Purifying and Characterizing Bacterially Expressed Soluble Lactate Dehydrogenase from Plasmodium knowlesi for the Development of Anti-Malarial Drugs
Abstract
1. Introduction
2. Results
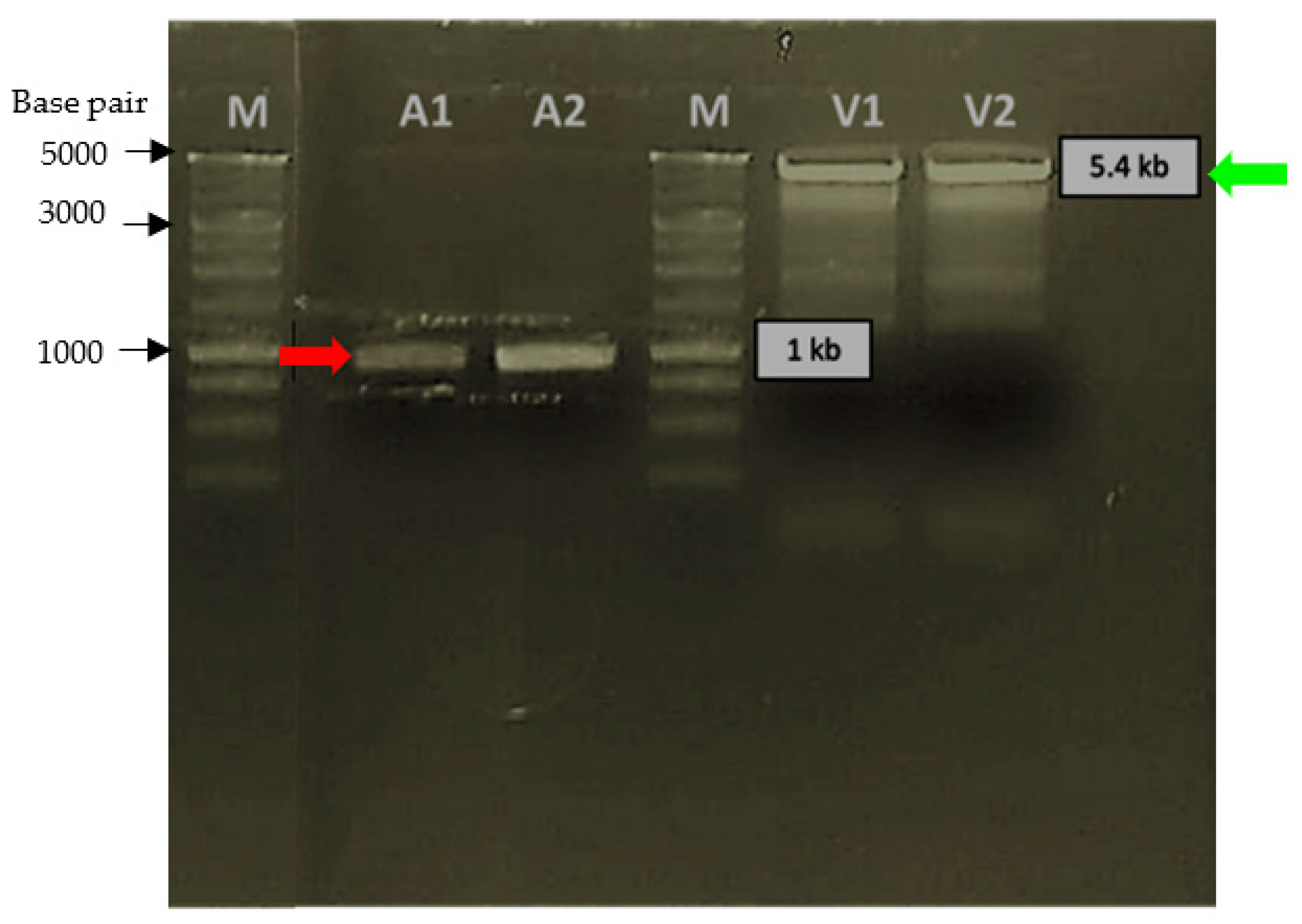
2.1. Sequence Analysis of Synthetic Pk-LDH, Cloning Strategy and Verification of Transformants
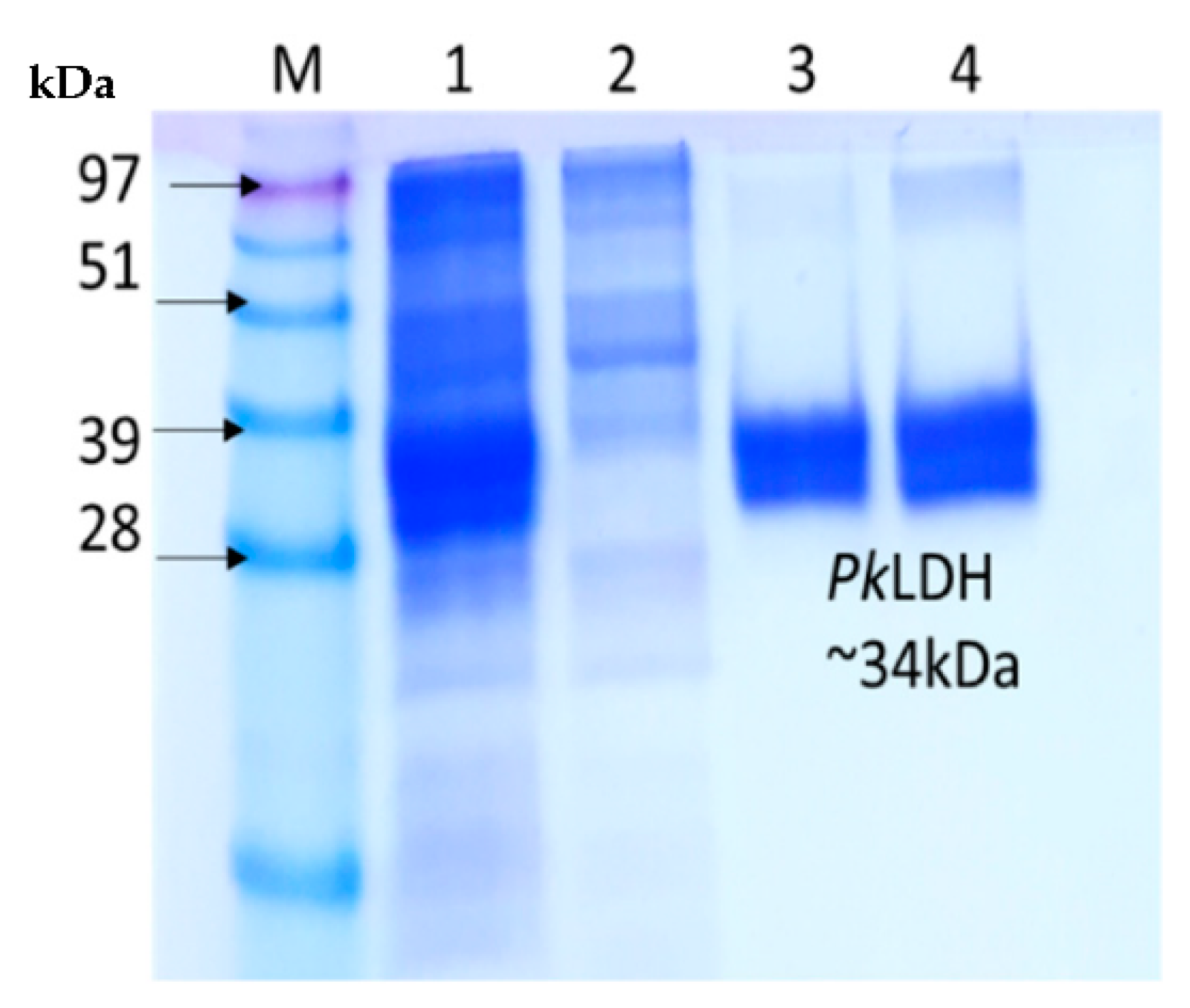
2.2. Expression of Recombinant Pk-LDH in E. coli BL21 (DE3)
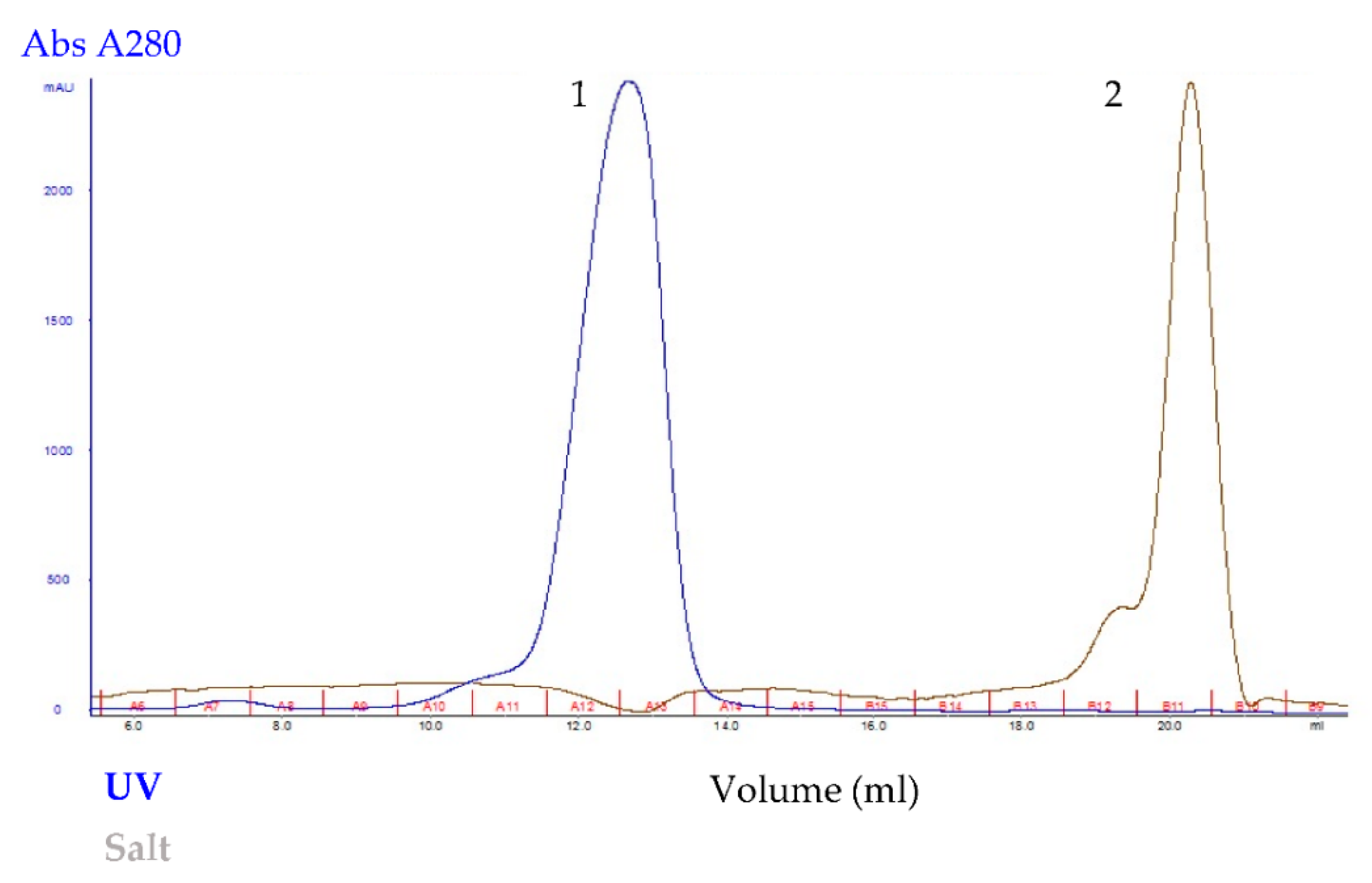
2.3. Purification of Recombinant Pk-LDH and Enzymatic Activity
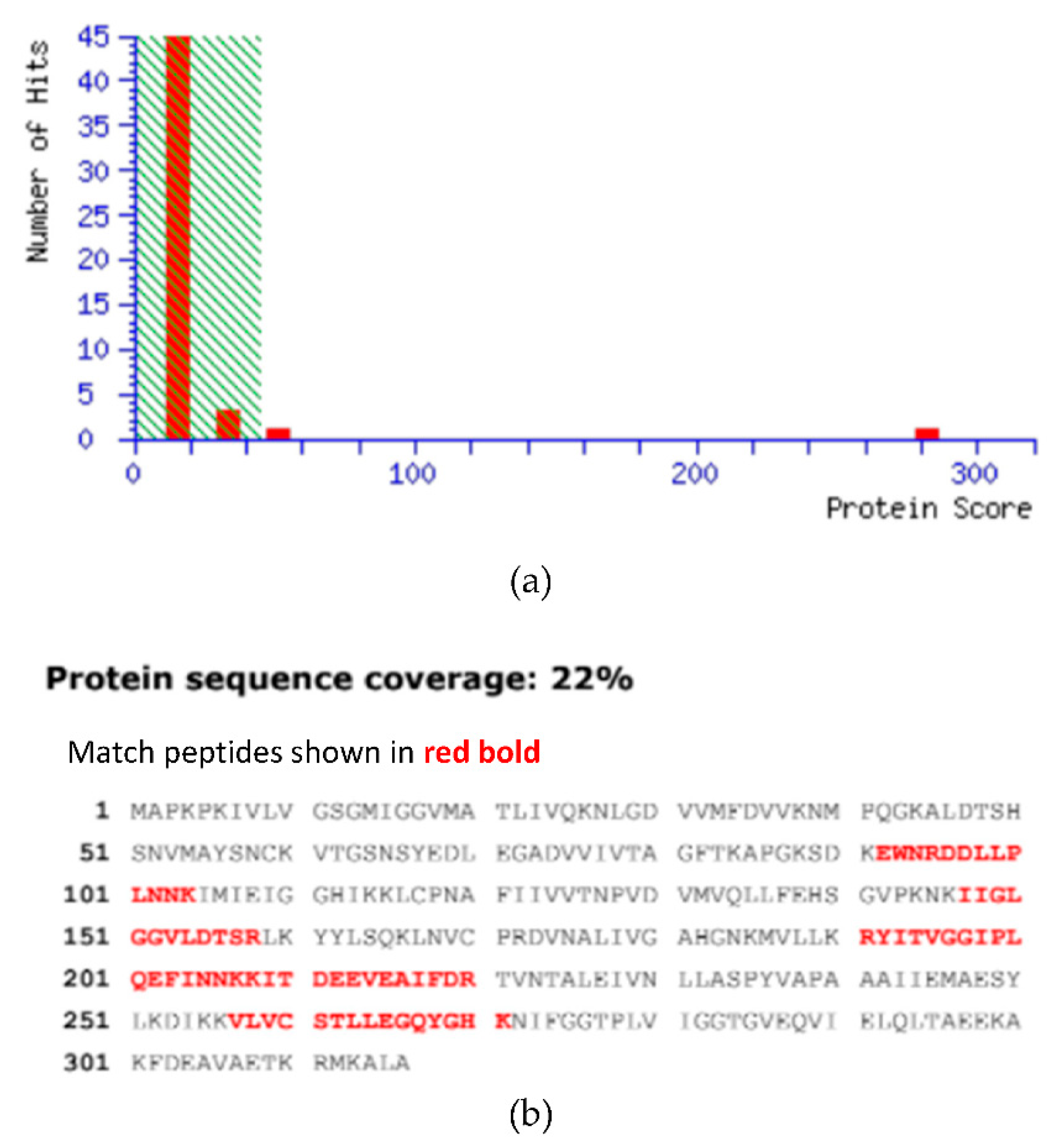
2.4. MALDI-TOF
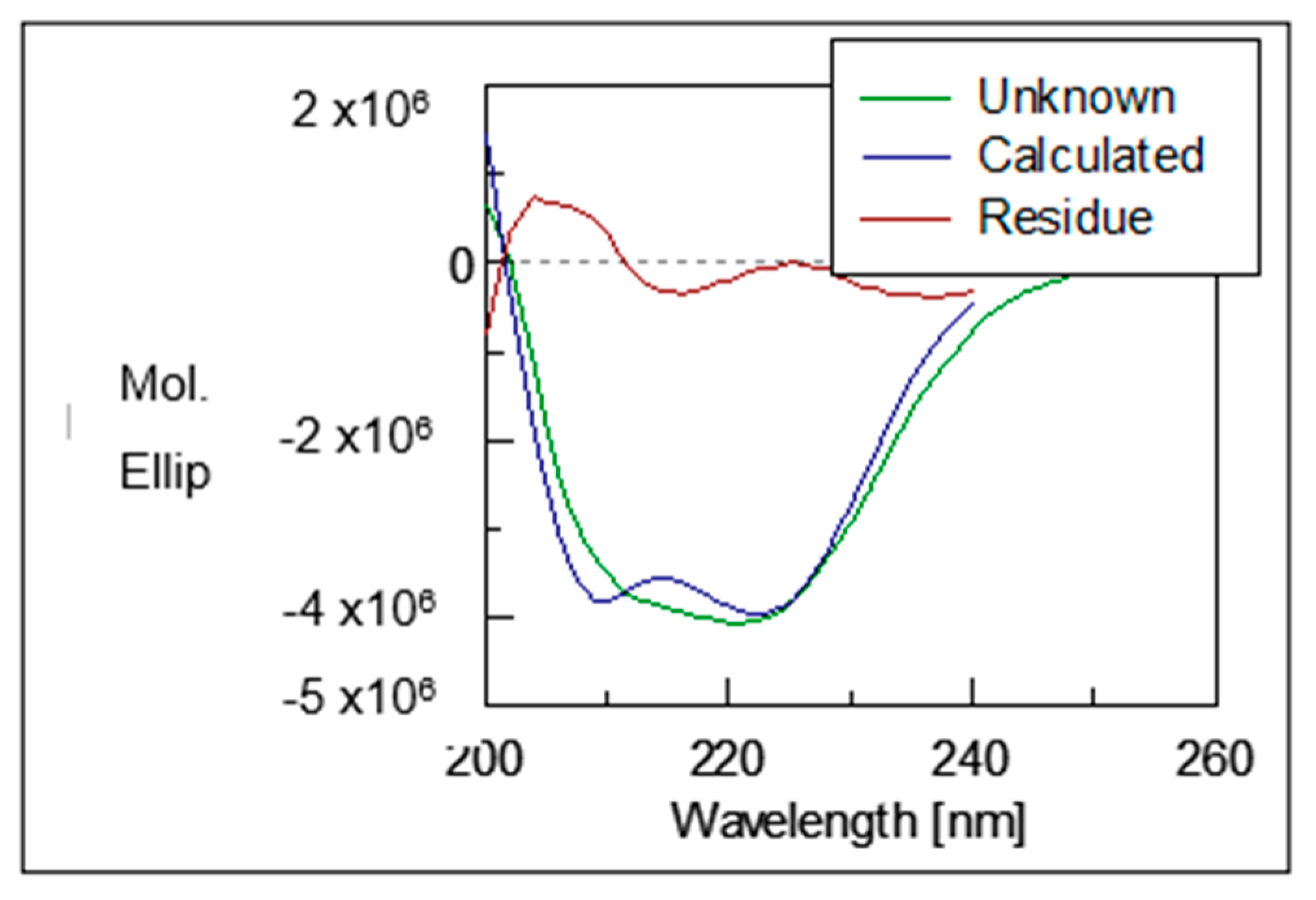
2.5. Circular Dichroisms of Recombinant Pk-LDH
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Amplification of Pk-LDH Gene, Cloning & Transformation into E. coli BL21 (DE3)
4.3. Expression of Recombinant Pk-LDH
4.4. Purification of Recombinant Pk-LDH and Protein Concentration
4.5. Pk-LDH Enzymatic Assays
4.6. In Gel Digestion and Matrix-Assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF) Mass Spectrometry
4.7. Circular Dichroisms Spectroscopy of Pk-LDH
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- World Malaria Report 2016; World Health Organizations: Geneva, Switzerland, 2017.
- Singh, B.; Sung, L.K.; Matusop, A.; Radhakrishnan, A.; Shamsul, S.S.; Cox-Singh, J.; Thomas, A.; Conway, D.J. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet 2004, 363, 1017–1024. [Google Scholar] [CrossRef]
- Kantele, A.; Jokiranta, T.S. Review of Cases with the Emerging Fifth Human Malaria Parasite, Plasmodium knowlesi. Clin. Infect. Dis. 2011, 52, 1356–1362. [Google Scholar] [CrossRef]
- Abeyasinghe, R. Plasmodium knowlesi current status and the request for review by an Evidence Review Group Malaria Policy Advisory Committee Geneva, Switzerland; 2016. Available online: http://www.who.int/malaria/mpac/mpac-sept2016-knowlesi.pdf (accessed on 2 September 2018).
- Cox-Singh, J.; Hiu, J.; Lucas, S.B.; Divis, P.C.; Zulkarnaen, M.; Chandran, P.; Wong, K.T.; Adem, P.; Zaki, S.R.; Singh, B.; et al. Severe malaria—A case of fatal Plasmodium knowlesi infection with post-mortem findings: A case report. Malar. J. 2010, 9, 10. [Google Scholar] [CrossRef]
- Dondorp, A.M.; Nosten, F.; Yi, P.; Das, D.; Phyo, A.P.; Tarning, J.; Lwin, K.M.; Ariey, F.; Hanpithakpong, W.; Lee, S.J.; et al. Artemisinin Resistance in Plasmodium falciparum Malaria. N. Engl. J. Med. 2009, 361, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Status Report on Artemisinin and ACT Resistance (April 2017). Geneva: World Health Organizations. 2018. Available online: https://www.who.int/malaria/publications/atoz/artemisinin-resistance-april2017/en/ (accessed on 12 December 2018).
- Nyunt, M.H.; Aye, K.M.; Kyaw, K.T.; Han, S.S.; Aye, T.T.; Wai, K.T.; Kyaw, M.P. Challenges encountered by local health volunteers in early diagnosis and prompt treatment of malaria in Myanmar artemisinin resistance containment zones. Malar. J. 2016, 15, 308. [Google Scholar] [CrossRef]
- Amir, A.; Cheong, F.W.; de Silva, J.R.; Liew, J.W.K.; Lau, Y.L. Plasmodium knowlesi malaria: Current research perspectives. Infect. Drug Resist. 2018, 11, 1145–1155. [Google Scholar] [CrossRef]
- Pal-Bhowmick, I.; Sadagopan, K.; Vora, H.K.; Sehgal, A.; Sharma, S.; Jarori, G.K. Cloning, over-expression, purification and characterization of Plasmodium falciparum enolase. Eur. J. Biochem. 2004, 271, 4845–4854. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, A.-K.; van Niekerk, D.D.; Adiels, C.B.; Kooi, B.; Goksör, M.; Snoep, J.L. Allosteric regulation of phosphofructokinase controls the emergence of glycolytic oscillations in isolated yeast cells. FEBS J. 2014, 281, 2784–2793. [Google Scholar] [CrossRef] [PubMed]
- Bruno, S.; Pinto, A.; Paredi, G.; Tamborini, L.; De Micheli, C.; La Pietra, V.; Marinelli, L.; Novellino, E.; Conti, P.; Mozzarelli, A. Discovery of Covalent Inhibitors of Glyceraldehyde-3-phosphate Dehydrogenase, A Target for the Treatment of Malaria. J. Med. Chem. 2014, 57, 7465–7471. [Google Scholar] [CrossRef]
- Ravindra, G.; Balaram, P. Plasmodium falciparum triosephosphate isomerase: New insights into an old enzyme. Pure Appl. Chem. 2005, 77, 281–289. [Google Scholar] [CrossRef]
- Penna-Coutinho, J.; Cortopassi, W.A.; Oliveira, A.A.; França, T.C.C.; Krettli, A.U. Antimalarial activity of potential inhibitors of Plasmodium falciparum lactate dehydrogenase enzyme selected by docking studies. PLoS ONE 2011, 6, e21237. [Google Scholar] [CrossRef]
- van Niekerk, D.D.; Penkler, G.P.; du Toit, F.; Snoep, J.L. Targeting glycolysis in the malaria parasite Plasmodium falciparum. FEBS J. 2016, 283, 634–646. [Google Scholar] [CrossRef]
- Nurhainis, O.S.; Fuad, F.A.A. Screening potential inhibitors of lactate dehydrogenase from Plasmodium knowlesi via virtual screening approaches. Trop. Biomed. 2017, 34, 841–854. [Google Scholar]
- Singh, V.; Kaushal, D.C.; Rathaur, S.; Kumar, N.; Kaushal, N.A. Cloning, overexpression, purification and characterization of Plasmodium knowlesi lactate dehydrogenase. Protein Expr. Purif. 2012, 84, 195–203. [Google Scholar] [CrossRef]
- Dunn, C.R.; Banfield, M.J.; Barker, J.J.; Higham, C.W.; Moreton, K.M.; Turgut-Balik, D.; Brady, R.L.; Holbrook, J.J. The structure of lactate dehydrogenase from Plasmodium falciparum reveals a new target for anti-malarial design. Nat. Struct. Biol. 1996, 3, 912–915. [Google Scholar] [CrossRef] [PubMed]
- Chaikuad, A.; Fairweather, V.; Conners, R.; Joseph-Horne, T.; Turgut-Balik, D.; Brady, R.L. Structure of lactate dehydrogenase from Plasmodium vivax: Complexes with NADH and APADH. Biochemistry 2005, 44, 16221–16228. [Google Scholar] [CrossRef]
- Roobsoong, W.; Roytrakul, S.; Kiatfuengfoo, R.; Nuchpramool, W.; Cui, L.; Udomsangpetch, R. Characterization of the Plasmodium vivax erythrocytic stage proteome and identification of a potent immunogenic antigen of the asexual stages. Malar. J. 2010, 9, P44. [Google Scholar] [CrossRef][Green Version]
- Pain, A.; Böhme, U.; Berry, A.E.; Mungall, K.; Finn, R.D.; Jackson, A.P.; Mourier, T.; Mistry, J.; Pasini, E.M.; Aslett, M.A.; et al. The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature 2008, 455, 799–803. [Google Scholar] [CrossRef] [PubMed]
- Berwal, R.; Gopalan, N.; Chandel, K.; Prasad, G.B.K.S.; Prakash, S. Plasmodium falciparum: Enhanced soluble expression, purification and biochemical characterization of lactate dehydrogenase. Exp. Parasitol. 2008, 120, 135–141. [Google Scholar] [CrossRef]
- Turgut-Balik, D.; Shoemark, D.K.; Sessions, R.B.; Moreton, K.M.; Holbrook, J.J. Mutagenic exploration of the active site of lactate dehydrogenase from Plasmodium falciparum. Biotechnol. Lett. 2001, 23, 923–927. [Google Scholar] [CrossRef]
- Schein, C.H. Production of Soluble Recombinant Proteins in Bacteria. Nat. Biotechnol. 1989, 7, 1141–1149. [Google Scholar] [CrossRef]
- Sørensen, H.P.; Mortensen, K.K. Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb. Cell Fact. 2005, 4, 1. [Google Scholar] [CrossRef]
- Baneyx, F.; Mujacic, M. Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 2004, 22, 1399–1408. [Google Scholar] [CrossRef] [PubMed]
- Augoff, K.; Hryniewicz-Jankowska, A.; Tabola, R. Lactate dehydrogenase 5: An old friend and a new hope in the war on cancer. Cancer Lett. 2015, 358, 1–7. [Google Scholar] [CrossRef]
- Mason, A.B.; He, Q.-Y.; Adams, T.E.; Gumerov, D.R.; Kaltashov, I.A.; Nguyen, V.; Macgillivray, R.T. Expression, purification, and characterization of recombinant nonglycosylated human serum transferrin containing a C-terminal hexahistidine tag. Protein Expr. Purif. 2001, 23, 142–150. [Google Scholar] [CrossRef]
- Peng, H.; Begg, G.E.; Harper, S.L.; Friedman, J.R.; Speicher, D.W.; Rauscher, F.J. Biochemical analysis of the Kruppel-associated box (KRAB) transcriptional repression domain: Spectral, kinetic, and stoichiometric properties of the KRAB·KAP-1 complex. J. Biol. Chem. 2000, 275, 18000–18010. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Huang, Z. Effect of His-Tag on Expression, Purification, and Structure of Zinc Finger Protein, ZNF191(243-368). Bioinorg. Chem. Appl. 2016, 2016, 8206854. [Google Scholar] [CrossRef]
- Shoemark, D.K.; Cliff, M.J.; Sessions, R.B.; Clarke, A.R. Enzymatic properties of the lactate dehydrogenase enzyme from Plasmodium falciparum. FEBS J. 2007, 274, 2738–2748. [Google Scholar] [CrossRef]
- Brown, W.M.; Yowell, C.A.; Hoard, A.; Vander Jagt, T.A.; Hunsaker, L.A.; Deck, L.M.; Royer, R.E.; Piper, R.C.; Dame, J.B.; Makler, M.T.; et al. Comparative structural analysis and kinetic properties of lactate dehydrogenases from the four species of human malarial parasites. Biochemistry 2004, 43, 6219–6229. [Google Scholar] [CrossRef] [PubMed]
- Mathema, V.B.; Na-Bangchang, K. A brief review on biomarkers and proteomic approach for malaria research. Asian Pac. J. Trop. Med. 2015, 8, 253–262. [Google Scholar] [CrossRef]
- Yergey, A.L.; Coorssen, J.R.; Backlund, P.S.; Blank, P.S.; Humphrey, G.A.; Zimmerberg, J.; Campbell, J.M.; Vestal, M.L. De novo sequencing of peptides using MALDI/TOF-TOF. J. Am. Soc. Mass Spectrom. 2002, 13, 784–791. [Google Scholar] [CrossRef]
- Hrabák, J.; Chudáčková, E.; Walková, R. Matrix-Assisted Laser Desorption Ionization–Time of Flight (MALDI-TOF) Mass Spectrometry for Detection of Antibiotic Resistance Mechanisms: From Research to Routine Diagnosis. Clin. Microbiol. Rev. 2013, 26, 103–114. [Google Scholar] [CrossRef]
- Dutta, S.; Ware, L.A.; Barbosa, A.; Ockenhouse, C.F.; Lanar, D.E. Purification, characterization, and immunogenicity of a disulfide cross-linked Plasmodium vivax vaccine candidate antigen, merozoite surface protein 1, expressed in Escherichia coli. Infect. Immun. 2001, 69, 5464–5470. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Kamath, K.S.; Srivastava, R.; Raghu, D.; Gollapalli, K.; Jain, R.; Gupta, S.V.; Ray, S.; Taur, S.; Dhali, S.; et al. Serum proteome analysis of vivax malaria: An insight into the disease pathogenesis and host immune response. J. Proteom. 2012, 75, 3063–3080. [Google Scholar] [CrossRef] [PubMed]
- Das, N.; Maity, S.; Chakraborty, J.; Pal, S.; Sardar, S.; Halder, U.C. Purification and characterization of a gelatinolytic serine protease from the seeds of ash gourd Benincasa hispida (Thunb.) Cogn. 2018; Volume 55. Available online: http://nopr.niscair.res.in/handle/123456789/44349 (accessed on 18 October 2021).
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta—Proteins Proteom. 2005, 1751, 119–139. [Google Scholar] [CrossRef]
- Matsuo, K.; Yonehara, R.; Gekko, K. Improved Estimation of the Secondary Structures of Proteins by Vacuum-Ultraviolet Circular Dichroism Spectroscopy. J. Biochem. 2005, 138, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, L.E.; Vera, R.; Valbuena, J.; Curtidor, H.; Garcia, J.; Puentes, A.; Ocampo, M.; Lopez, R.; Rosas, J.; Lopez, Y.; et al. Characterisation of Plasmodium falciparum RESA-like protein peptides that bind specifically to erythrocytes and inhibit invasion. Biol. Chem. 2007, 388, 15–24. [Google Scholar] [CrossRef]
- Pinzón, C.G.; Curtidor, H.; Reyes, C.; Méndez, D.; Patarroyo, M.E. Identification of Plasmodium falciparum RhopH3 protein peptides that specifically bind to erythrocytes and inhibit merozoite invasion. Protein Sci. 2008, 17, 1719–1730. [Google Scholar] [CrossRef]

| Purification Stages | Total Protein (mg) | Total Activity (units) | Specific Activity (µmol/min/mg) | Yield (%) | Fold Purification |
|---|---|---|---|---|---|
| Sonicated extract (crude) | 8.3 | 2505.77 | 301.9 | 100.0 | 1.0 |
| Immobilized metal affinity chromatography (IMAC) | 1.2 | 441.6 | 368.0 | 17.6 | 1.2 |
| Size exclusion chromatography (SEC) | 0.2 | 77.46 | 475.6 | 3.09 | 1.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salim, N.O.; Fuad, F.A.A.; Khairuddin, F.; Seman, W.M.K.W.; Jonet, M.A. Purifying and Characterizing Bacterially Expressed Soluble Lactate Dehydrogenase from Plasmodium knowlesi for the Development of Anti-Malarial Drugs. Molecules 2021, 26, 6625. https://doi.org/10.3390/molecules26216625
Salim NO, Fuad FAA, Khairuddin F, Seman WMKW, Jonet MA. Purifying and Characterizing Bacterially Expressed Soluble Lactate Dehydrogenase from Plasmodium knowlesi for the Development of Anti-Malarial Drugs. Molecules. 2021; 26(21):6625. https://doi.org/10.3390/molecules26216625
Chicago/Turabian StyleSalim, Nurhainis Ogu, Fazia Adyani Ahmad Fuad, Farahayu Khairuddin, Wan Mohd Khairulikhsan Wan Seman, and Mohd Anuar Jonet. 2021. "Purifying and Characterizing Bacterially Expressed Soluble Lactate Dehydrogenase from Plasmodium knowlesi for the Development of Anti-Malarial Drugs" Molecules 26, no. 21: 6625. https://doi.org/10.3390/molecules26216625
APA StyleSalim, N. O., Fuad, F. A. A., Khairuddin, F., Seman, W. M. K. W., & Jonet, M. A. (2021). Purifying and Characterizing Bacterially Expressed Soluble Lactate Dehydrogenase from Plasmodium knowlesi for the Development of Anti-Malarial Drugs. Molecules, 26(21), 6625. https://doi.org/10.3390/molecules26216625






