Antimicrobial and Anticancer Application of Silver(I) Dipeptide Complexes
Abstract
:1. Introduction
2. Results and Discussion
2.1. Syntheses of Complexes
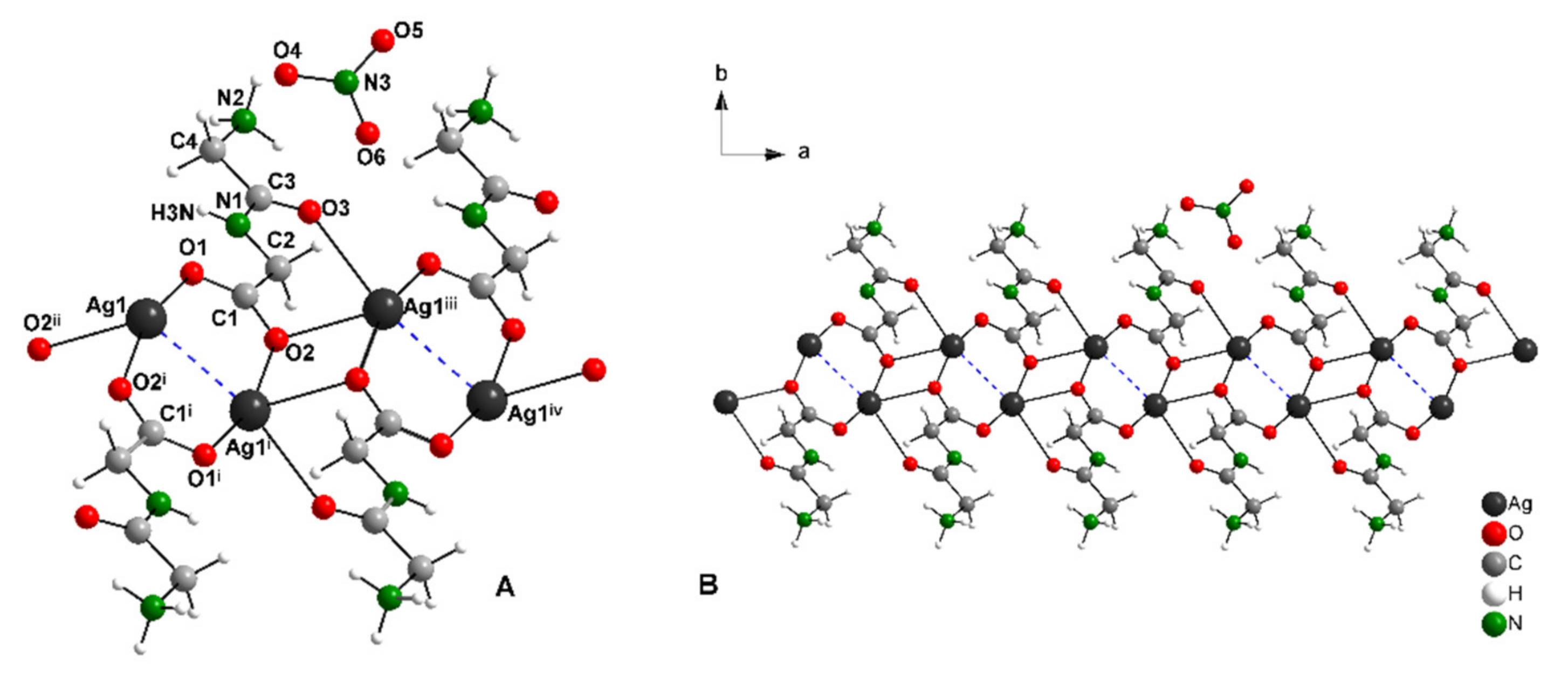
2.2. Description of Crystal Structures
2.3. IR Spectra
2.4. Thermogravimetric Analysis
2.5. Stability Study
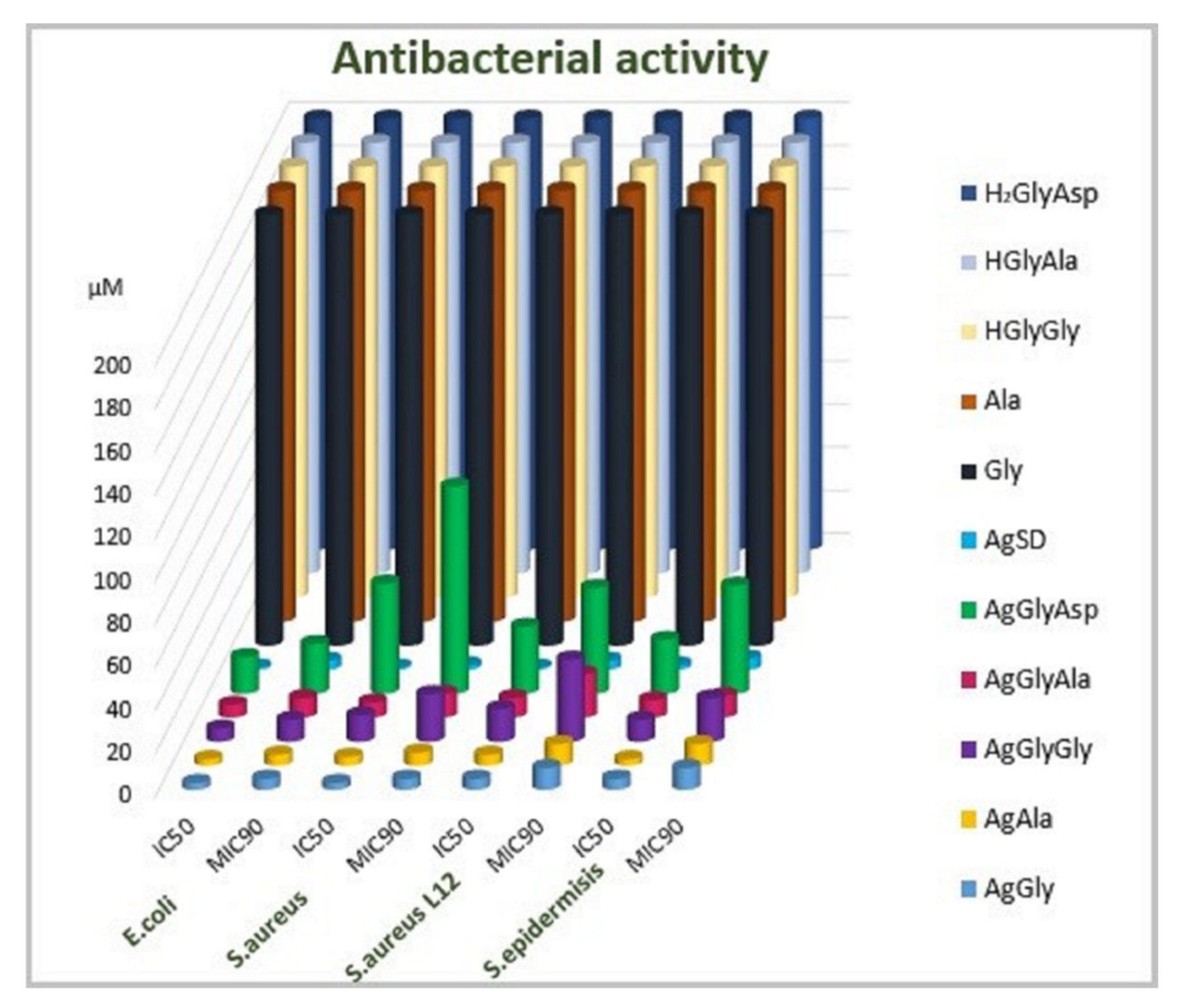
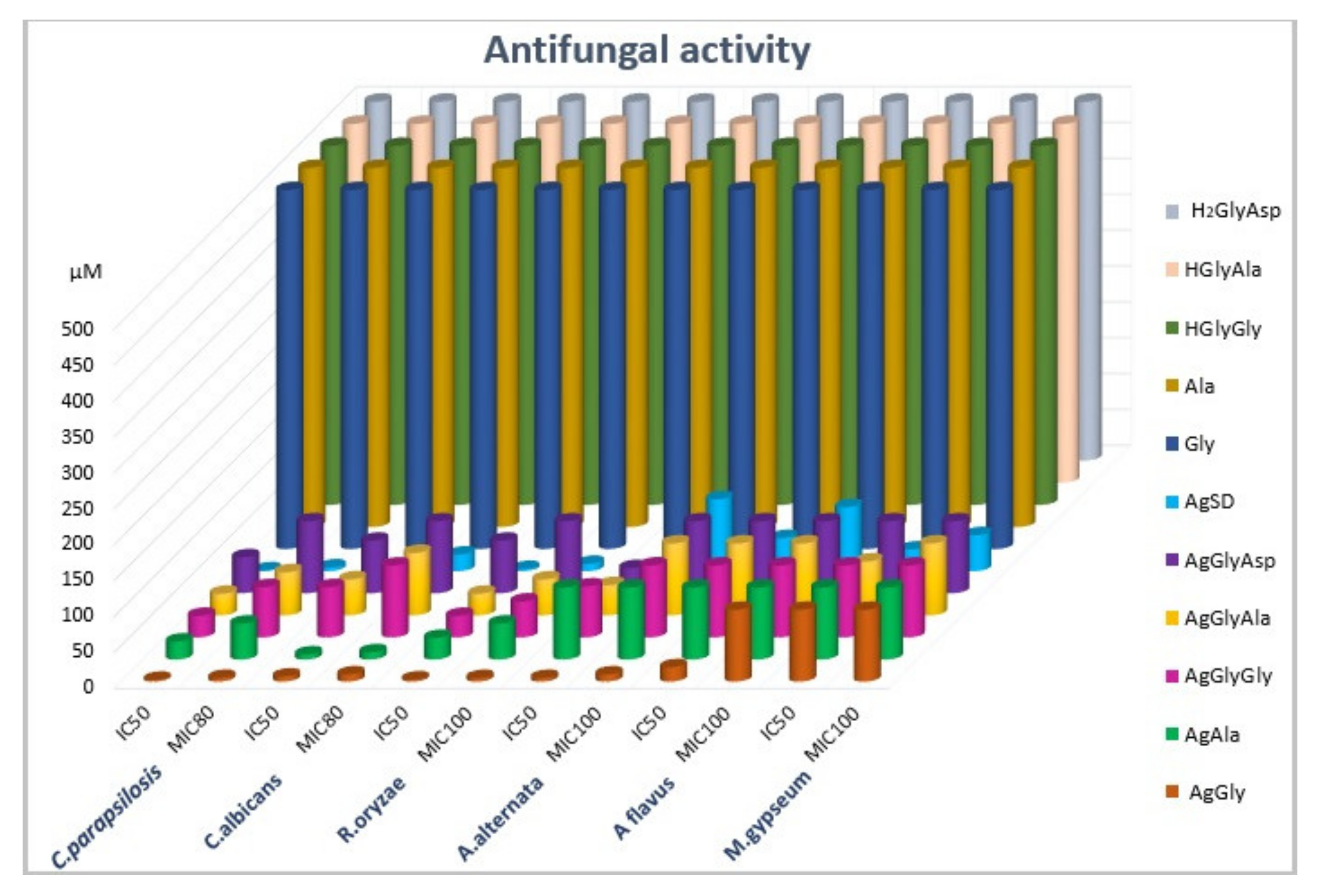
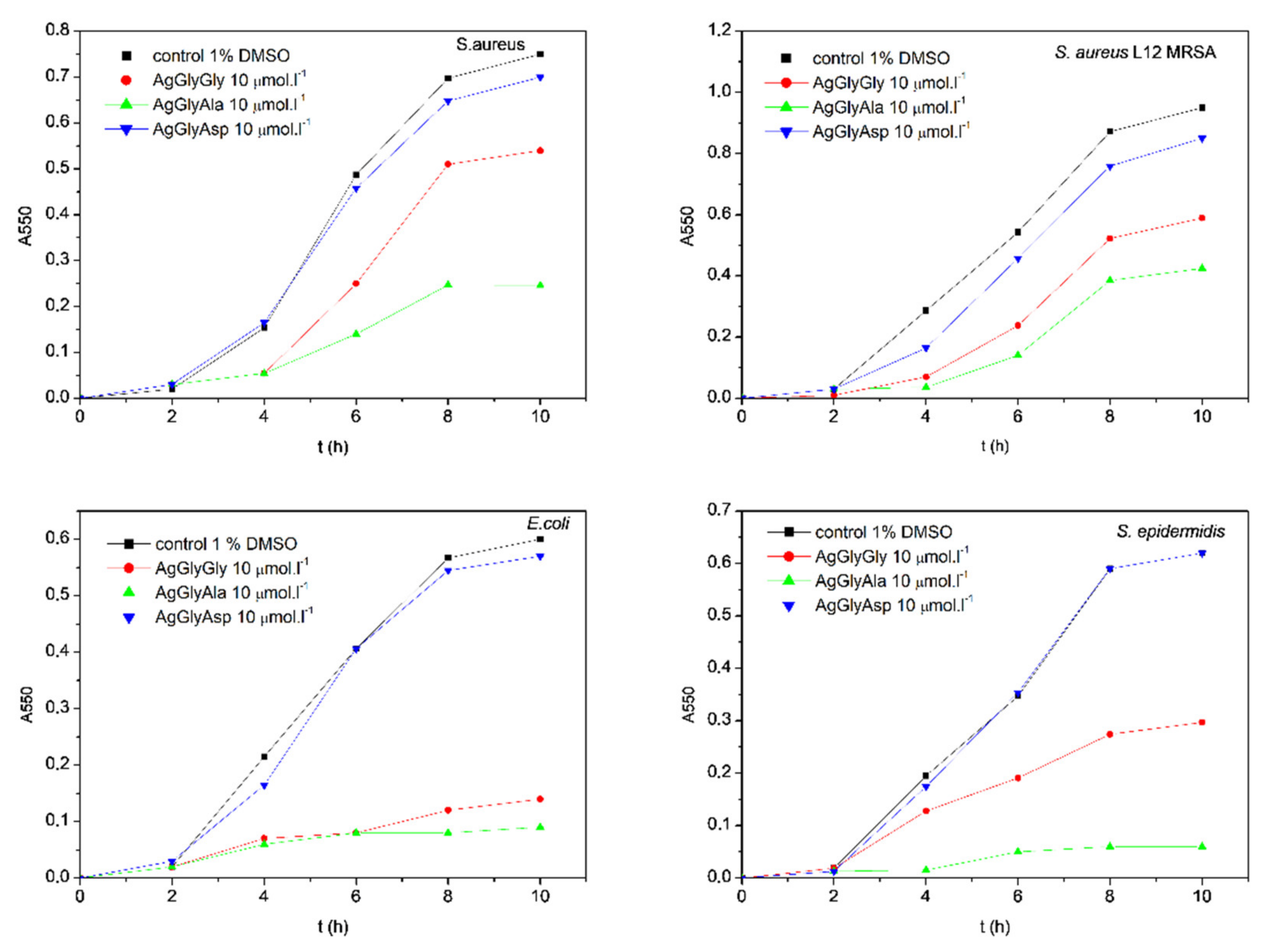
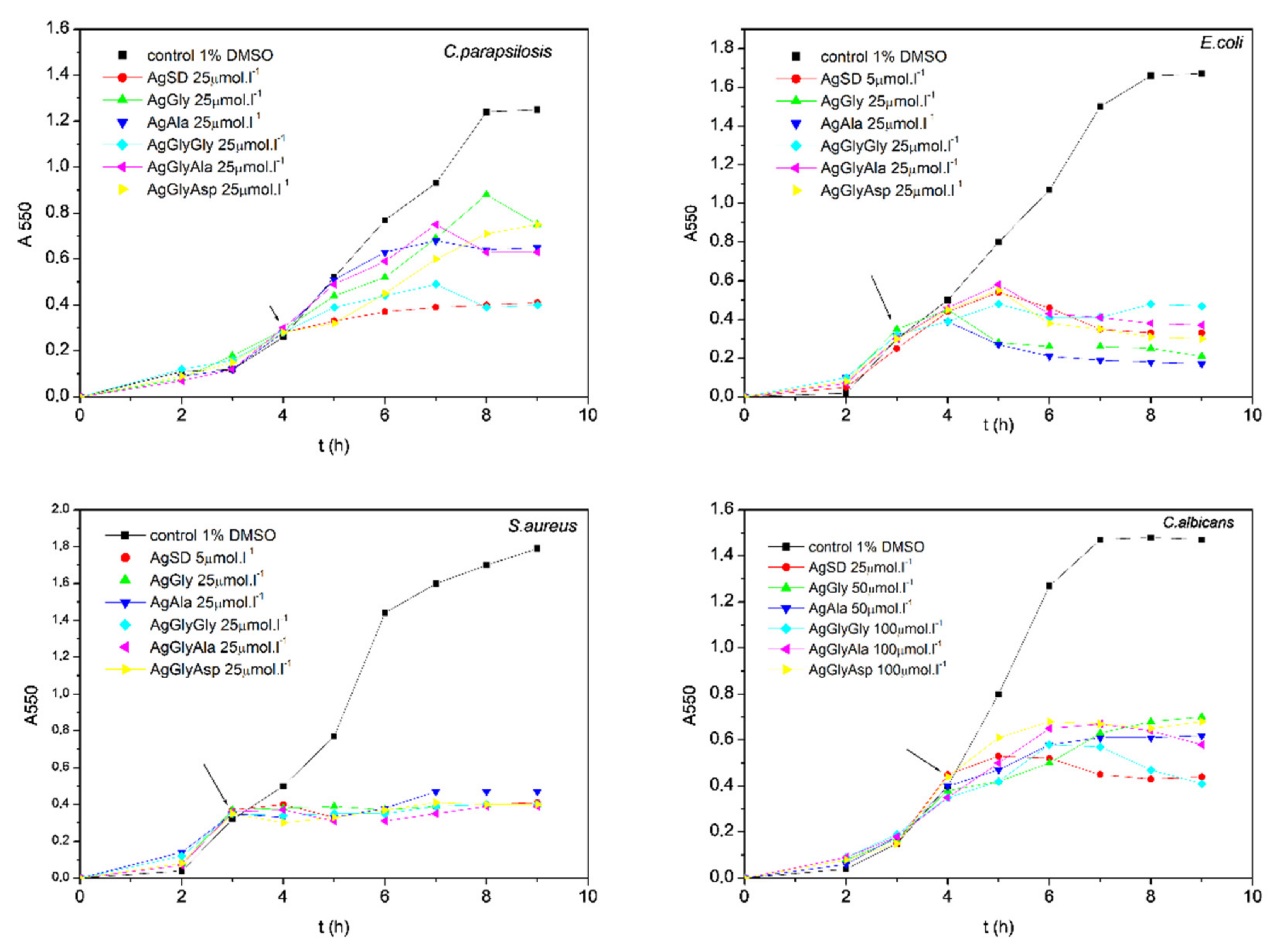
2.6. Antimicrobial Activity
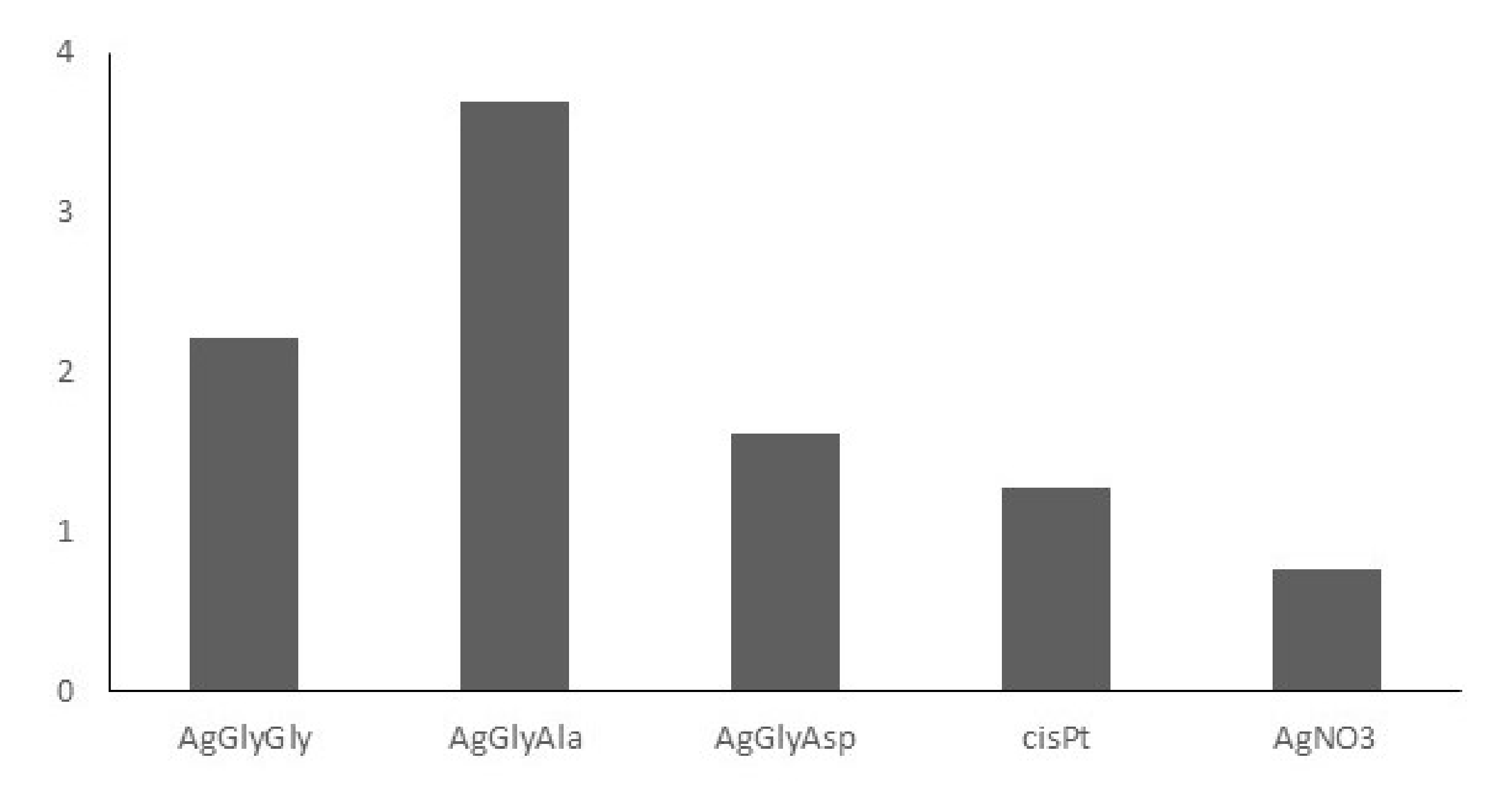
2.7. Anticancer Activity
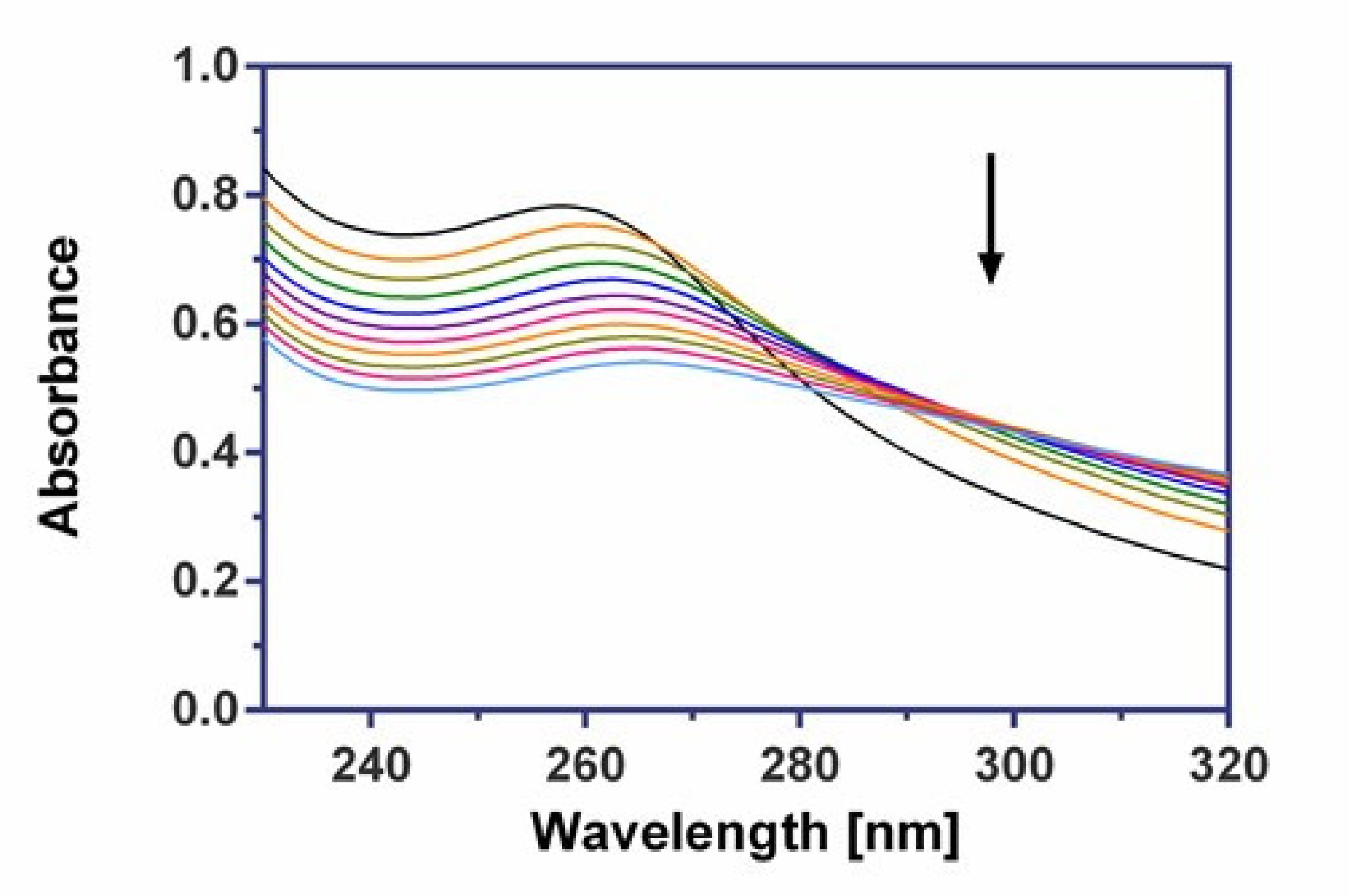
2.8. DNA Binding Properties
2.8.1. UV-Vis Spectroscopy
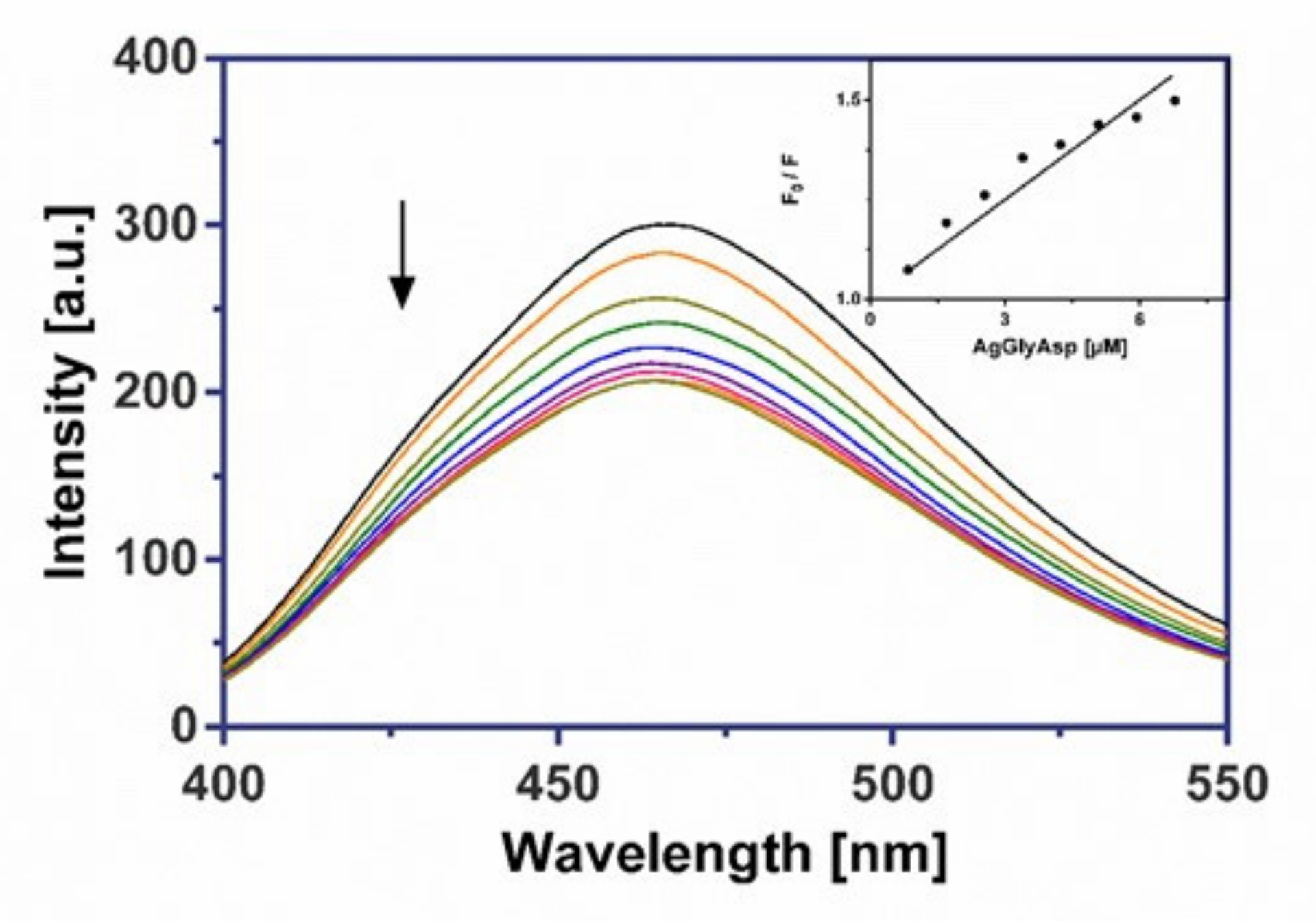
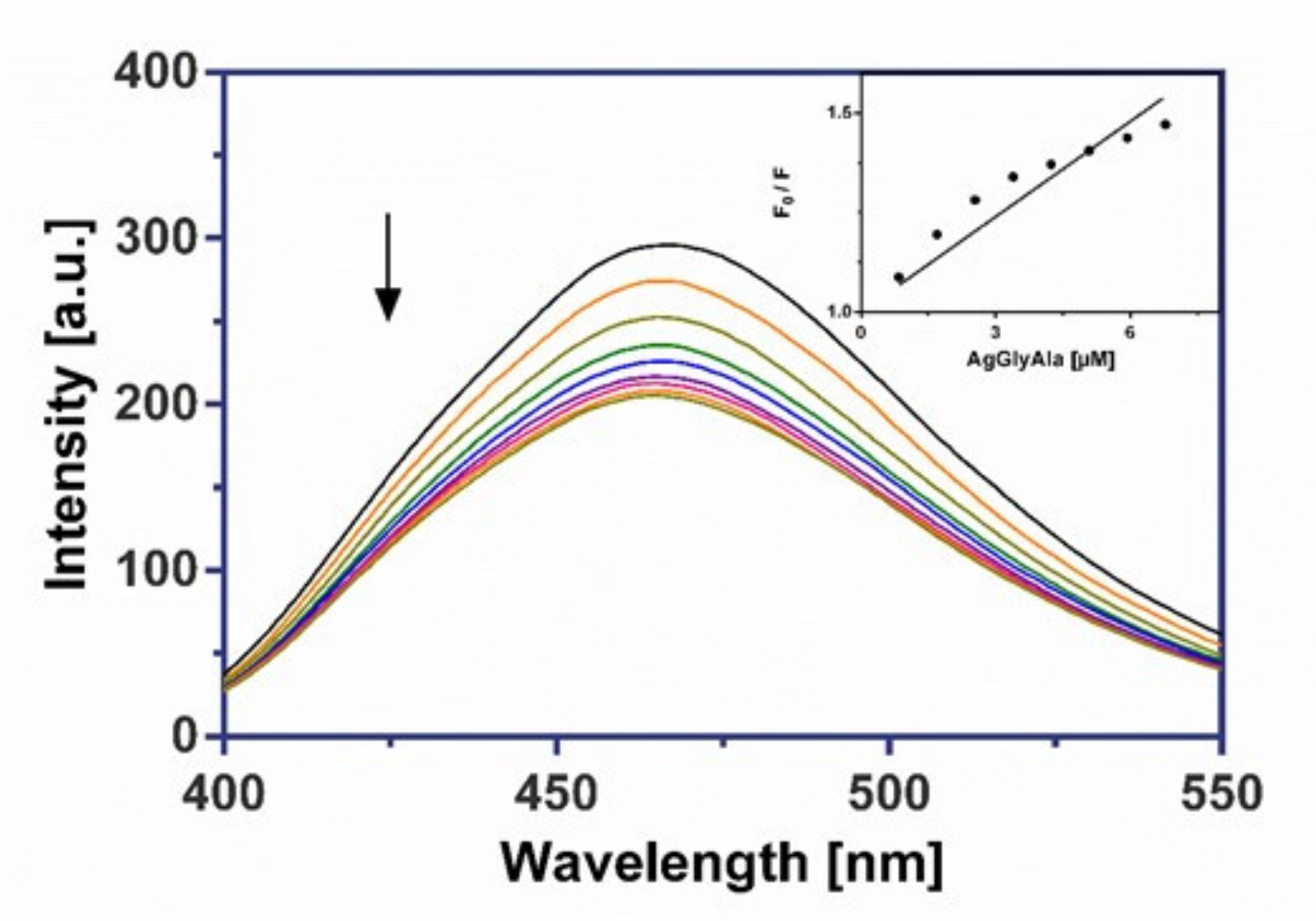
2.8.2. Fluorescence Measurements
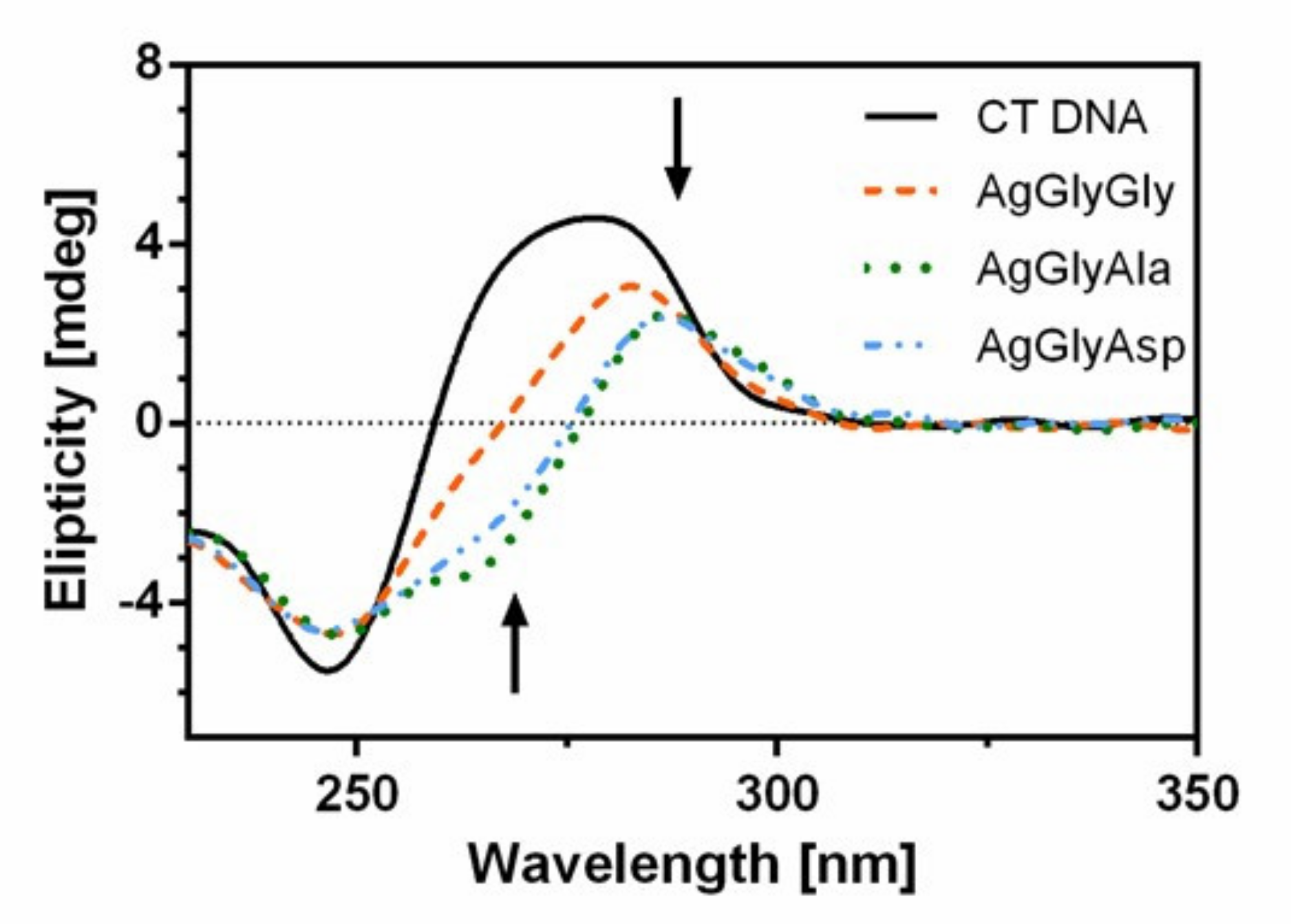
2.9. CD Spectroscopy
2.10. Viscosimetry
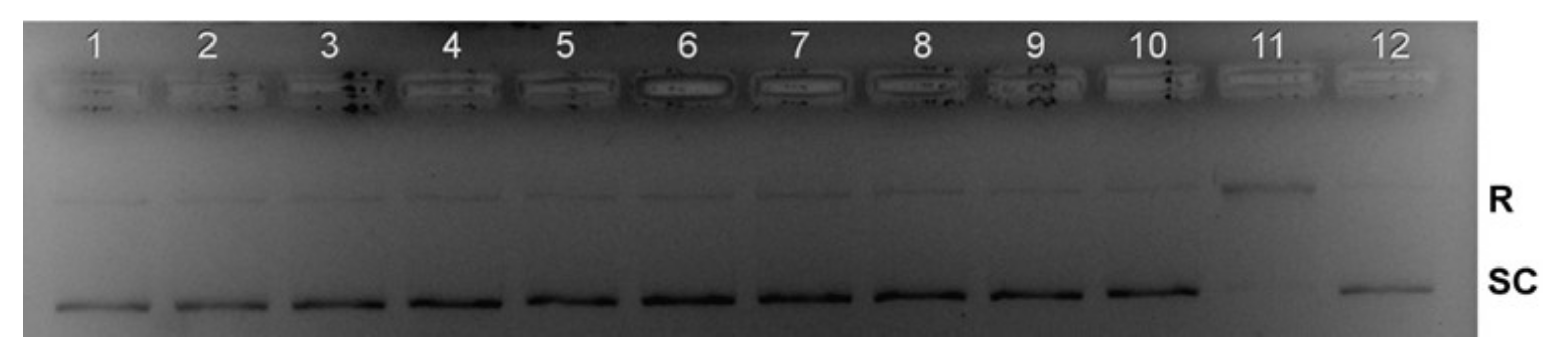
2.11. Topoisomerase I Inhibition
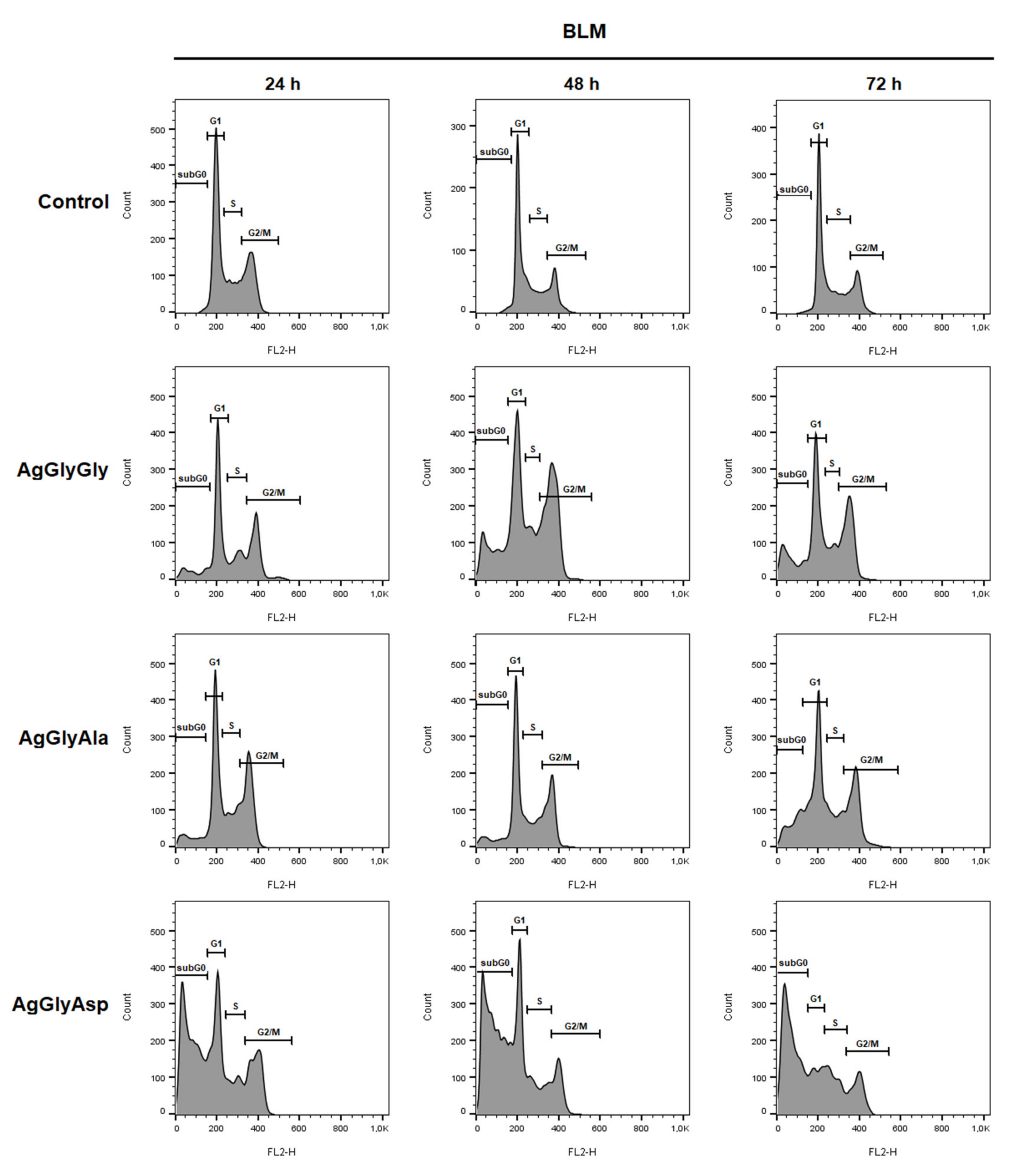
2.12. Cell Cycle Analysis
3. Materials and Methods
3.1. Materials
3.2. Synthesis
3.3. Physical Measurements
3.4. Antimicrobial Activity of Newly Prepared Complexes Assayed In Vitro
3.4.1. Assessment of Potential Mutagenic Activity
3.4.2. Influence of Silver Complexes on Microbial Biofilm Formation
3.5. Anticancer Activity
3.5.1. Cell Culture
3.5.2. MTS Assay
3.6. DNA Binding Properties
3.6.1. UV–Vis Absorption Measurements
3.6.2. Fluorescence Binding Study
3.6.3. Circular Dichroism Measurements
3.6.4. Viscosimetry
3.6.5. Topoisomerase I Inhibition Studies
3.7. Cell Cycle Analysis
Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Tornesello, A.L.; Borrelli, A.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Antimicrobial Peptides as Anticancer Agents: Functional Properties and Biological Activities. Molecules 2020, 25, 2850. [Google Scholar] [CrossRef]
- Raheem, N.; Straus, S.K. Mechanisms of Action for Antimicrobial Peptides with Antibacterial and Antibiofilm Functions. Front. Microbiol. 2019, 10, 2866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flemming, A.; Allison, V.D. Observations on a bacteriolytic substance (“lysozyme”) found in secretions and tissues. Br. J. Exp. Pathol. 1922, 3, 252–260. [Google Scholar]
- Hirsch, J.G. Phagocytin: A bactericidal substance from polymorphonuclear leucocytes. J. Exp. Med. 1956, 103, 589–611. [Google Scholar] [CrossRef] [PubMed]
- Bobone, S.; Stella, L. Selectivity of Antimicrobial Peptides: A Complex Interplay of Multiple Equilibria. Adv. Exp. Med. Biol. 2019, 1117, 175–214. [Google Scholar]
- Budagumpi, S.; Haque, R.A.; Endud, S.; Rehman, G.U.; Salman, A.W. Biologically Relevant Silver(I)-N-Heterocyclic Carbene Complexes: Synthesis, Structure, Intramolecular Interactions, and Applications. Eur. J. Inorg. Chem. 2013, 2013, 4367–4388. [Google Scholar] [CrossRef]
- Medici, S.; Peana, M.; Crisponi, G.; Nurchi, V.M.; Lachowicz, J.I.; Remelli, M.; Zoroddu, M.A. Silver coordination compounds: A new horizon in medicine. Coord. Chem. Rev. 2016, 327–328, 349–359. [Google Scholar] [CrossRef]
- Abarca, R.; Gomez, G.; Velasquez, C.; Paez, M.A.; Gulppi, M.; Arrieta, A.; Azocar, M.I. Antibacterial Behavior of Pyridinecarboxylatesilver(I) Complexes. Chin. J. Chem. 2012, 30, 1631–1635. [Google Scholar] [CrossRef]
- Homzová, K.; Gyoryová, K.; Bujdošová, Z.; Hudecová, D.; Ganajová, M.; Vargová, Z.; Kovářová, J. Synthesis, thermal, spectral and biological properties of zinc(II) 4-hydroxybenzoate complexes. J. Therm. Anal. Calorim. 2014, 116, 77–91. [Google Scholar] [CrossRef]
- Vargová, Z.; Almáši, M.; Hudecová, D.; Titková, D.; Rostášová, I.; Zeleňák, V.; Györyová, K. New silver(I) pyridinecarboxylate complexes: Synthesis, characterization, and antimicrobial therapeutic potential. J. Coord. Chem. 2014, 67, 1002–1021. [Google Scholar] [CrossRef]
- Almáši, M.; Vargová, Z.; Sabolová, D.; Kudláčová, J.; Hudecová, D.; Kuchár, J.; Očenášová, L.; Györyová, K. Ag(I) and Zn(II) isonicotinate complexes: Design, characterization, antimicrobial effect, and CT-DNA binding studies. J. Coord. Chem. 2015, 68, 4423–4443. [Google Scholar] [CrossRef]
- McCann, M.; Curran, R.; Ben-Shoshan, M.; McKee, V.; Devereux, M.; Kavanagh, K.; Kellett, A. Synthesis, structure and biological activity of silver(I) complexes of substituted imidazoles. Polyhedron 2013, 56, 180–188. [Google Scholar] [CrossRef] [Green Version]
- Sadek, B. Imidazole-substituted drugs and tendency for inhibition of cytochrome P450 isoenzymes: A review. Pharma Chem. 2011, 3, 410–419. [Google Scholar]
- Kalinowska-Lis, U.; Felczak, A.; Chęcińska, L.; Małecka, M.; Lisowska, K.; Ochocki, J. Influence of selected inorganic counter-ions on the structure and antimicrobial properties of silver(i) complexes with imidazole-containing ligands. New J. Chem. 2016, 40, 694–704. [Google Scholar] [CrossRef]
- Kalinowska-Lis, U.; Felczak, A.; Chęcińska, L.; Zawadzka, K.; Patyna, E.; Lisowska, K.; Ochocki, J. Synthesis, characterization and antimicrobial activity of water-soluble silver(i) complexes of metronidazole drug and selected counter-ions. Dalton Trans. 2015, 44, 8178–8189. [Google Scholar] [CrossRef] [PubMed]
- Rendošová, M.; Vargová, Z.; Kuchár, J.; Sabolová, D.; Levoča, Š.; Kudláčová, J.; Paulíková, H.; Hudecová, D.; Helebrandtová, V.; Almáši, M.; et al. New silver complexes with bioactive glycine and nicotinamide molecules—Characterization, DNA binding, antimicrobial and anticancer evaluation. J. Inorg. Biochem. 2017, 168, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kuzderová, G.; Rendošová, M.; Gyepes, R.; Almáši, M.; Sabolová, D.; Vilková, M.; Olejníková, P.; Hudecová, D.; Kello, M.; Vargová, Z. In vitro biological evaluation and consideration about structure-activity relationship of silver(I) aminoacidate complexes. J. Inorg. Biochem. 2020, 210, 11170. [Google Scholar] [CrossRef]
- Manzano, C.; Nakahata, D.H.; Corbi, P.P.; Tenorio, J.C.; Lustri, W.R.; Nogueira, F.A.R.; Aleixo, N.A.; Gomes, P.S.D.S.; Pavan, F.; Grecco, J.A.; et al. Silver complexes with fluoroanthranilic acid isomers: Spectroscopic characterization, antimycobacterial activity and cytotoxic studies over a panel of tumor cells. Inorg. Chim. Acta 2020, 502, 119293. [Google Scholar] [CrossRef]
- Samanta, T.; Munda, R.N.; Roymahapatra, G.; Nandy, A.; Saha, K.D.; Al-Deyab, S.S.; Dinda, J. Silver(I), Gold(I) and Gold(III)-N-Heterocyclic carbene complexes of naphthyl substituted annelated ligand: Synthesis, structure and cytotoxicity. J. Organomet. Chem. 2015, 791, 183–191. [Google Scholar] [CrossRef]
- Haque, R.A.; Choo, S.Y.; Budagumpi, S.; Iqbal, M.A.; Abdullah, A.A.-A. Silver(I) complexes of mono- and bidentate N-heterocyclic carbene ligands: Synthesis, crystal structures, and in vitro antibacterial and anticancer studies. Eur. J. Med. Chem. 2015, 90, 82–92. [Google Scholar] [CrossRef]
- Canakci, D.; Koyuncu, I.; Lolak, N.; Durgun, M.; Akocak, S.; Supuran, C.T. Synthesis and cytotoxic activities of novel copper and silver complexes of 1,3-diaryltriazene-substituted sulfonamides. J. Enzyme Inhib. Med. Chem. 2019, 34, 110–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akkoç, S.; Kayser, V.; Ilhan, I.Ö.; Hibbs, D.E.; Gök, Y.; Williams, P.A.; Hawkins, B.; Lai, F. New compounds based on a benzimidazole nucleus: Synthesis, characterization and cytotoxic activity against breast and colon cancer cell lines. J. Organomet. Chem. 2017, 839, 98–107. [Google Scholar] [CrossRef]
- Banti, C.N.; Papatriantafyllopoulou, C.; Manoli, M.; Tasiopoulos, A.J.; Hadjikakou, S.K. Nimesulide Silver Metallodrugs, Containing the Mitochondriotropic, Triaryl Derivatives of Pnictogen; Anticancer Activity against Human Breast Cancer Cells. Inorg. Chem. 2016, 55, 8681–8696. [Google Scholar] [CrossRef]
- Banti, C.N.; Hatzidimitriou, A.G.; Kourkoumelis, N.; Hadjikakou, S.K. Diclofenac conjugates with biocides through silver(I) ions (CoMeD’s); Development of a reliable model for the prediction of anti-proliferation of NSAID’s-silver formulations. J. Inorg. Biochem. 2019, 194, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Altay, A.; Caglar, S.; Caglar, B. Silver(I) complexes containing diclofenac and niflumic acid induce apoptosis in human-derived cancer cell lines. Arch. Physiol. Biochem. 2019, 1–11. [Google Scholar] [CrossRef]
- Li, S.; Zhang, S.; Jin, X.; Tan, X.; Lou, J.; Zhang, X.; Zhao, Y. Singly protonated dehydronorcantharidin silver coordination polymer induces apoptosis of lung cancer cells via reactive oxygen species-mediated mitochondrial pathway. Eur. J. Med. Chem. 2014, 86, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mahendiran, D.; Kumar, R.S.; Viswanathan, V.; Velmurugan, D.; Rahiman, A.Z. In vitro and in vivo anti-proliferative evaluation of bis(4′-(4-tolyl)-2,2′:6′,2″-terpyridine)copper(II) complex against Ehrlich ascites carcinoma tumors. J. Biol. Inorg. Chem. 2017, 22, 1109–1122. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-L.; Qin, Q.-P.; An, Y.-F.; Liu, Y.-C.; Huang, G.-B.; Luo, X.-J.; Zhang, G.-H. Study on potential antitumor mechanism of quinoline-based silver(I) complexes: Synthesis, structural characterization, cytotoxicity, cell cycle and caspase-initiated apoptosis. Inorg. Chem. Commun. 2014, 40, 73–77. [Google Scholar] [CrossRef]
- Liang, X.; Luan, S.; Yin, Z.; He, M.; He, C.; Yin, L.; Zou, Y.; Yuan, Z.; Li, L.; Song, X.; et al. Recent advances in the medical use of silver complex. Eur. J. Med. Chem. 2018, 157, 62–80. [Google Scholar] [CrossRef]
- Rendošová, M.; Gyepes, R.; Cingelova, M.I.; Mudronova, D.; Sabolová, D.; Kello, M.; Vargova, Z. In vitro selective inhibitory effect of silver(I) aminoacidates against bacteria and intestinal cell lines and elucidation of mechanism action by means of DNA binding properties, DNA cleavage and cell cycle arrest. Dalton Trans. 2021, 50, 936–953. [Google Scholar] [CrossRef]
- Flook, R.J.; Freeman, H.C.; Moore, C.J.; Scudder, M.L. Model compounds for metal–protein interaction: Crystal structures of seven cadmium(II) complexes of amino-acids and peptides. J. Chem. Soc. Chem. Commun. 1973, 753–754. [Google Scholar] [CrossRef]
- Takayama, T.; Ohuchida, S.; Koike, Y.; Watanabe, M.; Hashizume, D.; Ohashi, Y. Structural Analysis of Cadmium-Glycylglycine Complexes Studied by X-ray Diffraction and High Resolution 1l3Cd and 13C Solid State NMR. Bull. Chem. Soc. Jpn. 1996, 69, 1579–1586. [Google Scholar] [CrossRef]
- Acland, C.B.; Freeman, H.C. Model compounds for metal–protein interaction: Crystal structures of four silver(II) complexes with glycine, glycylglycine, and imidazole. J. Chem. Soc. D 1971, 17, 1016–1017. [Google Scholar] [CrossRef]
- Shiro, M.; Nakao, Y.; Yamauchi, O.; Nakahara, A. the crystal and molecular structure of chloroglycylglycinatocopper(II) monohydrate. Chem. Lett. 1972, 1, 123–124. [Google Scholar] [CrossRef]
- Rabone, J.; Yue, Y.-F.; Chong, S.Y.; Stylianou, K.C.; Bacsa, J.; Bradshaw, D.; Darling, G.R.; Berry, N.G.; Khimyak, Y.Z.; Ganin, A.Y.; et al. An Adaptable Peptide-Based Porous Material. Science 2010, 329, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Emami, S.; Paz, F.A.A.; Mendes, A.; Gales, L. Toward the Construction of 3D Dipeptide–Metal Frameworks. Cryst. Growth Des. 2014, 14, 4777–4780. [Google Scholar] [CrossRef] [Green Version]
- Koleva, B.B.; Zareva, S.; Kolev, T.; Spiteller, M. New Au(III), Pt(II) and Pd(II) complexes with glycyl-containing homopeptides. J. Coord. Chem. 2008, 61, 3534–3548. [Google Scholar] [CrossRef]
- Joseyphus, R.S.; Nair, M.S. Synthesis, characterization and biological studies of some Co(II), Ni(II) and Cu(II) complexes derived from indole-3-carboxaldehyde and glycylglycine as Schiff base ligand. Arab. J. Chem. 2010, 3, 195–204. [Google Scholar] [CrossRef]
- Fu, X.-B.; Liu, D.-D.; Lin, Y.; Hu, W.; Mao, Z.-W.; Le, X.-Y. Water-soluble DNA minor groove binders as potential chemotherapeutic agents: Synthesis, characterization, DNA binding and cleavage, antioxidation, cytotoxicity and HSA interactions. Dalton Trans. 2014, 43, 8721–8737. [Google Scholar] [CrossRef] [PubMed]
- Tabassum, S.; Al-Asbahy, W.M.; Afzal, M.; Shamsi, M.; Arjmand, F. DNA binding and cleavage studies of new sulfasalazine-derived dipeptide Zn(II) complex: Validation for specific recognition with 5′–TMP. J. Lumin. 2012, 132, 3058–3065. [Google Scholar] [CrossRef]
- Schmidbaur, H.; Schier, A. Argentophilic Interactions. Angew. Chem. Int. Ed. 2015, 54, 746–784. [Google Scholar] [CrossRef] [PubMed]
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge Structural Database. Acta Crystallogr. Sect. B Struct. Sci. Cryst. Eng. Mater. 2016, 72, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zou, Y.; Ni, C.-L.; Ni, Z.-P.; Li, Y.-Z.; Yao, Y.-G.; Meng, Q.-J. Synthesis and characterization of a dipeptide–copper(II)–2-aminomethylbenzimidazole ternary complex. J. Coord. Chem. 2004, 57, 899–906. [Google Scholar] [CrossRef]
- Buckingham, D.A.; Marzilli, P.A.; Maxwell, I.E.; Sargeson, A.M.; Fehlmann, M.; Freeman, H.C. The crystal structures of the glycylglycine O-ethyl ester and chloroaquo-complexes of β-(triethylenetetramine)cobalt(III). Chem. Commun. 1968, 9, 488–489. [Google Scholar] [CrossRef]
- Payne, J.W.; Smith, M.W. Peptide transport by microorganisms. Adv. Microb. Physiol. 1994, 36, 1–80. [Google Scholar] [CrossRef]
- Alves, R.A.; Payne, J.W. The number and nature of the peptide transport systems of E. coli: Characterization of specific transport mutants. Biochem. Soc. Trans. 1980, 8, 704–705. [Google Scholar] [CrossRef]
- Smith, M.W.; Tyreman, D.R.; Payne, G.M.; Marshall, N.J.; Payne, J.W. Substrate specificity of the periplasmic dipeptide-binding protein from Escherichia coli: Experimental basis for the design of peptide prodrugs. Microbiology 1999, 145, 2891–2901. [Google Scholar] [CrossRef] [Green Version]
- Perry, D.; Gilvarg, C. Spectrophotometric determination of affinities of peptides for their transport systems in Escherichia coli. J. Bacteriol. 1984, 160, 943–948. [Google Scholar] [CrossRef] [Green Version]
- Banti, C.N.; Raptopoulou, C.P.; Psycharis, V.; Hadjikakou, S.K. Novel silver glycinate conjugate with 3D polymeric intermolecular self-assembly architecture; an antiproliferative agent which induces apoptosis on human breast cancer cells. J. Inorg. Biochem. 2021, 216, 111351. [Google Scholar] [CrossRef]
- Kutlu, T.; Yıldırım, I.; Karabıyık, H.; Kılınçlı, A.; Tekedereli, I.; Gök, Y.; Dikmen, M.; Aktas, A. Cytotoxic activity and apoptosis induction by a series Ag(I)-NHC complexes on human breast cancer cells and non-tumorigenic epithelial cell line. J. Mol. Struct. 2021, 1228, 129462. [Google Scholar] [CrossRef]
- Satyanarayana, S.; Dabrowiak, J.C.; Chaires, J. Tris(phenanthroline)ruthenium(II) enantiomer interactions with DNA: Mode and specificity of binding. Biochemistry 1993, 32, 2573–2584. [Google Scholar] [CrossRef] [PubMed]
- Shahabadi, N.; Ghasemian, Z.; Hadidi, S. Binding Studies of a New Water-Soluble Iron(III) Schiff Base Complex to DNA Using Multispectroscopic Methods. Bioinorg. Chem. Appl. 2012, 2012, 126451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhadra, K.; Kumar, G.S. Interaction of berberine, palmatine, coralyne, and sanguinarine to quadruplex DNA: A comparative spectroscopic and calorimetric study. Biochim. Biophys. Acta 2011, 1810, 485–496. [Google Scholar] [CrossRef]
- Thakor, K.P.; Lunagariya, M.V.; Bhatt, B.S.; Patel, M.N. Fluorescence and absorption studies of DNA-Pd(II) complex interaction: Synthesis, spectroanalytical investigations and biological activities. Luminescence 2019, 34, 113–124. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.-M.; Chen, C.K.-M.; Hou, M.-H. Conformational Changes in DNA upon Ligand Binding Monitored by Circular Dichroism. Int. J. Mol. Sci. 2012, 13, 3394–3413. [Google Scholar] [CrossRef] [Green Version]
- Meenongwa, A.; Brissos, R.F.; Soikum, C.; Chaveerach, P.; Trongpanich, Y.; Chaveerach, U. Enhancement of biological activities of copper(II) complexes containing guanidine derivatives by enrofloxacin. J. Mol. Struct. 2021, 1241, 130645. [Google Scholar] [CrossRef]
- Shahabadi, N.; Fatahi, S.; Maghsudi, M. Synthesis of a new Pt(II) complex containing valganciclovir drug and calf-thymus DNA interaction study using multispectroscopic methods. J. Coord. Chem. 2018, 71, 258–270. [Google Scholar] [CrossRef]
- Brodie, C.R.; Collins, J.G.; Wright, J.R.A. DNA binding and biological activity of some platinum(II) intercalating compounds containing methyl-substituted 1,10-phenanthrolines. Dalton Trans. 2004, 8, 1145–1152. [Google Scholar] [CrossRef]
- Yildiz, U.; Coban, B. Chemical and photo-induced nuclease activity of a novel minor groove DNA binder Cu(II) complex. J. Serbian Chem. Soc. 2019, 84, 563–574. [Google Scholar] [CrossRef] [Green Version]
- Champoux, J.J. DNA Topoisomerases: Structure, Function, and Mechanism. Annu. Rev. Biochem. 2001, 70, 369–413. [Google Scholar] [CrossRef] [Green Version]
- Nitiss, J.L. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 2009, 9, 338–350. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Z.; Chen, S.; Chen, C.; Chen, J.; Chen, C.; Dai, Q.; Gao, C.; Jiang, Y. Design, synthesis and biological evaluation of 4-amidobenzimidazole acridine derivatives as dual PARP and Topo inhibitors for cancer therapy. Eur. J. Med. Chem. 2017, 138, 1135–1146. [Google Scholar] [CrossRef]
- Rendošová, M.; Vargová, Z.; Sabolová, D.; Imrichová, N.; Hudecová, D.; Gyepes, R.; Lakatoš, B.; Elefantová, K. Silver pyridine-2-sulfonate complex-its characterization, DNA binding, topoisomerase I inhibition, antimicrobial and anticancer response. J. Inorg. Biochem. 2018, 186, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Banti, C.N.; Papatriantafyllopoulou, C.; Tasiopoulos, A.J.; Hadjikakou, S.K. New metalo-therapeutics of NSAIDs against human breast cancer cells. Eur. J. Med. Chem. 2018, 143, 1687–1701. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.E.S.; Becceneri, A.B.; Solcia, M.C.; Santiago, J.V.B.; Moreira, M.B.; Neto, J.A.G.; Pavan, F.; Cominetti, M.R.; Pereira, J.C.M.; Netto, A.V.D.G. Cytotoxic and apoptotic effects of ternary silver(i) complexes bearing 2-formylpyridine thiosemicarbazones and 1,10-phenanthroline. Dalton Trans. 2020, 49, 5264–5275. [Google Scholar] [CrossRef]
- Ota, A.; Tajima, M.; Mori, K.; Sugiyama, E.; Sato, V.H.; Sato, H. The selective cytotoxicity of silver thiosulfate, a silver complex, on MCF-7 breast cancer cells through ROS-induced cell death. Pharmacol. Rep. 2021, 73, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Brandenburg, K.; Putz, H. DIAMOND–Crystal and Molecular Structure Visualization, v6.3; Crystal Impact GbR: Bonn, Germany, 2020. [Google Scholar]
- Jantová, S.; Hudecová, D.; Stankovský, Š.; Špirková, K.; Ružeková, L. Antibacterial effect of substituted 4-quinazolylhydrazines and their arylhydrazones determined by a modified microdilution method. Folia Microbiol. 1995, 40, 611–614. [Google Scholar] [CrossRef]
- Betina, V.; Mičeková, D. Antimicrobial properties of fungal macrolide antibiotics. J. Basic Microbiol. 1972, 12, 355–364. [Google Scholar] [CrossRef]
- Hudecova, D.; Jantova, S.; Melnik, M.; Uher, M. New azidometalkojates and their biological activity. Folia Microbiol. 1996, 41, 473–476. [Google Scholar] [CrossRef] [PubMed]
- Dudová, B.; Hudecová, D.; Pokorný, R.; Mičková, M.; Palicová, M.; Segl’a, P.; Melník, M. Copper complexes with bioactive ligands. Folia Microbiol. 2002, 47, 225–229. [Google Scholar] [CrossRef]
- Maron, D.M.; Ames, B.N. Revised methods for the Salmonella mutagenicity test. Mutat. Res. Mutagen. Relat. Subj. 1983, 113, 173–215. [Google Scholar] [CrossRef]
- Sherry, L.; Rajendran, R.; Lappin, D.F.; Borghi, E.; Perdoni, F.; Falleni, M.; Tosi, D.; Smith, K.; Williams, C.; Jones, B.; et al. Biofilms formed by Candida albicans bloodstream isolates display phenotypic and transcriptional heterogeneity that are associated with resistance and pathogenicity. BMC Microbiol. 2014, 14, 182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potočňák, I.; Vranec, P.; Farkasová, V.; Sabolová, D.; Vataščinová, M.; Kudláčová, J.; Radojević, I.D.; Čomić, L.R.; Markovic, B.S.; Volarevic, V.; et al. Low-dimensional compounds containing bioactive ligands. Part VI: Synthesis, structures, in vitro DNA binding, antibacterial and anticancer properties of first row transition metals complexes with 5-chloro-quinolin-8-ol. J. Inorg. Biochem. 2015, 154, 67–77. [Google Scholar] [CrossRef]
- Franich, A.A.; Živković, M.D.; Ilić-Tomić, T.; Đorđević, I.S.; Nikodinović-Runić, J.; Pavić, A.; Janjić, G.V.; Rajković, S. New minor groove covering DNA binding mode of dinuclear Pt(II) complexes with various pyridine-linked bridging ligands and dual anticancer-antiangiogenic activities. J. Biol. Inorg. Chem. 2020, 25, 395–409. [Google Scholar] [CrossRef] [PubMed]
- Smolková, R.; Zeleňák, V.; Gyepes, R.; Sabolová, D.; Imrichová, N.; Hudecová, D.; Smolko, L. Synthesis, characterization, DNA binding, topoisomerase I inhibition and antimicrobial activity of four novel zinc(II) fenamates. Polyhedron 2018, 141, 230–238. [Google Scholar] [CrossRef]



| HGlyGly | AgGlyGly | HGlyAla | AgGlyAla | H2GlyAsp | AgGlyAsp | |
|---|---|---|---|---|---|---|
| ν(NH3+) + ν(CH2) | 3350–2500 | 3350–2500 | 3350–2500 | 3350–2500 | 3350–2500 | 3350–2500 |
| ν(NH2) | 3283 | 3317 | 3199 | 3350 | 3327 | 3258 |
| 3162 | 3072 | |||||
| ν(CH) | - | - | 2813 | 2977 | 2996 | 2950 |
| - | - | 2958 | 2907 | |||
| δ(NH3+),δ(NH2)scissoring | 1653 | 1645 | 1684 | 1666 | 1666 | 1682 |
| 1512 | 1505 | |||||
| νas(COO−) | 1572 | 1522 | 1526 | 1550 | 1568 | 1515 |
| νs(COO−) | 1478 | 1446 | 1406 | 1437 | 1445 | 1436 |
| Δ(νas − νs) | 76 | 113 | 79 | |||
| δ(CCH) | - | 1396 | 1370 | 1365 | 1386 | 1368 |
| - | - | 1348 | 1335 | 1325 | ||
| ν(NO3-) | - | 1316 | - | 1291 | - | 1325 |
| ν(CCN)alif. | 1096 | 1077 | 1032 | 1032 | 1041 | 1039 |
| 1061 | 838 | 822 | ||||
| ν(CC) | 964,916 | 907 | 935 | 911 | 937 | 935 |
| 899 | 824 | 917 | 873,856 | 862 | 823 | |
| γ(COO−) | 733 | 716 | 774 | 781,762 | 763 | 764 |
| δ(CCO) | 588,528 | 598,548 | 562 | 545 | 564 | 553 |
| Chemical Shifts δ/ppm | ||||||
|---|---|---|---|---|---|---|
| Signal | HGlyGly | AgGlyGly | HGlyAla | AgGlyAla | H2GlyAsp | AgGlyAsp |
| A | 3.86 | 3.86 | 4.19 | 4.18 | 4.62 | 4.49 |
| B | 3.83 | 3.83 | 3.83 | 3.82 | 3.88 | 3.82 |
| C | - | - | 1.37 | 1.36 | 2.90 | 2.76; 2.52 |
| CC (µM) | AgSD | AgGly | AgAla | CC (µM) | AgGlyGly | AgGlyAla | AgGlyAsp | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A570 | I | A570 | I | A570 | I | A570 | I | A570 | I | A570 | I | |||
| S. aureus | 50 | 0.20 | medium | 0.28 | medium | 0.29 | medium | 100 | 0.28 | medium | 0.25 | medium | 0.25 | medium |
| 25 | 0.18 | medium | 0.27 | medium | 0.29 | medium | 50 | 0.22 | medium | 0.23 | medium | 0.24 | medium | |
| 10 | 0.20 | medium | 0.37 | strong | 0.39 | strong | 25 | 0.26 | medium | 0.25 | medium | 0.31 | medium | |
| 5 | 0.18 | medium | 0.45 | strong | 0.35 | strong | 10 | 0.39 | strong | 0.41 | strong | 0.35 | strong | |
| 1 | 0.29 | medium | 0.40 | strong | 0.47 | strong | 5 | 0.72 | strong | 0.51 | strong | 0.36 | strong | |
| control A570 = 0.47, I–strong | ||||||||||||||
| Complex Dose per Plate | Revertants CFU No. ± SD | |
|---|---|---|
| S. thypimurium TA100 | S. thypimurium TA98 | |
| AgSD | ||
| 20 ng | Toxicity | Toxicity |
| 10 ng | 167 ± 12 | 30 ± 8 |
| 5 ng | 165 ± 12 | 25 ± 7 |
| 2.5 ng | 125 ± 7 | 35 ± 7 |
| AgGly | ||
| 100 ng | Toxicity | Toxicity |
| 40 ng | 140 ± 18 | 32 ± 2 |
| 20 ng | 155 ± 7 | 36 ± 3 |
| 10 ng | 140 ± 14 | 35 ± 1 |
| 5 ng | 129 ± 12 | 29 ± 1 |
| AgAla | ||
| 100 ng | Toxicity | Toxicity |
| 40 ng | 187 ± 15 | 28 ± 2 |
| 20 ng | 172 ± 28 | 38 ± 4 |
| 10 ng | 157 ± 4 | 34 ± 1 |
| 5 ng | 175 ± 7 | 33 ± 5 |
| AgGlyGly | ||
| 200 ng | 135 ± 21 | 36 ± 2 |
| 100 ng | 138 ± 2 | 31 ± 1 |
| 50 ng | 162 ±17 | 36 ± 2 |
| 20 ng | 149 ± 12 | 29 ± 1 |
| AgGlyAla | ||
| 200 ng | Toxicity | Toxicity |
| 100 ng | 115 ± 7 | 28 ± 2 |
| 50 ng | 130 ± 14 | 34 ± 2 |
| 20 ng | 142 ± 8 | 27 ± 2 |
| AgGlyAsp | ||
| 200 ng | 149 ± 7 | 27 ± 1 |
| 100 ng | 141 ± 9 | 22 ± 3 |
| 50 ng | 129 ± 12 | 26 ± 3 |
| 20 ng | 137 ± 10 | 29 ± 1 |
| Negative control 1%DMSO | 130 ± 5 | 29 ± 2 |
| Spontaneous revertants | 160 ± 14 | 28 ± 3 |
| Positive control NFAA (100µg) | 1352± 124 | 28 ± 2 |
| HCT116 | HeLa | MCF-7 | MDA-MB-231 | Jurkat | BLM | BJ-5ta | |
|---|---|---|---|---|---|---|---|
| HGlyGly | >300 | >300 | >300 | >300 | >300 | >300 | >300 |
| HGlyAla | >300 | >300 | >300 | >300 | >300 | >300 | >300 |
| H2GlyAsp | >300 | >300 | >300 | >300 | >300 | >300 | >300 |
| AgGlyGly | 34.5 | 38.3 | 9.0 | 13.0 | 14.8 | 21.2 | 47.1 |
| AgGlyAla | 28.0 | 33.2 | 7.5 | 7.4 | 9.7 | 10.9 | 40.2 |
| AgGlyAsp | 35.5 | 33.3 | 33.8 | 33.6 | 36.8 | 35.6 | 57.6 |
| cisPt | 7.4 | 35.4 | 29.7 | 7.1 | 6.3 | 29.5 | 37.9 |
| AgNO3 | 21.8 | 45.3 | 13.2 | 25.9 | 6.6 | 31.5 | 24.3 |
| λmax (nm) | Δλmax (nm) | Hypochromism (%) | KSV (M−1) | |
|---|---|---|---|---|
| AgGlyAsp | 260 | 13 | 30.8 | 8.33 ± 0.07 × 104 |
| AgGlyAla | 268 | 9 | 40.7 | 7.97 ± 0.03 × 104 |
| AgGlyGly | 264 | 22 | 20.1 | 4.51 ± 0.05 × 104 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuzderová, G.; Rendošová, M.; Gyepes, R.; Sovová, S.; Sabolová, D.; Vilková, M.; Olejníková, P.; Bačová, I.; Stokič, S.; Kello, M.; et al. Antimicrobial and Anticancer Application of Silver(I) Dipeptide Complexes. Molecules 2021, 26, 6335. https://doi.org/10.3390/molecules26216335
Kuzderová G, Rendošová M, Gyepes R, Sovová S, Sabolová D, Vilková M, Olejníková P, Bačová I, Stokič S, Kello M, et al. Antimicrobial and Anticancer Application of Silver(I) Dipeptide Complexes. Molecules. 2021; 26(21):6335. https://doi.org/10.3390/molecules26216335
Chicago/Turabian StyleKuzderová, Gabriela, Michaela Rendošová, Róbert Gyepes, Simona Sovová, Danica Sabolová, Mária Vilková, Petra Olejníková, Ivana Bačová, Simonida Stokič, Martin Kello, and et al. 2021. "Antimicrobial and Anticancer Application of Silver(I) Dipeptide Complexes" Molecules 26, no. 21: 6335. https://doi.org/10.3390/molecules26216335
APA StyleKuzderová, G., Rendošová, M., Gyepes, R., Sovová, S., Sabolová, D., Vilková, M., Olejníková, P., Bačová, I., Stokič, S., Kello, M., & Vargová, Z. (2021). Antimicrobial and Anticancer Application of Silver(I) Dipeptide Complexes. Molecules, 26(21), 6335. https://doi.org/10.3390/molecules26216335