Abstract
Large volumes of fruit and vegetable production are lost during postharvest handling due to attacks by necrotrophic fungi. One of the promising alternatives proposed for the control of postharvest diseases is the induction of natural defense responses, which can be activated by recognizing molecules present in pathogens, such as chitin. Chitin is one of the most important components of the fungal cell wall and is recognized through plant membrane receptors. These receptors belong to the receptor-like kinase (RLK) family, which possesses a transmembrane domain and/or receptor-like protein (RLP) that requires binding to another RLK receptor to recognize chitin. In addition, these receptors have extracellular LysM motifs that participate in the perception of chitin oligosaccharides. These receptors have been widely studied in Arabidopsis thaliana (A. thaliana) and Oryza sativa (O. sativa); however, it is not clear how the molecular recognition and plant defense mechanisms of chitin oligosaccharides occur in other plant species or fruits. This review includes recent findings on the molecular recognition of chitin oligosaccharides and how they activate defense mechanisms in plants. In addition, we highlight some of the current advances in chitin perception in horticultural crops.
1. Introduction
One of the most important problems for the horticultural industry that has a negative impact on food security is the postharvest decay of fruits and vegetables [1]. Fruits and vegetables are highly susceptible to decay caused by necrotrophic fungi, such as Alternaria alternata (A. alternata), Botrytis cinerea (B. cinerea), and Colletotrichum gloeosporioides (C. gloeosporioides), among others, even under low temperature storage conditions [2,3]. The primary means to control fungal diseases is the use of synthetic fungicides; however, their use has caused concern due to their possible negative effects on human health, potentially toxic residues, long degradation period, and the induction of resistant strains [4]. In this sense, there is a need to control postharvest diseases. A promising alternative is the induction of natural plant defense mechanisms, which are a complex network of biochemical and molecular events that can limit the penetration and invasion of pathogens in plant tissue, thereby preventing or decreasing disease development [5,6].
The plant defense mechanism is very complex and genetically controlled. These defense responses begin when molecules associated with pathogens (pathogen-associated molecular patterns (PAMPs) or microbe-associated molecular patterns (MAMPs)) are recognized by plant membrane receptors (PRRs) and activate signal transduction to the nucleus and subsequently modify the transcriptional activity [7]. Chitin is present in the fungal cell wall, and it has been shown to participate in the plant–pathogen recognition phenomena [8,9] and in the activation of innate plant defense mechanisms [10]. The presence of receptors in plants that recognize chitin and chitin oligosaccharides (ChOs) has been widely demonstrated [11,12,13,14].
Currently, two families of chitin receptors have been reported in plants, namely receptor-like kinases (RLKs) and receptor-like proteins (RLPs), both of which are fundamental components for detecting MAMPs or PAMPs [15]. These membrane receptors that recognize chitin have been widely studied in O. sativa (rice) and A. thaliana (Arabidopsis). Chitin elicitor binding protein receptors, such as CEBiP, were the first chitin RLPs found in rice. Further, they are characterized by extracellular lysine domains that interact with an RLK for chitin recognition. CEBiP contains two extracellular LysM motifs and one transmembrane domain; however, it does not possess intracellular domains to initiate signal transduction and hence requires additional proteins [16,17]. On the other hand, RLKs, such as chitin elicitor receptor kinase (CERK1), are one of the main chitin receptors found in Arabidopsis. They include extracellular Lys domains that are probably involved in the interaction with PAMPs, a transmembrane domain, and a cytoplasmic kinase domain with basic auto phosphorylation/myelin protein (MBP) kinase activity, which can initiate a signaling cascade within the cell [18,19]. In addition to chitin receptor proteins, some studies have reported that there are other receptor proteins that are members of the lysine domain receptor-like kinase (LYK) family in Arabidopsis and play a role in the interaction with PAMPs [20]. When ChOs are recognized by PRRs, the signaling pathways starts and a complex defense response is activated, including the transcription of defense genes [21].
The recognition of ChOs by plant receptors has been reported mainly in Arabidopsis and rice; conversely, there are scarce studies in horticultural crops. According to some authors [22,23], ChOs are multicomponent transmembrane complexes that are related but differ in detail between species. Therefore, it is not clear how the molecular recognition of ChOs and the activation of downstream signaling occur in horticultural crops. The main objective of this review is to present an overview of recent findings regarding the molecular recognition of chitin oligosaccharides and how they activate defense mechanisms in plants. In addition, we highlight some of the current advances on how this phenomenon of recognition occurs in horticultural crops.
2. Molecular Mechanism of Plant Response to Pathogens
Over the course of evolution, plants have developed different strategies to resist and protect themselves from pathogen attacks. The first defense barrier in plants is the physical or structural barrier (waxes, cuticle, trichomes, etc.) that prevents pathogens and predators from invading the plant cell [23]. When plants become infected, several changes in the cuticle take place, which causes the activation of defense responses [24]. Once pathogens overcome the first line of defense, the second line of defense is initiated based on the cellular innate immunity, which allows the plant to resist or block the pathogen [25]. Among the main components that participate in this immune system are the PRRs found in the plasma membrane of plant cells, which recognize highly conserved molecular patterns called microbial-associated molecular patterns (MAMPs) or pathogen-associated molecular patterns (PAMPs). These are products secreted by microorganisms or released from their cell walls by hydrolytic enzymes during plant–pathogen interactions. MAMPs include peptidoglycans from Gram-positive bacteria, lipopolysaccharides from Gram-negative bacteria, glucans, proteins, or flagellin [26], while chitin and its oligosaccharides are considered PAMPs [27]. The perception of these molecular patterns during infection by pathogens triggers defense reactions known as PAMP-triggered immunity (PTI). In this regard, chitin and its ChOs are considered general elicitor compounds capable of inducing a defense response in plant cells [28,29].
When MAMPs or PAMPs are recognized by PRRs, the defense responses are induced through the signaling cascade by mitogen-activated protein kinases (MAPKs) and calcium-dependent protein kinase (CDPK). The message induces the production of signaling hormones, such as salicylic acid (SA), jasmonic acid (JA), ethylene (ET), abscisic acid (ABA), auxins (AUX), cytokinin (CK), gibberellins (GB), and brassinosteroids (BR) [6,30,31]. Through these signaling pathways, a complex defense response is activated, including modifications to create structural defenses (random creation of bonds between cell wall polymers and lignification), the induction of reactive oxygen species (ROS), and nitric oxide (NO) (Figure 1). This activation has a role in transcriptional reprogramming and induction of high expression of genes related to early defense [32,33]. These genes encode pathogenesis-related (PR) proteins, such as glucanases, chitinases, peroxidases, and enzymes, such as phenylalanine ammonia-lyase, a key enzyme involved in the synthesis of phytoalexins [34,35], which play important roles in the defense against pathogens [6,36,37,38]. In this context, a successful plant defense mechanism should inhibit pathogen infection and protect the plant against future attacks.
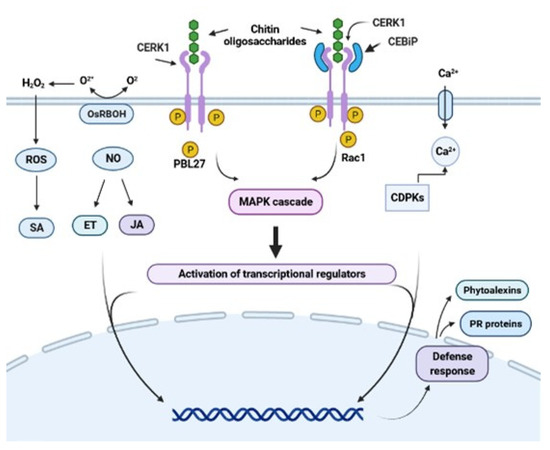
Figure 1.
Representative scheme of chitin oligosaccharide perception by the chitin receptors RLK and RLP. Structural differences between the RLP (CEBiP) and RLK (CERK1) receptors are shown. Chitin oligosaccharides bind to CERK1 in Arabidopsis. This complex sends the signal to the intermediate protein, which will begin to phosphorylate and trigger the MAP kinase pathway, inducing the accumulation of SA, JA, ET, NO, and ROS and activating the defense responses. The CEBiP receptor in rice binds to CERK1, forming a heterodimer complex that will allow it to recognize chitin oligosaccharides and activate its defense mechanism in the same way. Figure created using BioRender (https://biorender.com/ accessed on 22 July 2021).
3. Cellular Recognition of Fungal Molecules: Chitin
The fungal cell wall is made up of glycoproteins, glucans, and chitin, among other molecules. There is growing interest in chitin as this molecule has been shown to be essential in plant–pathogen recognition [9] and can induce activation of the innate plant defense mechanism [10]. Chitin molecules are composed of sugars linked by glycosidic bonds β (1→4), forming a linear chain of N-acetyl-2-amino-2-deoxy-D-glucose units, which are the second most abundant polymers in nature after cellulose [39,40]. Microscopically, it is in crystalline or semicrystalline form [41], making this polysaccharide a rigid and resistant material that acts as a very powerful barrier in the fungal cell wall and protects the organisms themselves [10]. Chitin is an insoluble polysaccharide when extracted by alkaline treatment and partially soluble when extracted by enzymatic treatment, from which other compounds of high biological significance are derived, such as chitosan and ChOs [42]. According to Buendia et al. [17] and Zhan et al. [43], ChOs are soluble depending on the pH and degree of polymerization (dp).
At the level of molecular recognition, some studies indicate the relationship of chitin and ChOs with the induction of plant defense mechanisms. Recent studies relate chitin as an enhancer of the biological control by Rhodotorula mucilaginosa (R. mucilaginosa) yeast against Rhizopus infection in peach crops. The addition of 0.5% of chitin reduced the postharvest disease of fruits by blue mold by up to half. In these fruits, differential expression of proteins was observed concerning the untreated fruits, including glucosidases, hydrolases, NADH dehydrogenases, and kinases with serine/threonine domains, which are decisive in the plant defense response against pathogen [44]. Similarly, the addition of chitin isolated from the yeast cell wall (Saccharomyces cerevisiae) in tomato fruit was correlated with increased expression levels of enzymes of oxidative metabolisms, such as the enzymes superoxide dismutase, peroxidase, and catalase, as well as enzymes inherent to the response to fungal infection, such as glucanase and chitinase [45].
4. Plant Membrane Receptors That Recognize Chitin and ChOs
Currently, two families of chitin receptors are reported in plants, namely RLKs and RLPs, which are fundamental components for detecting MAMPs or PAMPs [46,47]. These are related multicomponent transmembrane complexes, but they showed differences between species [22].
RLKs are a group of membrane proteins with an extracellular domain, including a sensor domain, a transmembrane region, and an intracellular region containing a Ser/Thr kinase domain with homology to protein kinases involved in signal transduction. Furthermore, they are divided into subfamilies according to the amount of lysine residues in their extracellular region [47]. On the other hand, RLP receptors lack an intracellular domain and, in some cases, are anchored to the membrane through a transmembrane domain of another protein or a glycosyl-phosphatidyl inositol group [17,29,48].
These membrane receptors of the RLK or RLP types have been widely studied and characterized in rice and Arabidopsis. In rice plants, chitin elicitor binding protein (CEBiP), which recognizes the N-acetyl groups of chitin by the lysine domains (LysM) of the receptor [21,49], was the first chitin receptor identified [16]. The molecular weight of this protein is estimated at 75 kDa, and it has 11 glycosylation sites through which N-acetyl glucosamine (GlcNAc) can bind. According to some authors, in rice plants, two receptors are involved in chitin-activated immunity [50]. LysM-RLP (OsCEBiP) binds to N-acetylated chitin fragments, initiating receptor homodimerization and further heterodimerization with OsCERK1, initiating PTI [51]. Additionally, the authors showed that in the OsCEBiP receptor, the central lysine motif is essential for recognizing ChOs with a dp of 8. CEBiP is dimerized with the OsCERK1 protein for binding to these oligosaccharides (Figure 1). In this sense, the chitin receptor system in rice requires both CEBiP and OsCERK1 for chitin sensing and signaling.
In Arabidopsis, a family of proteins related to chitin perception is found, and the CERK1 receptor is the most important within the RLK receptor family. LysM-RLK in Arabidopsis (AtCERK1) binds to N-acetylated chitin fragments with three LysM motifs and mediates the plant defenses induced by chitin through the formation of homodimers [12,18]. Important findings to understand the function of the CERK1 receptor in Arabidopsis were made by Petutsching et al. [13]. It was confirmed that in the absence of CERK1, no defense response was induced in Arabidopsis exposed to chitin and CERK1 showed the highest affinity for chitin. Additionally, the authors highlighted the importance of the lysine domains and the phosphorylation of CERK1 in the ectodomain for the recognition process of ChOs and the intracellular domain to initiate signal transduction.
According to some studies, the recognition of ChOs is more effective depending on their polymerization and acetylation degrees. For instance, in the cork tree Hevea brasiliensis, a linear relationship of 1:1 for (GlcNac)5 with the receptor was shown, and for (GlcNac)8, the ratio was 2:1 protein–oligosaccharide, indicating dimerization of receptors for the recognition of ChOs with higher dp [11]. (GlcNac)6-8 was recognized by membrane receptors in Arabidopsis [52]. In another study, (GlcNac)5-9 was recognized with great efficiency by the CERK1 receptor of Arabidopsis [12], while ChOs with a lower dp did not show affinity for the CERK1 receptor [13]. Later, Liu et al. [53] described the X-ray crystal structure of the AtCERK1 ectodomain and predicted the interaction of ChOs with the second LysM motif of the extracellular domain. These authors suggested that long chain chitin oligomers (dp ≥ 6) bind to LysM domains in two monomers, resulting in the homodimerization of AtCERK1. This dimerization was shown to activate the intracellular kinase domain [13,53]. However, there is a possibility that, as in rice, the active chitin receptor in Arabidopsis can be a complex of more than one protein [22].
In addition to chitin receptor proteins, members of the lysine domain receptor-like kinase (LYK) family have been reported in Arabidopsis. The plant LysM proteins mostly function as pattern recognition receptors (PRRs) that recognize chitin to induce the plant’s immunity [26]. Cao et al. [20] identified a LysM-RLK in Arabidopsis (AtLYK5) that binds to chitin with a higher affinity than AtCERK1. The authors proposed that AtLYK5 functions as the major chitin receptor, recruiting AtCERK1 to form a chitin-inducible receptor complex, which agrees with the results reported by other authors [54]. Erwig et al. [55] demonstrated that CERK1 is essential for recognizing ChOs in Arabidopsis and reported other receptor proteins involved in chitin perception. Specifically, LYK5 was shown to be required for the phosphorylation and dimerization processes of the CERK1 receptor. The authors concluded that CERK1, LYK4, and LYK5 were inducible proteins and that their presence in the cytoplasmic membrane was due to the presence of chitin and depended on a complex network of vesicular trafficking. Therefore, there are proteins that bind the intracellular domains of receptors and activate the signaling cascade, and kinase activity is required to activate immune responses [22,53]. For example, PBL27 is the downstream component of AtCERK1 after chitin perception, and it can be phosphorylated by AtCERK1 [56]. PBL19 and PBL27 phosphorylate MAPKKK to activate MAPK cascades, which in turn activates defense genes in the nucleus [57]. In summary, the receptors for these chitin patterns, such as CEBiP/CERK1, are activated by ligand binding and trigger various immune responses.
5. Recognition of Chitin and Its Oligosaccharides in Horticultural Crops
The chitin receptor complex and the relationship among the components have been mostly studied in rice and Arabidopsis model systems. However, few studies have been performed to investigate the presence of chitin receptors and the mode of action in defense activation in horticultural crops. For instance, Zhang et al. [58] made important contributions to understanding the biological function of a chitin receptor-binding protein in banana (Musa acuminata). The authors confirmed the presence of the MaLYK1 protein in the plasma membrane. This protein is a LysM domain chitin receptor that belongs to a subclade represented by AtCERK1 (AtLYK1) and OsCERK1 (OsLYK9). The expression of MaLYK1 was found to be higher in banana roots, while it was lower in yellow fruit. In addition, MaLYK1 expression was induced by Fusarium oxysporum f. sp. cubense race 4 (Foc4), and its function was essential not only in recognizing the fungus but also in recognizing the fungus in the symbiotic relationship established between plants and mycorrhizae. Besides, chitin treatment (GlcNAc)8 induced the expression of defense genes, such as phenylalanine ammonia-lyase, β-1,3-glucanase, and pathogenesis-related protein 1. Similar studies reported the activation of the MdCERK1 receptor in apple (Malus domestica) during Rhizoctonia solani infection, observing that MdCERK1 expression increased in roots and leaves. This protein is found in the plasma membrane and includes three lysine-rich domains in its extracellular zone, a transmembrane domain, and an intracellular domain with serine/threonine motifs [59]. Recently, another CERK gene, designated as MdCERK1–2, was identified in shoot barks of apples. It encodes a protein with high similarity with the previously reported MdCERK1 and AtCERK1. Results revealed that MdCERK1–2 expression in apple was induced by Botryosphaeria dothidea and Glomerella cingulate. Additionally, it was observed that the MdCERK1–2 overexpression in transgenic Nicotiana benthamiana plants improved their resistance to A. alternata infection. These results suggest that MdCERK1–2 is involved in apple defense responses against pathogenic fungi [60].
In tomato (Solanum lycopersicum), Bti9 interacts with the AvrPtoB protein from Pseudomonas syringae [61]. Bti9 is a protein kinase with 76% similarity with LysM CERK1 found in Arabidopsis, which belongs to the same clade as SlLYK11, SlLYK12, and SlLYK13. The authors suggested that these proteins are involved in the response to PAMPs that trigger the signaling cascade. Liao et al. [62] showed the relationship between homologous CERK1 receptors in tomato (SlLYK1, SlLYK12, and SlLYK13) and colonization of arbuscular mycorrhizae (AMs). The authors found that SlLYK1 gene expression was upregulated after chitin oligosaccharide treatment, whereas SlLYK12 was involved in AM symbiosis. Another interesting piece of data to elucidate the phenomena of chitin recognition showed that cell death was induced by overexpressing SlLYK13, suggesting that the chitin receptor genes have distinct functions in plants depending on the specific plant tissue or PAMPs applied. As well as in Arabidopsis, when ChOs are perceived in tomato, there are proteins that bind the intracellular domains of the chitin receptor and activate the signaling cascade, which includes kinase activity required for the activation of immune responses [22]. In tomato, the TPK1b protein is probably an ortholog of BIK1 in Arabidopsis, which is essential in the defense response of Arabidopsis against B. cinerea. The susceptibility to B. cinerea was increased when TPK1b was inhibited by RNAi [63]. Another study found that in the defense response mediated by systemin, TPK1b was phosphorylated by the ortholog of RLK1 (PORK1). In the same study, when PORK1 was silenced, the levels of TPK1b phosphorylation decreased to the same extent, which affected the defense response through this pathway [64]. These studies further support the essential role of TPK1b in the transduction of the RLK receptor signal in various plant defense responses.
In summary, it was found that Arabidopsis thaliana and Oryza sativa have a group of receptors with high affinity for ChOs and that the interactions for this recognition is fairly well understood. In the same way, one of these receptors (RLK type) was described in certain horticultural crops in which the structural patterns are preserved. Additionally, the role of ChOs in the activation of the defense mechanism was observed in horticultural crops.
6. Concluding Remarks and Future Perspectives
Great advances have been made to elucidate the molecular recognition of chitin oligosaccharides by plant membrane receptors and the functional receptor complex based on studies performed in the model plants Arabidopsis thaliana and Oryza sativa. According to the few studies reported in horticultural crops, it is possible to suggest that the molecular mechanism of chitin oligosaccharide perception occurs through RLK-type receptors with lysine domains, such as the CERK1 receptor reported in Arabidopsis. This suggests that the chitin receptors reported in model plants such as Arabidopsis and rice could be conserved in other plant species. Nevertheless, details of the receptor complex formation induced by chitin oligosaccharides and the activation of signaling and defense responses within the cell are not clear. Therefore, future studies are required to fully elucidate the molecular mechanism of the receptor role in the response to pathogen attack.
Understanding the molecular mechanism of pathogen recognition and the activation of the defense mechanism would help with the problem of horticultural postharvest losses and food supply by providing safe and effective alternatives for the development of disease-resistant crops and/or food preservation without using synthetic chemical compounds.
Author Contributions
Y.H.G., O.R.Z. and R.T.-R., conceived and designed the idea and wrote the manuscript; M.E.B.-F., M.E.T.-H., E.C.-M. and A.R.-C. wrote and reviewed the manuscript; M.E.B.-F. and M.E.T.-H. provided technical assistance, scientific correction, and language revision for the final versions of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received financial support from the Sectoral Research Fund for Education of the National Council for Science and Technology (CONACyT) from Mexico (CB 2016-01, Grant No. 287254).
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not Applicable.
Data Availability Statement
Not Applicable.
Acknowledgments
Financial support from the CONACyT (Mexico) is greatly appreciated. The authors Y.H.G. and O.R.Z. thank CONACyT for the PhD scholarship. We are grateful to Alfonso Aguilar Valenzuela for the computer drawing enhancement of graphical abstract and Figure 1 and to the Research Center of Food and Development (CIAD, AC) for all the equipment facilities.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ala, F.T.; Askarne, L.; Boubaker, H.; Bo, E.H.; Ben Ao, A.A. Control of Gray Mold Disease of Tomato by Postharvest Application of Organic Acids and Salts. Plant Pathol. J. 2017, 16, 62–72. [Google Scholar] [CrossRef] [Green Version]
- Troncoso-Rojas, R.; Tiznado-Hernández, M. Alternaria alternata (black rot, black spot). In Postharvest Decay of Fruits and Vegetables: Control Strategies; Bautista-Baños, S., Ed.; Elsevier Inc.: Cambridge, MA, USA, 2014; pp. 147–187. [Google Scholar]
- Takao, K.; Akagi, Y.; Tsuge, T.; Kodama, M. Functional characterization of putative G protein-coupled receptors in the tomato pathotype of Alternaria alternata. J. Gen. Plant Pathol. 2016, 82, 82–88. [Google Scholar] [CrossRef]
- Yang, L.-N.; He, M.-H.; Ouyang, H.-B.; Zhu, W.; Pan, Z.-C.; Sui, Q.-J.; Shang, L.-P.; Zhan, J. Cross-resistance of the pathogenic fungus Alternaria alternata to fungicides with different modes of action. BMC Microbiol. 2019, 19, 205. [Google Scholar] [CrossRef] [Green Version]
- Sudheeran, P.K.; Sela, N.; Carmeli-Weissberg, M.; Ovadia, R.; Panda, S.; Feygenberg, O.; Maurer, D.; Oren-Shamir, M.; Aharoni, A.; Alkan, N. Induced defense response in red mango fruit against Colletotrichum gloeosporioides. Hortic. Res. 2021, 8, 1–11. [Google Scholar] [CrossRef]
- Alkan, N.; Friedlander, G.; Ment, D.; Prusky, D.; Fluhr, R. Simultaneous transcriptome analysis of Colletotrichum gloeosporioides and tomato fruit pathosystem reveals novel fungal pathogenicity and fruit defense strategies. New Phytol. 2015, 205, 801–815. [Google Scholar] [CrossRef]
- Delteil, A.; Gobbato, E.; Cayrol, B.; Estevan, J.; Michel-Romiti, C.; Dievart, A.; Kroj, T.; Morel, J.-B. Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol. 2016, 16, 17. [Google Scholar] [CrossRef]
- Costa-De-Oliveira, S.; Silva, A.P.; Miranda, I.M.; Salvador, A.; Azevedo, M.M.; Munro, C.A.; Rodrigues, A.G.; Pina-Vaz, C. Determination of chitin content in fungal cell wall: An alternative flow cytometric method. Cytom. Part. A 2013, 83A, 324–328. [Google Scholar] [CrossRef] [Green Version]
- Vallet, A.S.; Mesters, J.R.; Thomma, B.P. The battle for chitin recognition in plant-microbe interactions. FEMS Microbiol. Rev. 2015, 39, 171–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pusztahelyi, T. Chitin and chitin-related compounds in plant–fungal interactions. Mycology 2018, 9, 189–201. [Google Scholar] [CrossRef]
- Asensio, J.L.; Cañada, F.J.; Siebert, H.-C.; Laynez, J.; Poveda, A.; Nieto, P.M.; Soedjanaamadja, U.; Gabius, H.-J.; Jiménez-Barbero, J. Structural basis for chitin recognition by defense proteins: GlcNAc residues are bound in a multivalent fashion by extended binding sites in hevein domains. Chem. Biol. 2000, 7, 529–543. [Google Scholar] [CrossRef] [Green Version]
- Iizasa, E.; Mitsutomi, M.; Nagano, Y. Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro. J. Biol. Chem. 2010, 285, 2996–3004. [Google Scholar] [CrossRef] [Green Version]
- Petutschnig, E.K.; Jones, A.M.; Serazetdinova, L.; Lipka, U.; Lipka, V. The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 2010, 285, 28902–28911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinya, T.; Nakagawa, T.; Kaku, H.; Shibuya, N. Chitin-mediated plant–fungal interactions: Catching, hiding and handshaking. Curr. Opin. Plant Biol. 2015, 26, 64–71. [Google Scholar] [CrossRef]
- Huang, C.; Yan, Y.; Zhao, H.; Ye, Y.; Cao, Y. Arabidopsis CPK5 phosphorylates the chitin receptor LYK5 to regulate plant innate immunity. Front. Plant Sci. 2020, 11, 702. [Google Scholar] [CrossRef]
- Kaku, H.; Nishizawa, Y.; Ishii-Minami, N.; Akimoto-Tomiyama, C.; Dohmae, N.; Takio, K.; Minami, E.; Shibuya, N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 11086–11091. [Google Scholar] [CrossRef] [Green Version]
- Buendia, L.; Girardin, A.; Wang, T.; Cottret, L.; Lefebvre, B. LysM receptor-like kinase and lysM receptor-like protein families: An update on phylogeny and functional characterization. Front. Plant Sci. 2018, 9, 1531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef] [Green Version]
- Weibing, W.; Zhou, Z.; Wang, W.; Li, L.; Rao, S.; Wu, Y.; Zhang, X.; Menke, F.L.; Chen, S.; Zhou, J.-M. Receptor-Like cytoplasmic kinases directly link diverse pattern recognition receptors to the activation of mitogen-activated protein kinase cascades in Arabidopsis. Plant Cell 2018, 30, 1543–1561. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Liang, Y.; Tanaka, K.; Nguyen, C.; Jedrzejczak, R.P.; Joachimiak, A.; Stacey, G. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 2014, 3, e03766. [Google Scholar] [CrossRef]
- Desaki, Y.; Miyata, K.; Suzuki, M.; Shibuya, N.; Kaku, H. Plant immunity and symbiosis signaling mediated by LysM receptors. Innate Immun. 2017, 24, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Gubaeva, E.; Gubaev, A.; Melcher, R.L.J.; Cord-Landwehr, S.; Singh, R.; El Gueddari, N.E.; Moerschbacher, B.M. ‘Slipped sandwich’ model for chitin and chitosan perception in Arabidopsis. Mol. Plant-Microbe Interact. 2018, 31, 1145–1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alkan, N.; Fortes, A.M. Insights into molecular and metabolic events associated with fruit response to post-harvest fungal pathogens. Front. Plant Sci. 2015, 6, 889. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.; Qin, D.; Din, G.M.U.; Liu, T.; Chen, W.; Gao, L. Characterization of histological changes at the tillering stage (Z21) in resistant and susceptible wheat plants infected by Tilletia controversa Kühn. BMC Plant Biol. 2021, 21, 49. [Google Scholar] [CrossRef]
- Kim, W.; Prokchorchik, M.; Tian, Y.; Kim, S.; Jeon, H.; Segonzac, C. Perception of unrelated microbe-associated molecular patterns triggers conserved yet variable physiological and transcriptional changes in Brassica rapa ssp. pekinensis. Hortic. Res. 2020, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.-P.; Li, J.-J.; Dhar, N.; Li, J.-P.; Chen, J.-Y.; Jian, W.; Dai, X.-F.; Yang, X.-Y. Lysin motif (LysM) proteins: Interlinking manipulation of plant immunity and fungi. Int. J. Mol. Sci. 2021, 22, 3114. [Google Scholar] [CrossRef] [PubMed]
- Shinya, T.; Motoyama, N.; Ikeda, A.; Wada, M.; Kamiya, K.; Hayafune, M.; Kaku, H.; Shibuya, N. Functional characterization of CEBiP and CERK1 homologs in Arabidopsis and rice reveals the presence of different chitin receptor systems in plants. Plant Cell Physiol. 2012, 53, 1696–1706. [Google Scholar] [CrossRef] [Green Version]
- Egusa, M.; Matsui, H.; Urakami, T.; Okuda, S.; Ifuku, S.; Nakagami, H.; Kaminaka, H. Chitin Nanofiber Elucidates the Elicitor Activity of Polymeric Chitin in Plants. Front. Plant Sci. 2015, 6, 1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdul Malik, N.A.; Kumar, I.S.; Nadarajah, K. Elicitor and Receptor Molecules: Orchestrators of Plant Defense and Immunity. Int. J. Mol. Sci. 2020, 21, 963. [Google Scholar] [CrossRef] [Green Version]
- Yuan, M.; Huang, Y.; Ge, W.; Jia, Z.; Song, S.; Zhang, L.; Huang, Y. Involvement of jasmonic acid, ethylene and salicylic acid signaling pathways behind the systemic resistance induced by Trichoderma longibrachiatum H9 in cucumber. BMC Genom. 2019, 20, 144. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-L.; Yuan, F.; Li, X.-Y.; Ma, R.-C.; Xie, H. Jasmonic acid and ethylene signaling pathways participate in the defense response of Chinese cabbage to Pectobacterium carotovorum infection. J. Integr. Agric. 2021, 20, 1314–1326. [Google Scholar] [CrossRef]
- Skelly, M.J.; Furniss, J.J.; Grey, H.; Wong, K.-W.; Spoel, S.H. Dynamic ubiquitination determines transcriptional activity of the plant immune coactivator NPR1. eLife 2019, 8, 8. [Google Scholar] [CrossRef]
- Gao, S.; Wang, F.; Niran, J.; Li, N.; Yin, Y.; Yu, C.; Jiao, C.; Yao, M. Transcriptome analysis reveals defense-related genes and pathways against Xanthomonas campestris pv. vesicatoria in pepper (Capsicum annuum L.). PLoS ONE 2021, 16, e0240279. [Google Scholar] [CrossRef]
- Sánchez-Estrada, A.; Tiznado-Hernández, M.E.; Ojeda-Contreras, A.J.; Valenzuela-Quintanar, A.I.; Troncoso-Rojas, R. Induction of enzymes and phenolic compounds related to the natural defence response of netted melon fruit by a bio-elicitor. J. Phytopathol. 2009, 157, 24–32. [Google Scholar] [CrossRef]
- Troncoso-Rojas, R.; Sanchez-Estrada, A.; Carvallo, T.; González-León, A.; Ojeda-Contreras, J.; Aguilar-Valenzuela, A.; Tiznado-Hernández, M.-E. A fungal elicitor enhances the resistance of tomato fruit to Fusarium oxysporum infection by activating the phenylpropanoid metabolic pathway. Phytoparasitica 2012, 41, 133–142. [Google Scholar] [CrossRef]
- Kishimoto, K.; Kouzai, Y.; Kaku, H.; Shibuya, N.; Minami, E.; Nishizawa, Y. Perception of the chitin oligosaccharides contributes to disease resistance to blast fungus Magnaporthe oryzae in rice. Plant J. 2010, 64, 343–354. [Google Scholar] [CrossRef]
- Malinovsky, F.G.; Fangel, J.U.; Willats, W.G.T. The role of the cell wall in plant immunity. Front. Plant Sci. 2014, 5, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matilla, M.A. Metabolic responses of plants upon different plant–pathogen interactions. In Plant Metabolites and Regulation under Environmental Stress; Ahmad, P., Ahanger, M.A., Singh, V.P., Tripathi, D.K., Alam, P., Alyemeni, M.N., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 195–214, Chapter 10. [Google Scholar]
- El Knidri, H.; Belaabed, R.; Addaou, A.; Laajeb, A.; Lahsini, A. Extraction, chemical modification and characterization of chitin and chitosan. Int. J. Biol. Macromol. 2018, 120, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Malerba, M.; Cerana, R. Recent applications of chitin- and chitosan-based polymers in plants. Polymers 2019, 11, 839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pillai, C.K.S.; Paul, W.; Sharma, C.P. Chitin and chitosan polymers: Chemistry, solubility and fiber formation. Prog. Polym. Sci. 2009, 34, 641–678. [Google Scholar] [CrossRef]
- García, Y.H.; Troncoso-Rojas, R.; Tiznado-Hernández, M.E.; Báez-Flores, M.E.; Carvajal-Millan, E.; Rascon-Chu, A.; Lizardi-Mendoza, J.; Martínez-Robinson, K.G. Enzymatic treatments as alternative to produce chitin fragments of low molecular weight from Alternaria alternata. J. Appl. Polym. Sci. 2019, 136, 47339. [Google Scholar] [CrossRef]
- Zhan, J.; Qin, Y.; Gao, K.; Fan, Z.; Wang, L.; Xing, R.; Liu, S.; Li, P. Efficacy of a chitin-based water-soluble derivative in inducing Purpureocillium lilacinum against nematode disease (Meloidogyne incognita). Int. J. Mol. Sci. 2021, 22, 6870. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, Q.; Ge, L.; Zhang, G.; Zhang, X.; Zhang, X. Chitin enhances biocontrol of Rhodotorula mucilaginosa to postharvest decay of peaches. Int. J. Biol. Macromol. 2016, 88, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Fu, D.; Jin, L.; Chen, M.; Zheng, X.; Yu, T. Chitin isolated from yeast cell wall induces the resistance of tomato fruit to Botrytis cinerea. Carbohydr. Polym. 2018, 199, 341–352. [Google Scholar] [CrossRef]
- Monaghan, J.; Zipfel, C. Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 2012, 15, 349–357. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Wu, Y. Receptor-like kinases and regulation of plant innate immunity. Enzymes 2016, 40, 105–142. [Google Scholar] [CrossRef]
- Liebrand, T.W.H.; Berg, G.C.M.V.D.; Zhang, Z.; Smit, P.; Cordewener, J.H.G.; America, A.; Sklenar, J.; Jones, A.; Tameling, W.I.L.; Robatzek, S.; et al. Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. USA 2013, 110, 10010–10015. [Google Scholar] [CrossRef] [Green Version]
- Richards, S.; Rose, L.E. The evolutionary history of LysM-RLKs (LYKs/LYRs) in wild tomatoes. BMC Evol. Biol. 2019, 19, 141. [Google Scholar] [CrossRef]
- Shimizu, T.; Nakano, T.; Takamizawa, D.; Desaki, Y.; Ishii-Minami, N.; Nishizawa, Y.; Minami, E.; Okada, K.; Yamane, H.; Kaku, H.; et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010, 64, 204–214. [Google Scholar] [CrossRef] [Green Version]
- Hayafune, M.; Berisio, R.; Marchetti, R.; Silipo, A.; Kayama, M.; Desaki, Y.; Arima, S.; Squeglia, F.; Ruggiero, A.; Tokuyasu, K.; et al. Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc. Natl. Acad. Sci. USA 2014, 111, E404–E413. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Ramonell, K.; Somerville, S.; Stacey, G. Characterization of early, chitin-induced gene expression in Arabidopsis. Mol. Plant-Microbe Interact. 2002, 15, 963–970. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Liu, Z.; Song, C.; Hu, Y.; Han, Z.; She, J.; Fan, F.; Wang, J.; Jin, C.; Chang, J.; et al. Chitin-induced dimerization activates a plant immune receptor. Science 2012, 336, 1160–1164. [Google Scholar] [CrossRef] [Green Version]
- Volk, H.; Marton, K.; Flajšman, M.; Radišek, S.; Tian, H.; Hein, I.; Podlipnik, Č.; Thomma, B.P.H.J.; Košmelj, K.; Javornik, B.; et al. Chitin-binding protein of Verticillium nonalfalfae disguises fungus from plant chitinases and suppresses chitin-triggered host immunity. Mol. Plant-Microbe Interact. 2019, 32, 1378–1390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erwig, J.; Ghareeb, H.; Kopischke, M.; Hacke, R.; Matei, A.; Petutschnig, E.; Lipka, V. Chitin-induced and chitin elicitor receptor kinase1 (CERK1) phosphorylation-dependent endocytosis of Arabidopsis thaliana Lysin motif-containing receptor-like kinase5 (LYK5). New Phytol. 2017, 215, 382–396. [Google Scholar] [CrossRef] [Green Version]
- Yamada, K.; Yamaguchi, K.; Shirakawa, T.; Nakagami, H.; Mine, A.; Ishikawa, K.; Fujiwara, M.; Narusaka, M.; Narusaka, Y.; Ichimura, K.; et al. The Arabidopsis CERK 1-associated kinase PBL 27 connects chitin perception to MAPK activation. EMBO J. 2016, 35, 2468–2483. [Google Scholar] [CrossRef]
- Gong, B.-Q.; Wang, F.-Z.; Li, J.-F. Hide-and-Seek: Chitin-triggered plant immunity and fungal counterstrategies. Trends Plant Sci. 2020, 25, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yuan, L.; Staehelin, C.; Li, Y.; Ruan, J.; Liang, Z.; Xie, Z.; Wang, W.; Xie, J.; Huang, S. The lysin motif-containing receptor-like kinase 1 protein of banana is required for perception of pathogenic and symbiotic signals. New Phytol. 2019, 223, 1530–1546. [Google Scholar] [CrossRef]
- Zhou, Z.; Tian, Y.; Cong, P.; Zhu, Y. Functional characterization of an apple (Malus × domestica) LysM domain receptor encoding gene for its role in defense response. Plant Sci. 2018, 269, 56–65. [Google Scholar] [CrossRef]
- Chen, Q.; Dong, C.; Sun, X.; Zhang, Y.; Dai, H.; Bai, S. Overexpression of an apple LysM-containing protein gene, MdCERK1–2, confers improved resistance to the pathogenic fungus, Alternaria alternata, in Nicotiana benthamiana. BMC Plant Biol. 2020, 20, 146. [Google Scholar] [CrossRef]
- Zeng, L.; Velásquez, A.C.; Munkvold, K.R.; Zhang, J.; Martin, G.B. A tomato LysM receptor-like kinase promotes immunity and its kinase activity is inhibited by AvrPtoB. Plant J. 2012, 69, 92–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, D.; Sun, X.; Wang, N.; Song, F.; Liang, Y. Tomato LysM receptor-like kinase SlLYK12 is involved in arbuscular mycorrhizal symbiosis. Front. Plant Sci. 2018, 9, 1004. [Google Scholar] [CrossRef] [Green Version]
- AbuQamar, S.; Chai, M.-F.; Luo, H.; Song, F.; Mengiste, T. Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. Plant Cell 2008, 20, 1964–1983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, S.; Liao, C.-J.; Jaiswal, N.; Lee, S.; Yun, D.-J.; Lee, S.Y.; Garvey, M.; Kaplan, I.; Mengiste, T. Tomato PEPR1 ortholog receptor-like kinase1 regulates responses to systemin, necrotrophic fungi, and insect herbivory. Plant Cell 2018, 30, 2214–2229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).