 1205–1214. [Google Scholar] [CrossRef]
- Crosbie, P.A.J.; Crosbie, E.J.; Aspinall-O’Dea, M.; Walker, M.; Harrison, R.; Pernemalm, M.; Shah, R.; Joseph, L.; Booton, R.; Pierce, A.; et al. ERK and AKT phosphorylation status in lung cancer and emphysema using nanocapillary isoelectric focusing. BMJ Open Respir. Res. 2016, 3, e000114. [Google Scholar] [CrossRef]
- Ebi, H.; Costa, C.; Faber, A.C.; Nishtala, M.; Kotani, H.; Juric, D.; Pelle, P.D.; Song, Y.C.; Yano, S.J.; Mino-Kenudson, M.; et al. PI3K regulates MEK/ERK signaling in breast cancer via the Rac-GEF, P-Rex1. Proc. Natl. Acad. Sci. USA 2013, 110, 21124–21129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, D.P.; Zhang, Y.M.; Hu, X.C.; Li, J.J.; Zhang, W. Activation of AKT/ERK confers non-small cell lung cancer cells resistance to vinorelbine. Int. J. Clin. Exp. Pathol. 2014, 7, 134–143. [Google Scholar]
- Koga, F.; Xu, W.P.; Karpova, T.S.; McNally, J.G.; Baron, R.; Neckers, L. Hsp90 inhibition transiently activates Src kinase and promotes Src-dependent Akt and Erk activation. Proc. Natl. Acad. Sci. USA 2006, 103, 11318–11322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.M.; Han, L.W.; Zhang, S.S.; Li, X.B.; He, Q.X.; Han, J.; Wang, X.M.; Liu, K.C. Targeted discovery and identification of novel nucleoside biomarkers in Apostichopus japonicus viscera using metabonomics. Nucl. Nucl. Nucl. Acids 2018, 38, 1–15. [Google Scholar] [CrossRef]
- Zhang, X.M.; Shi, Y.P.; Wang, L.Z.; Li, X.B.; Zhang, S.S.; Wang, X.M.; Jin, M.; Hsiao, C.D.; Lin, H.W.; Han, L.W.; et al. Metabolomics for biomarker discovery in fermented black garlic and potential bioprotective responses against cardiovascular diseases. J. Agric. Food Chem. 2019, 67, 12191–12198. [Google Scholar] [CrossRef]
- Sun, X.; Dai, Y.S.; Tan, G.L.; Liu, Y.Q.; Li, N. Integration analysis of m6A-SNPs and eQTLs associated with sepsis reveals platelet degranulation and Staphylococcus aureus infection are mediated by m6A mRNA methylation. Front. Genet. 2020, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, W.G.; Zeng, X.M.; Tang, X.M.; Zhang, S.; Zhong, F.Y.; Peng, X.N.; Zhong, Y.; Rosol, T.J.; Deng, X.Y.; et al. The role of microRNA-148a and downstream DLGAP1 on the molecular regulation and tumor progression on human glioblastoma. Oncogene 2019, 38, 7234–7248. [Google Scholar] [CrossRef] [PubMed]
- Helgren, T.R.; Hagen, T.J. Demonstration of AutoDock as an educational tool for drug discovery. J. Chem. Educ 2017, 94, 345–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiwari, A.; Saxena, S.; Pant, A.B.; Srivastava, P. Protein-ligand interaction studies of retinol-binding protein 3 with herbal molecules using AutoDock for the management of Eales’ disease. J. Ocul. Biol. Inf. 2012, 5, 40–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mothay, D.; Ramesh, K.V. Binding site analysis of potential protease inhibitors of COVID-19 using AutoDock. VirusDisease 2020, 31, 194–199. [Google Scholar] [CrossRef]
- Chan, Q.K.Y.; Lam, H.M.; Ng, C.F.; Lee, A.Y.Y.; Chan, E.S.Y.; Ng, H.K.; Ho, S.M.; Lau, K.M. Activation of GPR30 inhibits the growth of prostate cancer cells through sustained activation of Erk1/2, c-jun/c-fos-dependent upregulation of p21, and induction of G2 cell-cycle arrest. Cell Death Differ. 2010, 17, 1511–1523. [Google Scholar] [CrossRef]
- Sun, Y.; Du, C.; Wang, B.; Zhang, Y.; Liu, X.; Ren, G. Upregulation of eEF1A2 promotes proliferation and inhibits apoptosis in prostate cancer. BioChem. Bioph Res. 2014, 450, 1–6. [Google Scholar] [CrossRef]

| Concentrations | Cells Viability | ||
|---|---|---|---|
| Ginsenoside Rc | Ginsenoside Rb3 | Notoginsenoside R1 | |
| 0.1 mM | 92.05% | 87.24% | 86.40% |
| 0.5 mM | 91.41% | 78.58% | 65.91% |
| 1 mM | 75.33% | 72.22% | 42.73% |
| 1.5 mM | 72.04% | 64.27% | 14.24% |
| 2 mM | 59.84% | 43.01% | 4.52% |
| 2.5 mM | 49.18% | 24.11% | 1.50% |
| 3 mM | 40.53% | 1.66% | 0.61% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Han, L.; Li, P.; Zhang, S.; Zhang, M.; Li, X.; Chu, J.; Wang, L.; Tu, P.; Zhang, Y.; et al. Region-Specific Biomarkers and Their Mechanisms in the Treatment of Lung Adenocarcinoma: A Study of Panax quinquefolius from Wendeng, China. Molecules 2021, 26, 6829. https://doi.org/10.3390/molecules26226829
Zhang X, Han L, Li P, Zhang S, Zhang M, Li X, Chu J, Wang L, Tu P, Zhang Y, et al. Region-Specific Biomarkers and Their Mechanisms in the Treatment of Lung Adenocarcinoma: A Study of Panax quinquefolius from Wendeng, China. Molecules. 2021; 26(22):6829. https://doi.org/10.3390/molecules26226829
Chicago/Turabian StyleZhang, Xuanming, Liwen Han, Peihai Li, Shanshan Zhang, Mengqi Zhang, Xiaobin Li, Jie Chu, Lizhen Wang, Pengfei Tu, Yun Zhang, and et al. 2021. "Region-Specific Biomarkers and Their Mechanisms in the Treatment of Lung Adenocarcinoma: A Study of Panax quinquefolius from Wendeng, China" Molecules 26, no. 22: 6829. https://doi.org/10.3390/molecules26226829
APA StyleZhang, X., Han, L., Li, P., Zhang, S., Zhang, M., Li, X., Chu, J., Wang, L., Tu, P., Zhang, Y., & Liu, K. (2021). Region-Specific Biomarkers and Their Mechanisms in the Treatment of Lung Adenocarcinoma: A Study of Panax quinquefolius from Wendeng, China. Molecules, 26(22), 6829. https://doi.org/10.3390/molecules26226829








