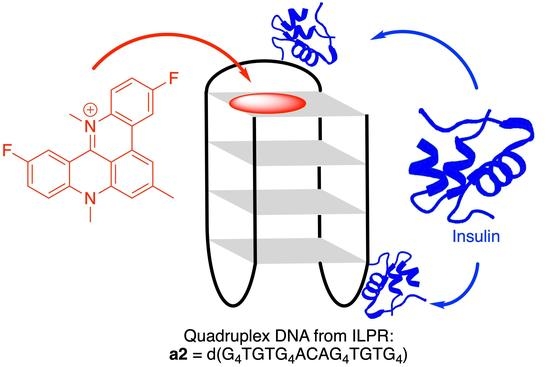
Studies on the Interactions of 3,11-Difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium and Insulin with the Quadruplex-Forming Oligonucleotide Sequence a2 from the Insulin-Linked Polymorphic Region
Abstract
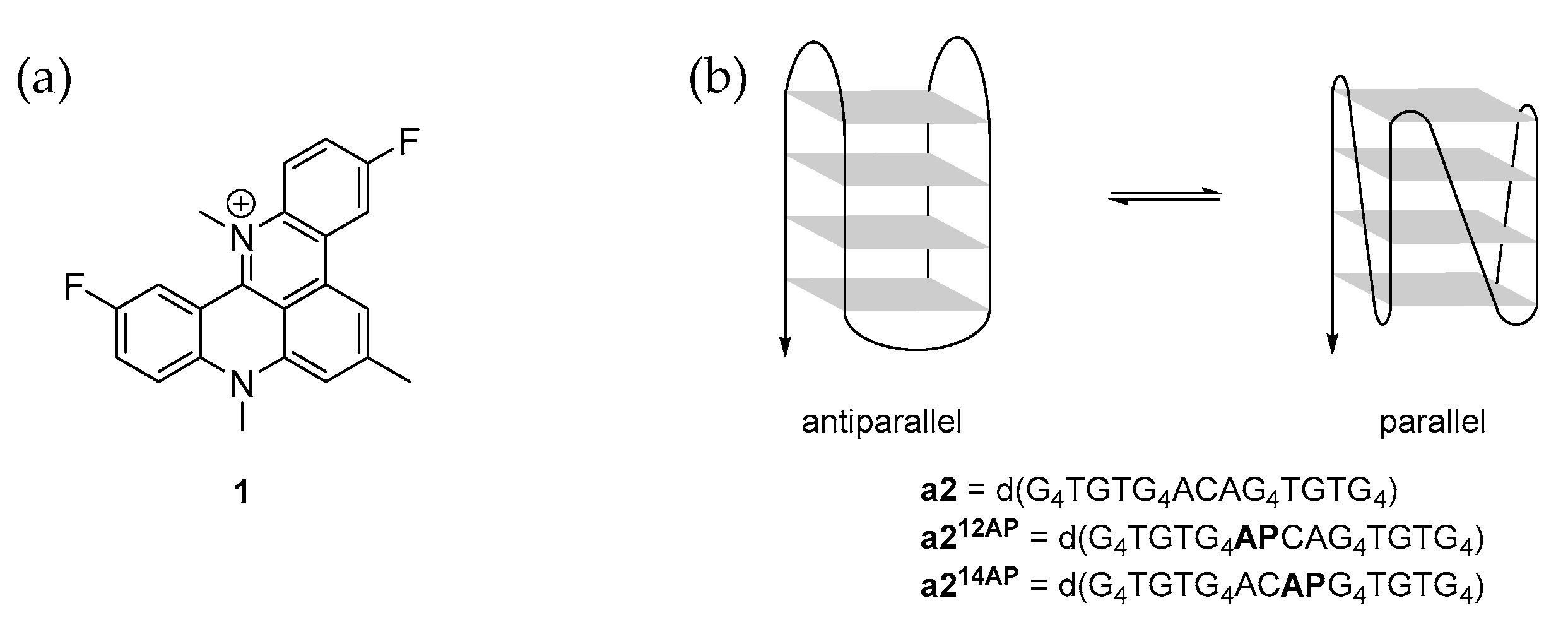
:1. Introduction
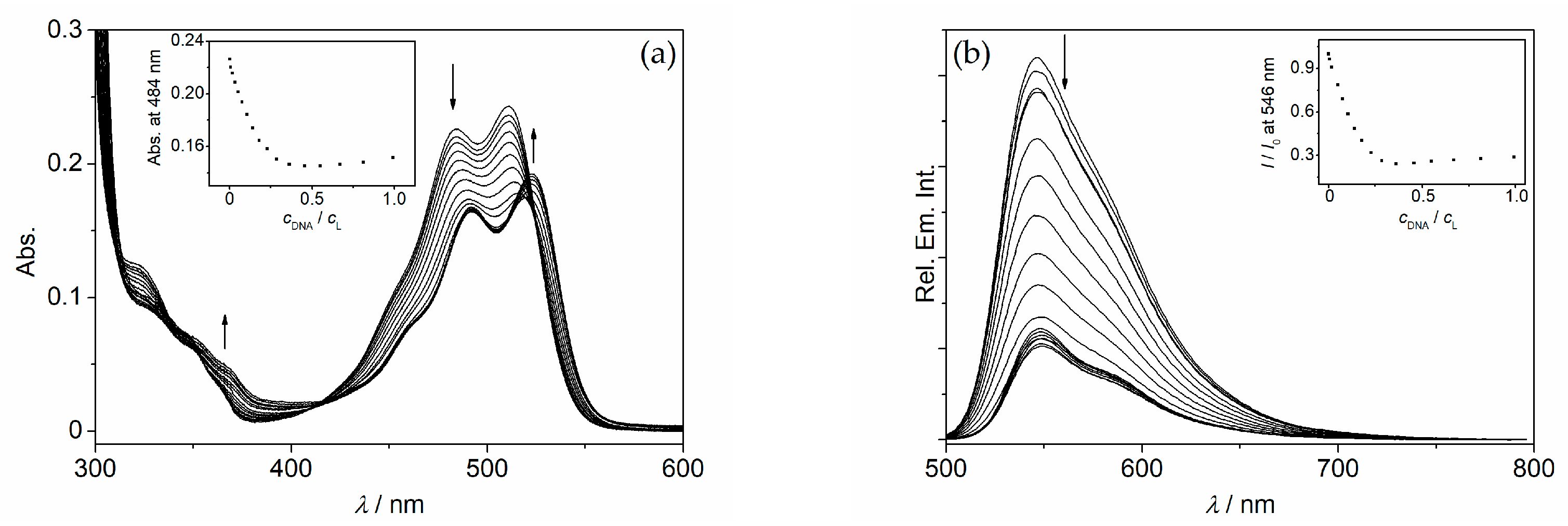
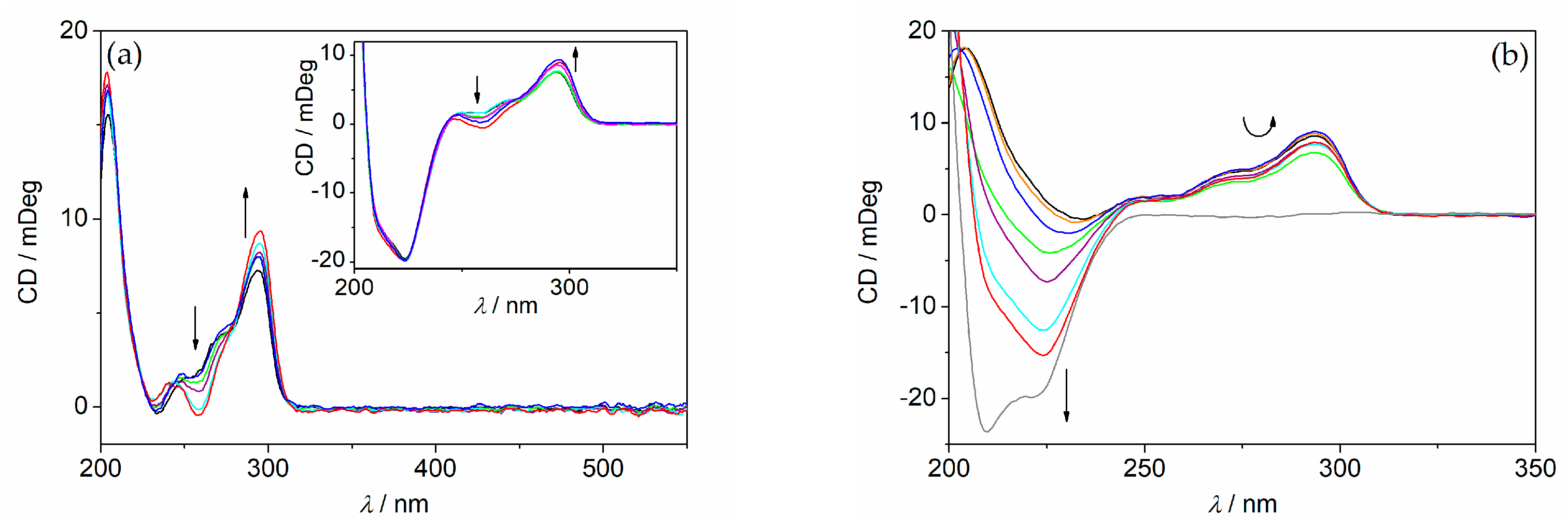
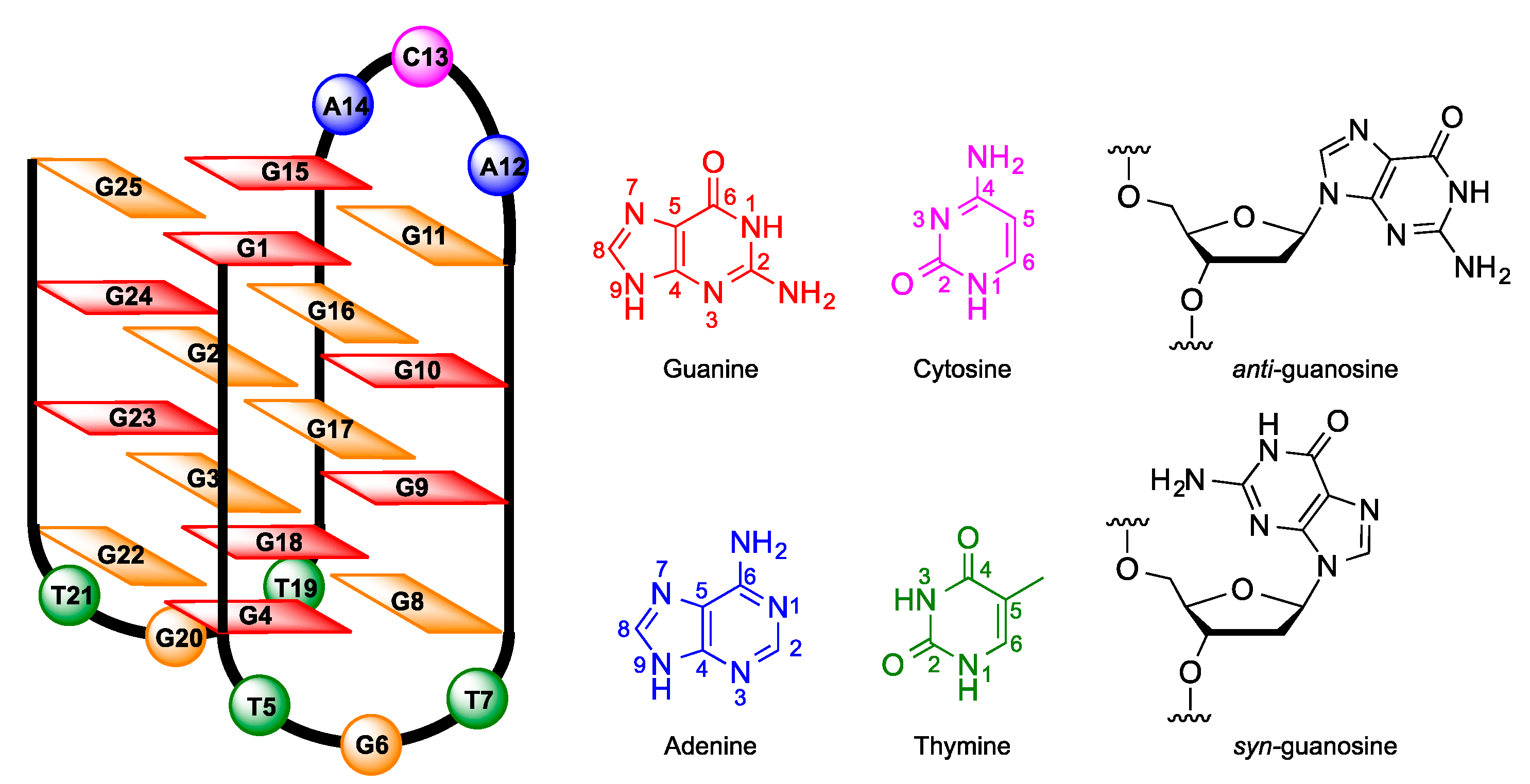
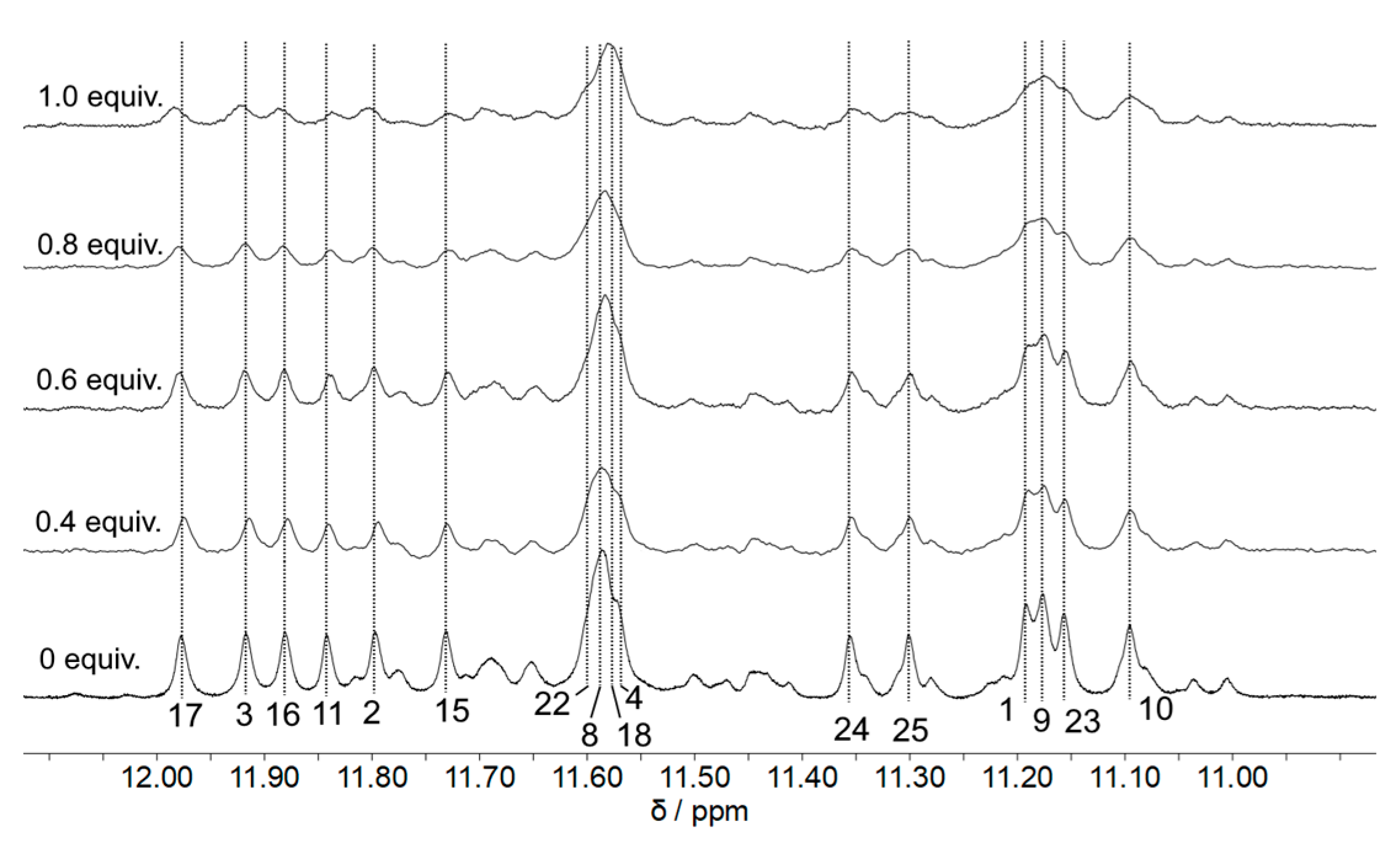
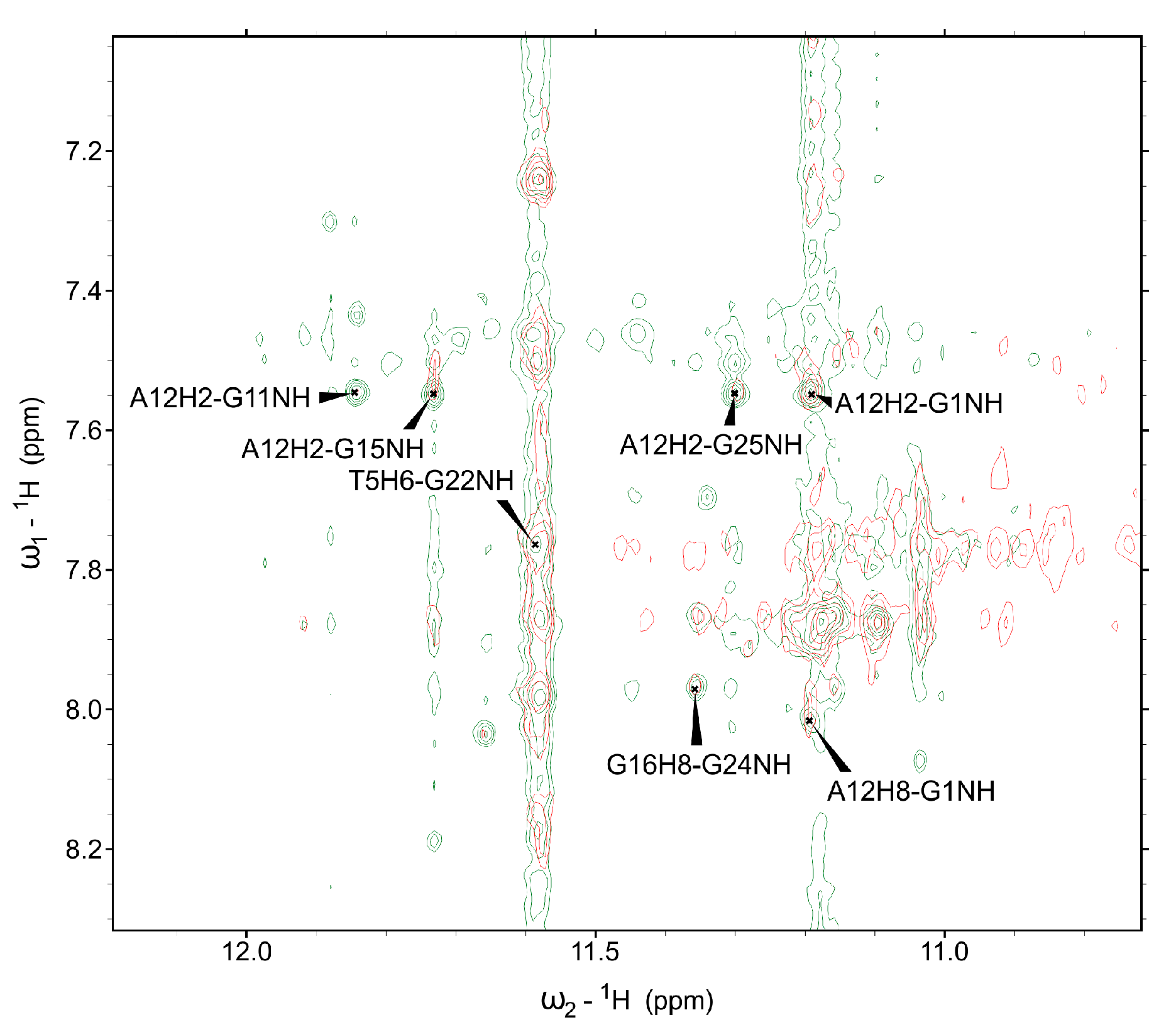
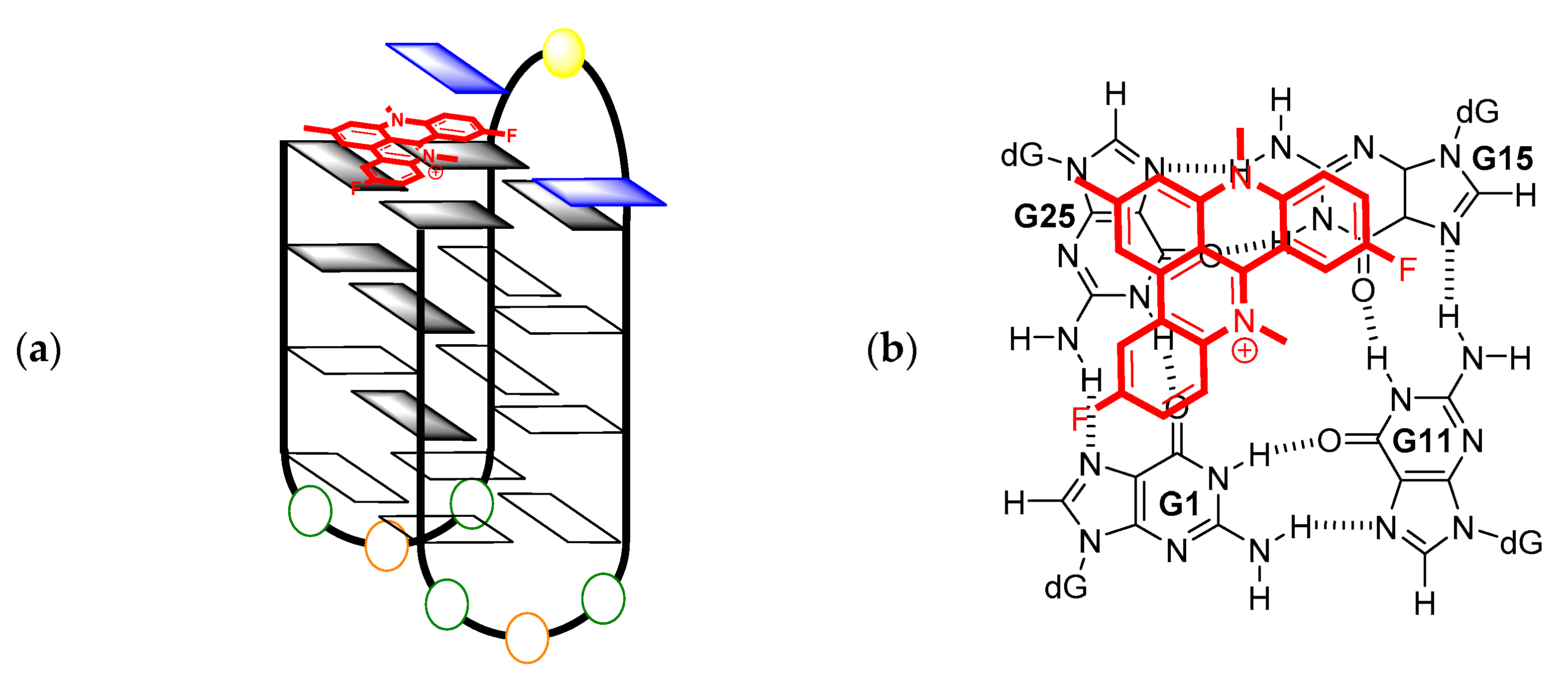
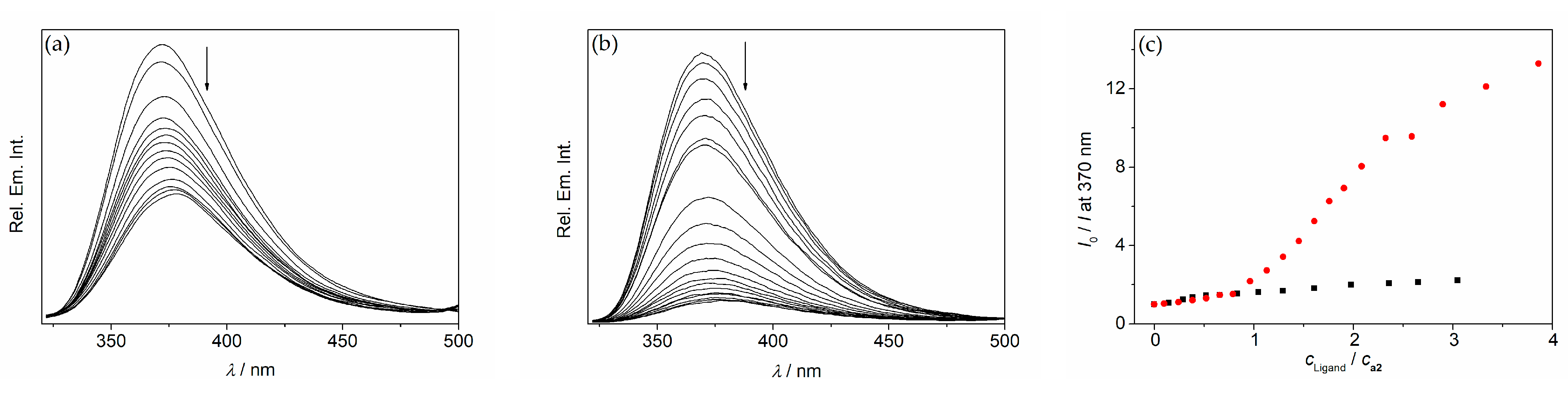
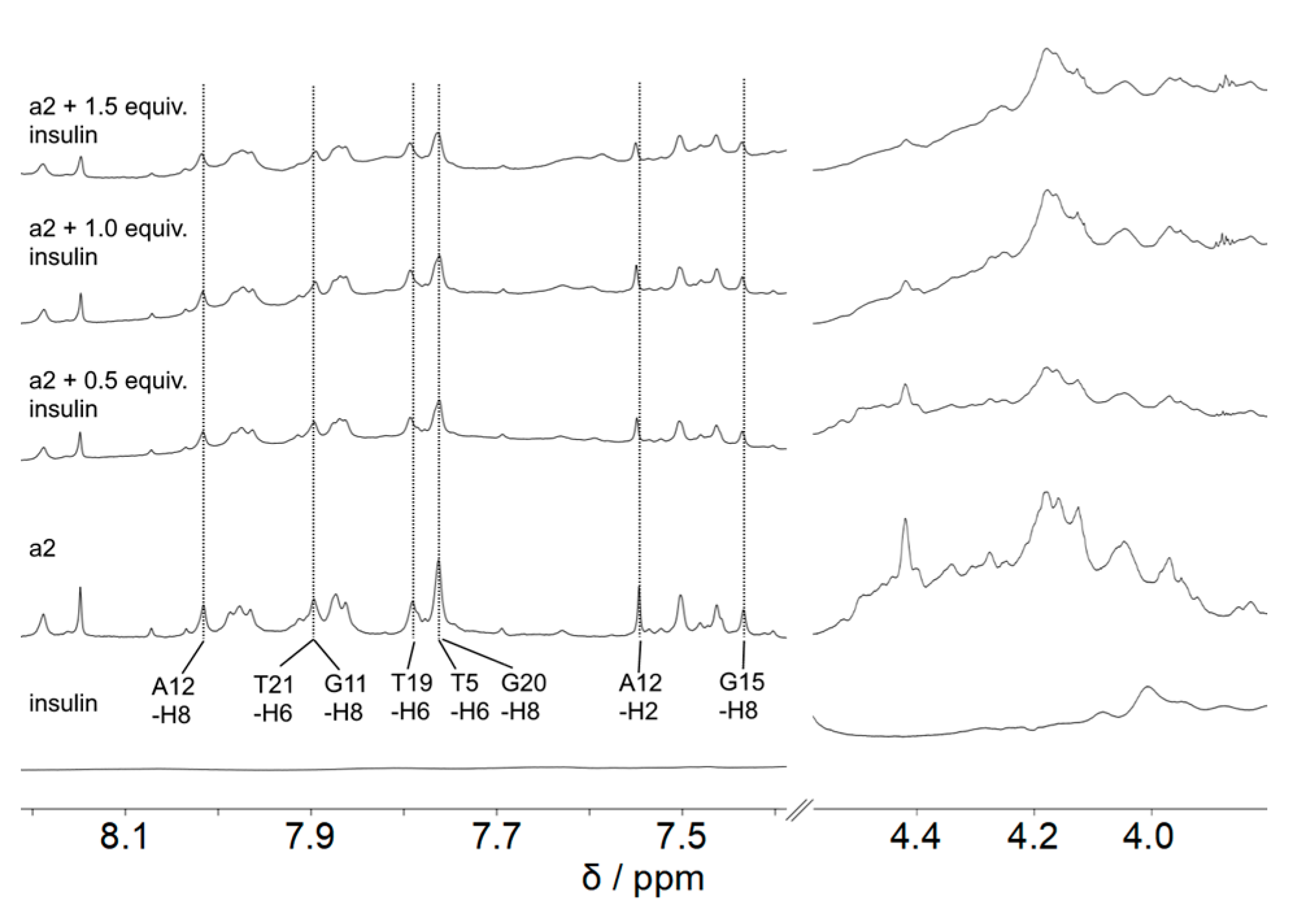
2. Results
3. Materials and Methods
3.1. Equipment
3.2. Materials
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Kim, N. The Interplay between G-quadruplex and Transcription. Curr. Med. Chem. 2019, 26, 2898–2917. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.-Q.; Chen, C.-Z.; Xue, Y.; Hao, Y.-H.; Tan, Z. G-Quadruplex Hinders Translocation of BLM Helicase on DNA: A Real-Time Fluorescence Spectroscopic Unwinding Study and Comparison with Duplex Substrates. J. Am. Chem. Soc. 2010, 132, 10521–10527. [Google Scholar] [CrossRef]
- González, V.; Guo, K.; Hurley, L.; Sun, D. Identification and Characterization of Nucleolin as a c-myc G-quadruplex-binding Protein. J. Biol. Chem. 2009, 284, 23622–23635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.-B.; Dickerhoff, J.; Wu, G.; Yang, D. PDGFR-β Promoter Forms a Vacancy G-Quadruplex that Can Be Filled in by dGMP: Solution Structure and Molecular Recognition of Guanine Metabolites and Drugs. J. Am. Chem. Soc. 2020, 142, 5204–5211. [Google Scholar] [CrossRef]
- Ray, S.; Tillo, D.; Boer, R.E.; Assad, N.; Barshai, M.; Wu, G.; Orenstein, Y.; Yang, D.; Schneekloth, J.S., Jr.; Vinson, C. Custom DNA Microarrays Reveal Diverse Binding Preferences of Proteins and Small Molecules to Thousands of G-Quadruplexes. ACS Chem. Biol. 2020, 15, 925–935. [Google Scholar] [CrossRef]
- Thakur, R.K.; Kumar, P.; Halder, K.; Verma, A.; Kar, A.; Parent, J.-L.; Basundra, R.; Kumar, A.; Chowdhury, S. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res. 2009, 37, 172–183. [Google Scholar] [CrossRef] [Green Version]
- Fry, M.; Loeb, L.A. Human Werner Syndrome DNA Helicase Unwinds Tetrahelical Structures of the Fragile X Syndrome Repeat Sequence d(CGG). J. Biol. Chem. 1999, 274, 12797–12802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohaghegh, P. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001, 29, 2843–2849. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Xing, Z.; Tran, E.J.; Yang, D. DDX5 helicase resolves G-quadruplex and is involved in MYC gene transcriptional activation. Proc. Natl. Acad. Sci. USA 2019, 116, 20453–20461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, G.C.; German, M.S.; Rutter, W.J. The minisatellite in the diabetes susceptibility locus IDDM2 regulates insulin transcription. Nat. Genet. 1995, 9, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Rotwein, P.; Yokoyama, S.; Didier, D.K.; Chirgwin, J.M. Genetic analysis of the hypervariable region flanking the human insulin gene. Am. J. Hum. Genet. 1986, 39, 291–299. [Google Scholar]
- Yu, Z.; Schonhoft, J.D.; Dhakal, S.; Bajracharya, R.; Hegde, R.; Basu, S.; Mao, H. ILPR G-quadruplexes formed in seconds demonstrate high mechanical stabilities. J. Am. Chem. Soc. 2009, 131, 1876–1882. [Google Scholar] [CrossRef]
- Gerasimov, J.Y.; Schaefer, C.S.; Yang, W.; Grout, R.L.; Lai, R.Y. Development of an electrochemical insulin sensor based on the insulin-linked polymorphicregion. Biosens. Bioelectron. 2013, 42, 62–68. [Google Scholar] [CrossRef]
- Xiao, J.; Carter, J.A.; Frederick, K.A.; McGown, L.B. A genome-inspired DNA ligand for the affinity capture of insulin and insulin-like growth factor-2. J. Sep. Sci. 2009, 32, 1654–1664. [Google Scholar] [CrossRef] [Green Version]
- Timmer, C.M.; Michmerhuizen, N.L.; Witte, A.B.; Van Winkle, M.; Zhou, D.; Sinniah, K. An Isothermal Titration and Differential Scanning Calorimetry Study of the G-Quadruplex DNA–Insulin Interaction. J. Phys. Chem. B 2014, 118, 1784–1790. [Google Scholar] [CrossRef]
- Rafati, A.; Zarrabi, A.; Abediankenari, S.; Aarabi, M.; Gill, P. Sensitive colorimetric assay using insulin G-quadruplex aptamer arrays on DNA nanotubes coupled with magnetic nanoparticles. R. Soc. Open Sci. 2018, 5, 171835. [Google Scholar] [CrossRef] [Green Version]
- Schonhoft, J.D.; Das, A.; Achamyeleh, F.; Samdani, S.; Sewell, A.; Mao, H.; Basu, S. ILPR repeats adopt diverse G-quadruplex conformations that determine insulin binding. Biopolymers 2010, 93, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Chen, Y.-Q.; Wang, S.-R.; Zhou, X. G-Quadruplex: A Regulator of Gene Expression and Its Chemical Targeting. Chem 2018, 4, 1314–1344. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA. 2002, 99, 11593–11598. [Google Scholar] [CrossRef] [Green Version]
- Kosiol, N.; Juranek, S.; Brossart, P.; Heine, A.; Paeschke, K. G-quadruplexes: A promising target for cancer therapy. Mol. Cancer 2021, 20, 40. [Google Scholar] [CrossRef]
- Mazzini, S.; Gargallo, R.; Musso, L.; De Santis, F.; Aviñó, A.; Scaglioni, L.; Eritja, R.; Di Nicola, M.; Zunino, F.; Amatulli, A.; et al. Stabilization of c-KIT G-Quadruplex DNA Structures by the RNA Polymerase I Inhibitors BMH-21 and BA-41. Int. J. Mol. Sci. 2019, 20, 4927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dzubiel, D.; Ihmels, H.; Mahmoud, M.M.A.; Thomas, L. A comparative study of the interactions of cationic hetarenes with quadruplex-DNA forming oligonucleotide sequences of the insulin-linked polymorphic region (ILPR). Beilstein J. Org. Chem. 2014, 10, 2963–2974. [Google Scholar] [CrossRef] [Green Version]
- Catasti, P.; Chen, X.; Moyzis, R.K.; Bradbury, E.M.; Gupta, G. Structure-function correlations of the insulin-linked polymorphic region. J. Mol. Biol. 1996, 264, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Phatak, P.; Cookson, J.C.; Dai, F.; Smith, V.; Gartenhaus, R.B.; Stevens, M.F.G.; Burger, A.M. Telomere uncapping by the G-quadruplex ligand RHPS4 inhibits clonogenic tumour cell growth in vitro and in vivo consistent with a cancer stem cell targeting mechanism. Br. J. Cancer 2007, 96, 1223–1233. [Google Scholar] [CrossRef] [PubMed]
- Zizza, P.; Cingolani, C.; Artuso, S.; Salvati, E.; Rizzo, A.; D’Angelo, C.; Porru, M.; Pagano, B.; Amato, J.; Randazzo, A.; et al. Intragenic G-quadruplex structure formed in the human CD133 and its biological and translational relevance. Nucleic Acids Res. 2016, 44, 1579–1590. [Google Scholar] [CrossRef]
- Berardinelli, F.; Tanori, M.; Muoio, D.; Buccarelli, M.; Di Masi, A.; Leone, S.; Ricci-Vitiani, L.; Pallini, R.; Mancuso, M.; Antoccia, A. G-quadruplex ligand RHPS4 radiosensitizes glioblastoma xenograft in vivo through a differential targeting of bulky differentiated- and stem-cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 311. [Google Scholar] [CrossRef] [PubMed]
- Dalloul, Z.; Chenuet, P.; Dalloul, I.; Boyer, F.; Aldigier, J.-C.; Laffleur, B.; El Makhour, Y.; Ryffel, B.; Quesniaux, V.F.J.; Togbé, D.; et al. G-quadruplex DNA targeting alters class-switch recombination in B cells and attenuates allergic in-flammation. J. Allergy Clin. Immunol. 2018, 142, 1352–1355. [Google Scholar] [CrossRef] [PubMed]
- Falabella, M.; Kolesar, J.E.; Wallace, C.; De Jesus, D.; Sun, L.; Taguchi, Y.V.; Wang, C.; Wang, T.; Xiang, I.M.; Alder, J.K.; et al. G-quadruplex dynamics contribute to regulation of mitochondrial gene expression. Sci. Rep. 2019, 9, 5605. [Google Scholar] [CrossRef]
- Smith, J.A.; George, M.W.; Kelly, J.M. Transient spectroscopy of dipyridophenazine metal complexes which undergo photo-induced electron transfer with DNA. Coord. Chem. Rev. 2011, 255, 2666–2675. [Google Scholar] [CrossRef]
- Viola, G.; Bressanini, M.; Gabellini, N.; Vedaldi, D.; Dall’Acqua, F.; Ihmels, H. Naphthoquinolizinium derivatives as a novel platform for DNA-binding and DNA-photodamaging chromophores. Photochem. Photobiol. Sci. 2002, 1, 882–889. [Google Scholar] [CrossRef]
- Torimura, M.; Kurata, S.; Yamada, K.; Yokomaku, T.; Kamagata, Y.; Kanagawa, T.; Kurane, R. Fluorescence-quenching phenomenon by photoinduced electron transfer between a fluorescent dye and a nucleotide base. Anal. Sci. 2001, 17, 155–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patro, B.S.; Bhattacharyya, R.; Gupta, P.; Bandyopadhyay, S.; Chattopadhyay, S. Mechanism of coralyne-mediated DNA photo-nicking process. J. Photochem. Photobiol. 2019, 194, 140–148. [Google Scholar] [CrossRef]
- Stootman, F.H.; Fisher, D.M.; Rodger, A.; Aldrich-Wright, J.R. Improved curve fitting procedures to determine equilibrium binding constants. Analyst 2006, 131, 1145–1151. [Google Scholar] [CrossRef]
- Mulholland, K.; Siddiquei, F.; Wu, C. Binding modes and pathway of RHPS4 to human telomeric G-quadruplex and duplex DNA probed by all-atom molecular dynamics simulations with explicit solvent. Phys. Chem. Chem. Phys. 2017, 19, 18685–18694. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Xu, J.; Chen, Y.; Wang, Q.; Lu, Z.; Chen, Y.; Chen, K.; Han, S.; Fang, Z.; Wang, P.; et al. Photoactive G-Quadruplex Ligand Identifies Multiple G-Quadruplex-Related Proteins with Extensive Sequence Tolerance in the Cellular Environment. J. Am. Chem. Soc. 2021, 143, 1917–1923. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Duchambon, P.; Masson, V.; Loew, D.; Bombard, S.; Teulade-Fichou, M.-P. Nucleolin Discriminates Drastically between Long-Loop and Short-Loop Quadruplexes. Biochemistry 2020, 59, 1261–1272. [Google Scholar] [CrossRef]
- Carvalho, J.; Queiroz, J.A.; Cruz, C. Circular Dichroism of G-Quadruplex: A Laboratory Experiment for the Study of Topology and Ligand Binding. J. Chem. Educ. 2017, 94, 1547–1551. [Google Scholar] [CrossRef]
- Garner, T.P.; Williams, H.E.L.; Gluszyk, K.I.; Roe, S.; Oldham, N.J.; Stevens, M.F.G.; Moses, J.E.; Searle, M.S. Selectivity of small molecule ligands for parallel and anti-parallel DNA G-quadruplex structures. Org. Biomol. Chem. 2009, 7, 4194–4200. [Google Scholar] [CrossRef]
- Šmidlehner, T.; Piantanida, I.; Pescitelli, G. Polarization spectroscopy methods in the determination of interactions of small molecules with nucleic acids–tutorial. Beilstein J. Org. Chem. 2018, 14, 84–105. [Google Scholar] [CrossRef] [PubMed]
- Nordén, B.; Kurucsev, T. Analysing DNA complexes by circular and linear dichroism. J. Mol. Recognit. 1994, 7, 141–155. [Google Scholar] [CrossRef]
- Yamashita, T.; Uno, T.; Ishikawa, Y. Stabilization of guanine quadruplex DNA by the binding of porphyrins with cationic side arms. Bioorg. Med. Chem. 2005, 13, 2423–2430. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Tang, Y.; Xiang, J.; Xu, G.; Zhang, Y.; Zhang, H.; Xu, L. Spectroscopic studies of the interaction between quercetin and G-quadruplex DNA. Bioorg. Med. Chem. Lett. 2006, 16, 3586–3589. [Google Scholar] [CrossRef]
- Cary, P.D.; Kneale, G.G. Circular dichroism for the analysis of protein-DNA interactions. Methods Mol. Biol. 2009, 543, 613–625. [Google Scholar] [PubMed]
- Ambrus, A.; Chen, D.; Dai, J.; Bialis, T.; Jones, R.A.; Yang, D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006, 34, 2723–2735. [Google Scholar] [CrossRef] [Green Version]
- Phan, A.T.; Kuryavyi, V.; Burge, S.; Neidle, S.; Patel, D.J. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007, 129, 4386–4392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adrian, M.; Heddi, B.; Phan, A.T. NMR spectroscopy of G-quadruplexes. Methods 2012, 57, 11–24. [Google Scholar] [CrossRef]
- Lin, C.; Dickerhoff, J.; Yang, D. NMR Studies of G-Quadruplex Structures and G-Quadruplex-Interactive Compounds. Methods Mol. Biol. 2019, 2035, 157–176. [Google Scholar]
- Greene, K.L.; Wang, Y.; Live, D. Influence of the glycosidic torsion angle on 13 C and 15 N shifts in guanosine nucleotides: Investigations of G-tetrad models with alternating syn and anti bases. J. Biomol. NMR 1995, 5, 333–338. [Google Scholar] [CrossRef]
- Furukawa, A.; Konuma, T.; Yanaka, S.; Sugase, K. Quantitative analysis of protein–ligand interactions by NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2016, 96, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Gavathiotis, E.; Heald, R.A.; Stevens, M.F.G.; Searle, M.S. Recognition and Stabilization of Quadruplex DNA by a Potent New Telomerase Inhibitor: NMR Studies of the 2:1 Complex of a Pentacyclic Methylacridinium Cation with d(TTAGGGT)4. Angew. Chem. Int. Ed. 2001, 40, 4749–4751. [Google Scholar] [CrossRef]
- Gray, R.D.; Petraccone, L.; Buscaglia, R.; Chaires, J.B. 2-Aminopurine as a Probe for Quadruplex Loop Structures. Methods Mol. Biol. 2010, 608, 121–136. [Google Scholar]
- Ihmels, H.; Löhl, K.; Paululat, T.; Uebacha, S. NMR-spectroscopic investigation of the complex between tetraazoniapentapheno[6,7-h]pentaphene and quadruplex DNA Tel26. New J. Chem. 2018, 42, 13813–13818. [Google Scholar] [CrossRef]
- Schnarr, L.; Jana, J.; Preckwinkel, P.; Weisz, K. Impact of a Snap-Back Loop on Stability and Ligand Binding to a Parallel G-Quadruplex. J. Phys. Chem. B 2020, 124, 2778–2787. [Google Scholar] [CrossRef]
- Lee, W.; Tonelli, M.; Markley, J.L. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 2015, 31, 1325–1327. [Google Scholar] [CrossRef] [Green Version]
- Bortolozzi, R.; Ihmels, H.; Thomas, L.; Tian, M.; Viola, G. 9-(4-Dimethylaminophenyl)-benzo[b]quinolizinium: A Near-Infrared Fluorophore for the Multicolor Analysis of Proteins and Nucleic Acids in Living Cells. Chem. Eur. J. 2013, 19, 8736–8741. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wickhorst, P.J.; Ihmels, H.; Paululat, T. Studies on the Interactions of 3,11-Difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium and Insulin with the Quadruplex-Forming Oligonucleotide Sequence a2 from the Insulin-Linked Polymorphic Region. Molecules 2021, 26, 6595. https://doi.org/10.3390/molecules26216595
Wickhorst PJ, Ihmels H, Paululat T. Studies on the Interactions of 3,11-Difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium and Insulin with the Quadruplex-Forming Oligonucleotide Sequence a2 from the Insulin-Linked Polymorphic Region. Molecules. 2021; 26(21):6595. https://doi.org/10.3390/molecules26216595
Chicago/Turabian StyleWickhorst, Peter Jonas, Heiko Ihmels, and Thomas Paululat. 2021. "Studies on the Interactions of 3,11-Difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium and Insulin with the Quadruplex-Forming Oligonucleotide Sequence a2 from the Insulin-Linked Polymorphic Region" Molecules 26, no. 21: 6595. https://doi.org/10.3390/molecules26216595
APA StyleWickhorst, P. J., Ihmels, H., & Paululat, T. (2021). Studies on the Interactions of 3,11-Difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium and Insulin with the Quadruplex-Forming Oligonucleotide Sequence a2 from the Insulin-Linked Polymorphic Region. Molecules, 26(21), 6595. https://doi.org/10.3390/molecules26216595