Lipid A-Mediated Bacterial–Host Chemical Ecology: Synthetic Research of Bacterial Lipid As and Their Development as Adjuvants
Abstract
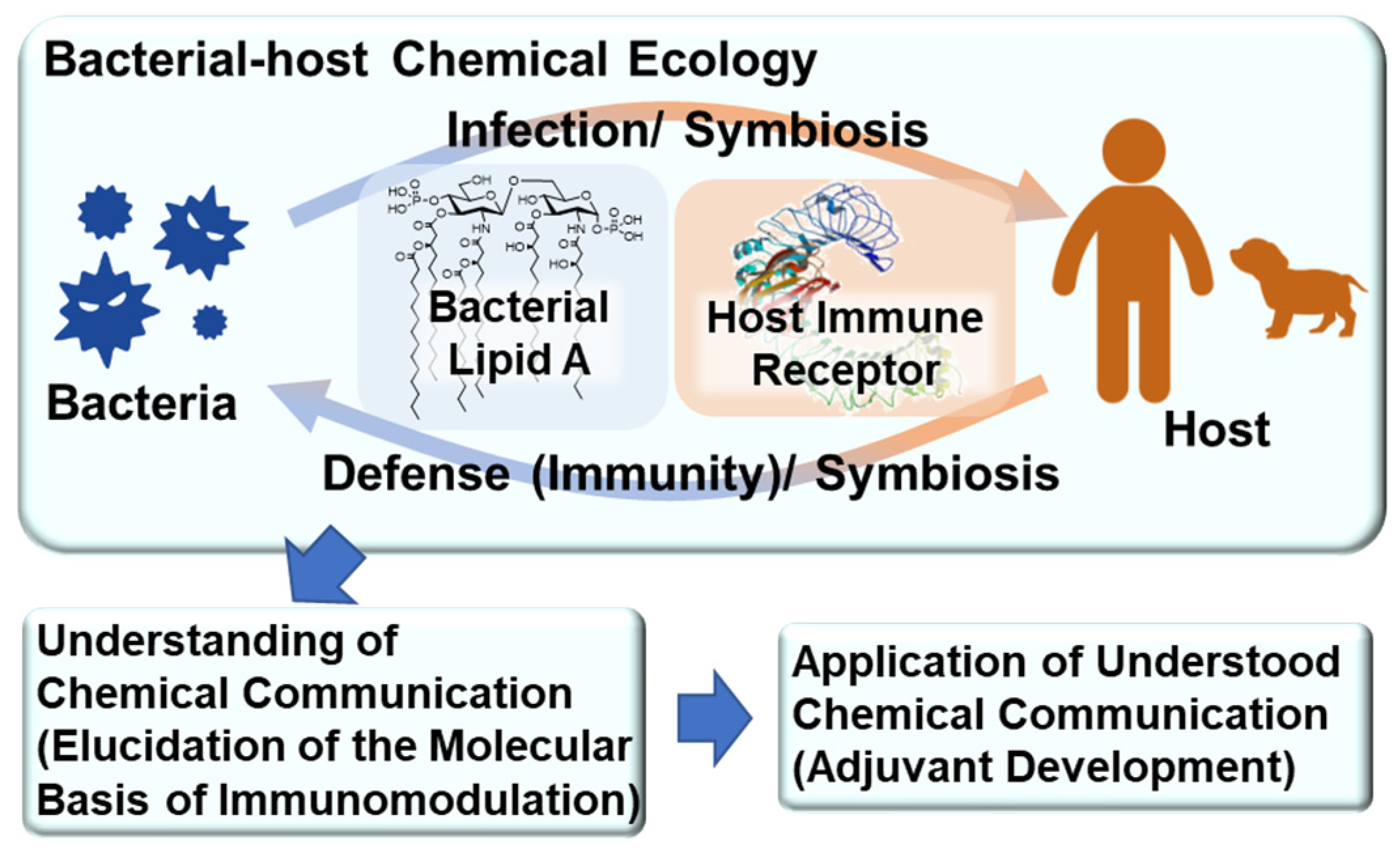
1. Introduction
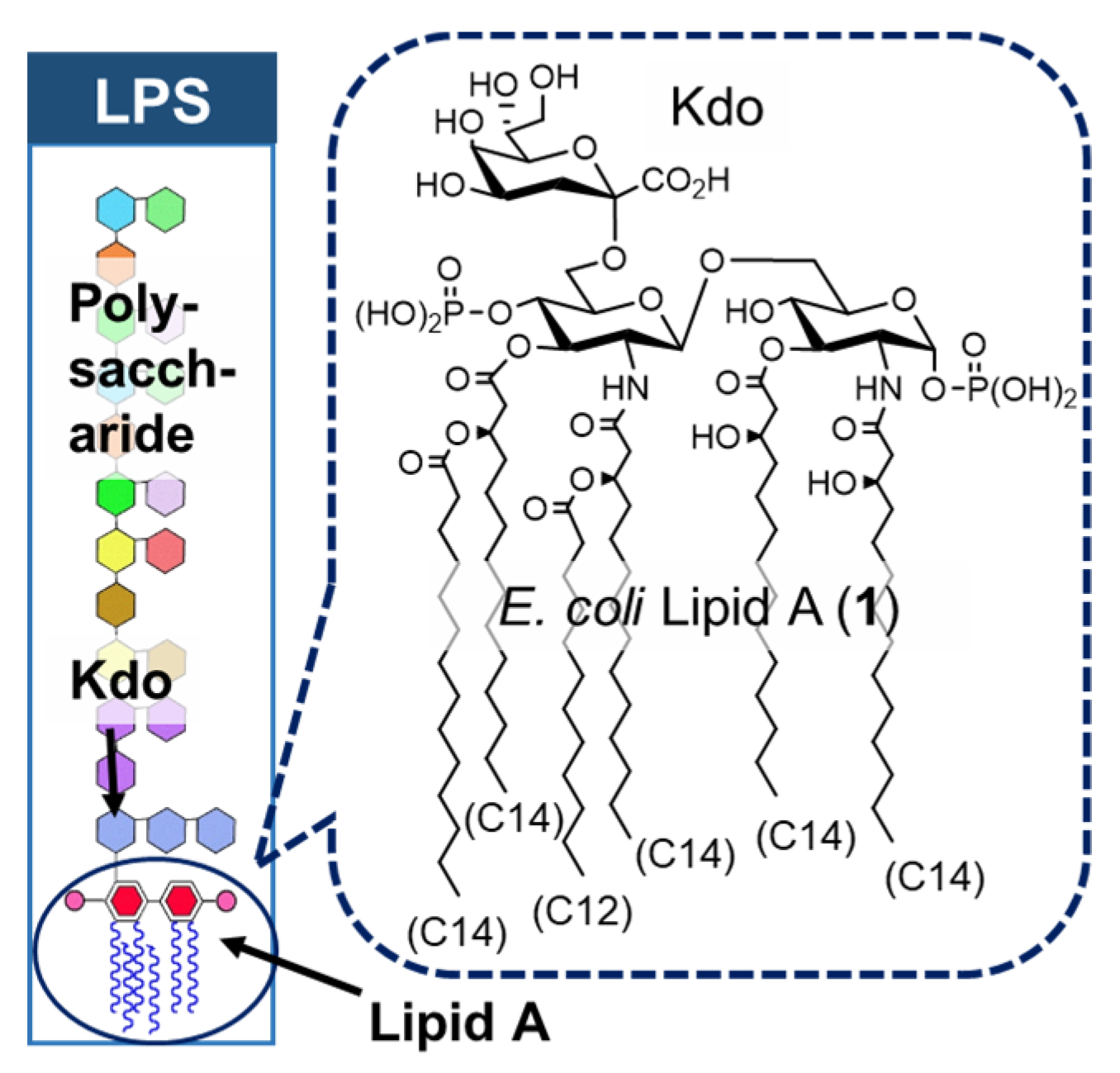
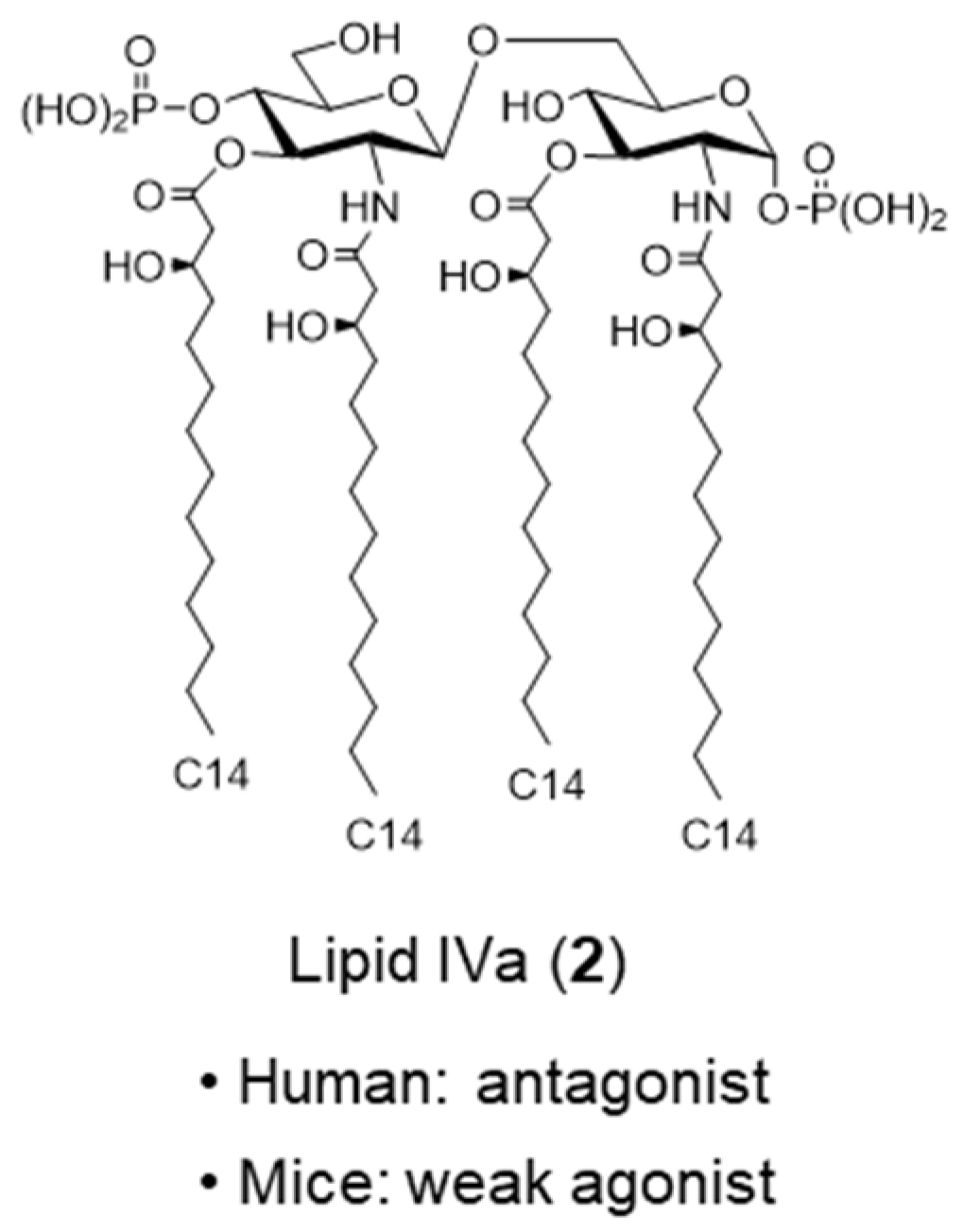
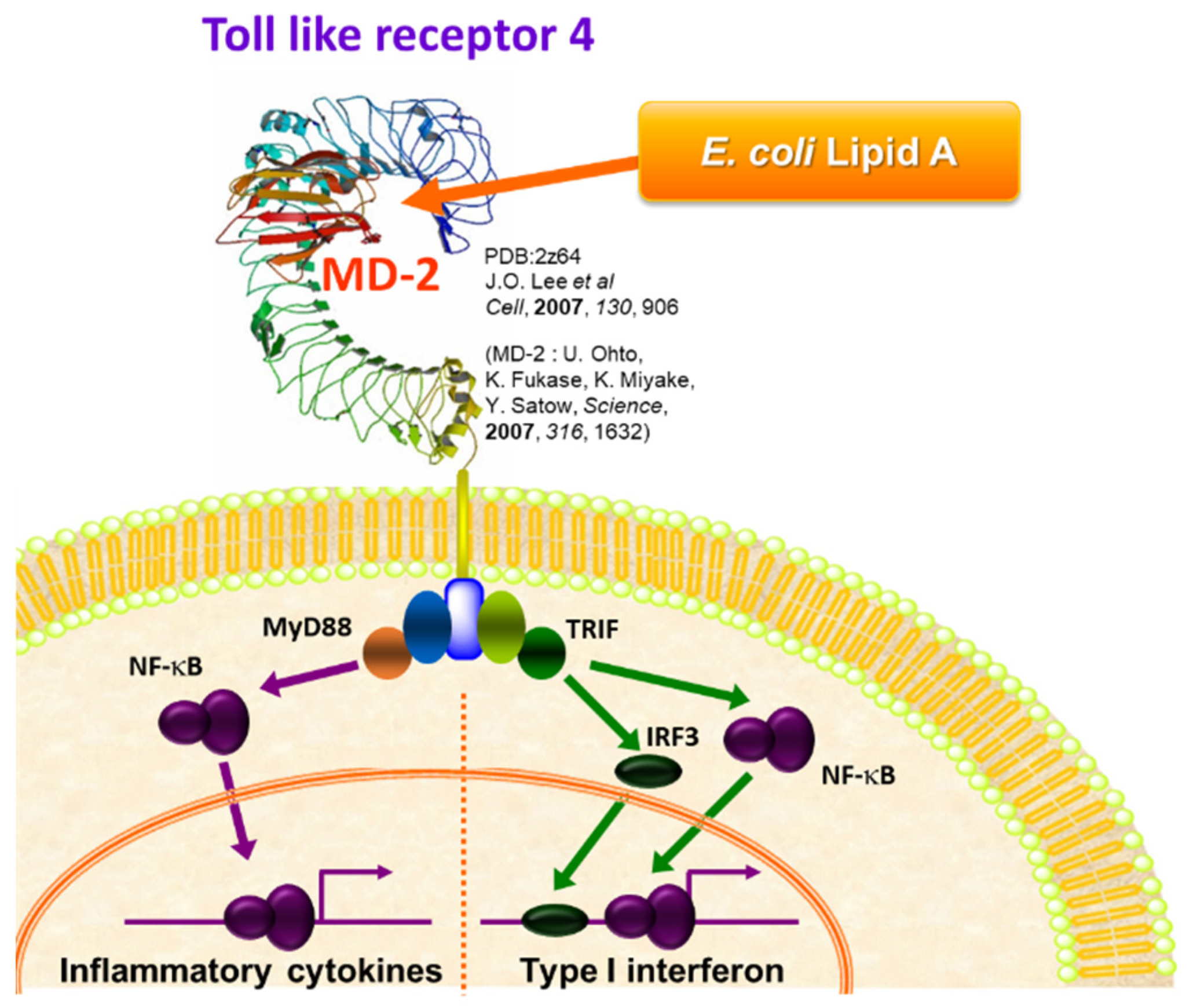
2. Bacterial LPS and Its Active Principle Lipid A, an Innate Immune Stimulator
3. Synthetic and Semi-Synthetic Lipid A Adjuvants in Practical Use
4. Lipid A Adjuvants Development Based on Bacterial–Host Chemical Ecology
4.1. Parasitic Bacterial Lipid A
4.2. Symbiotic Bacterial Lipid A
5. Lipid A in the Environment and Fermented Foods, Their Potential as Adjuvants
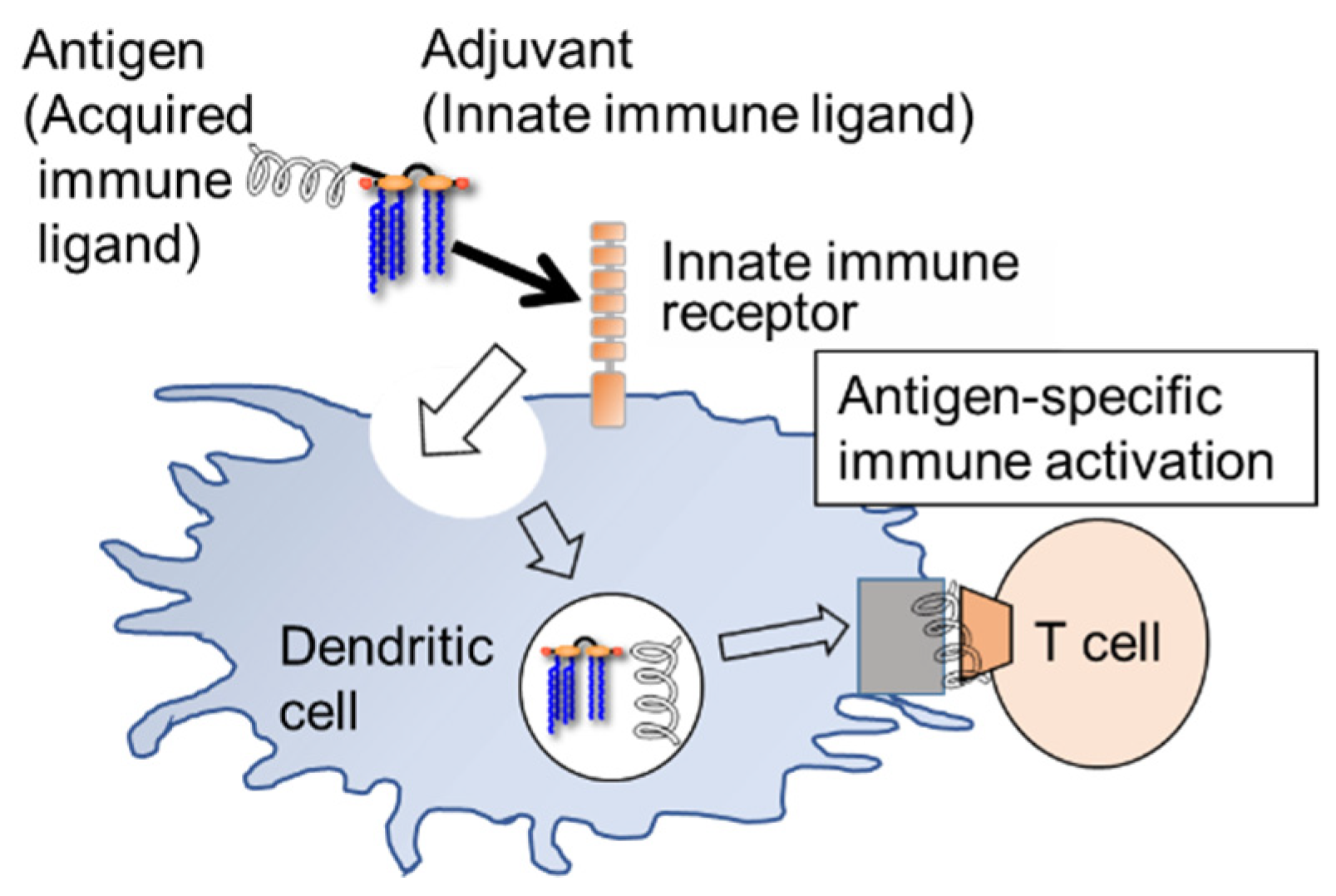
6. Self-Adjuvating Vaccines Based on Lipid A
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Kusumoto, S.; Fukase, K.; Shiba, T. Key structures of bacterial peptidoglycan and lipopolysaccharide triggering the innate immune system of higher animals: Chemical synthesis and functional studies. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010, 86, 322–337. [Google Scholar] [CrossRef]
- Wei, M.Q.; Mengesha, A.; Good, D.; Anne, J. Bacterial targeted tumour therapy-dawn of a new era. Cancer Lett. 2008, 259, 16–27. [Google Scholar] [CrossRef]
- Coley, W.B. The treatment of malignant tumors by repeated inoculations of erysipelas. With a report of ten original cases. 1893. Clin. Orthop. Relat. Res. 1991, 262, 3–11. [Google Scholar]
- Shimoyama, A.; Saeki, A.; Tanimura, N.; Tsutsui, H.; Miyake, K.; Suda, Y.; Fujimoto, Y.; Fukase, K. Chemical synthesis of Helicobacter pylori lipopolysaccharide partial structures and their selective proinflammatory responses. Chemistry 2011, 17, 14464–14474. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, Y.; Shimoyama, A.; Saeki, A.; Kitayama, N.; Kasamatsu, C.; Tsutsui, H.; Fukase, K. Innate immunomodulation by lipophilic termini of lipopolysaccharide; synthesis of lipid As from Porphyromonas gingivalis and other bacteria and their immunomodulative responses. Mol. Biosyst. 2013, 9, 9876–9896. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, Y.; Shimoyama, A.; Suda, Y.; Fukase, K. Synthesis and immunomodulatory activities of Helicobacter pylori lipophilic terminus of lipopolysaccharide including lipid A. Carbohydr. Res. 2012, 356, 37–43. [Google Scholar] [CrossRef] [PubMed]
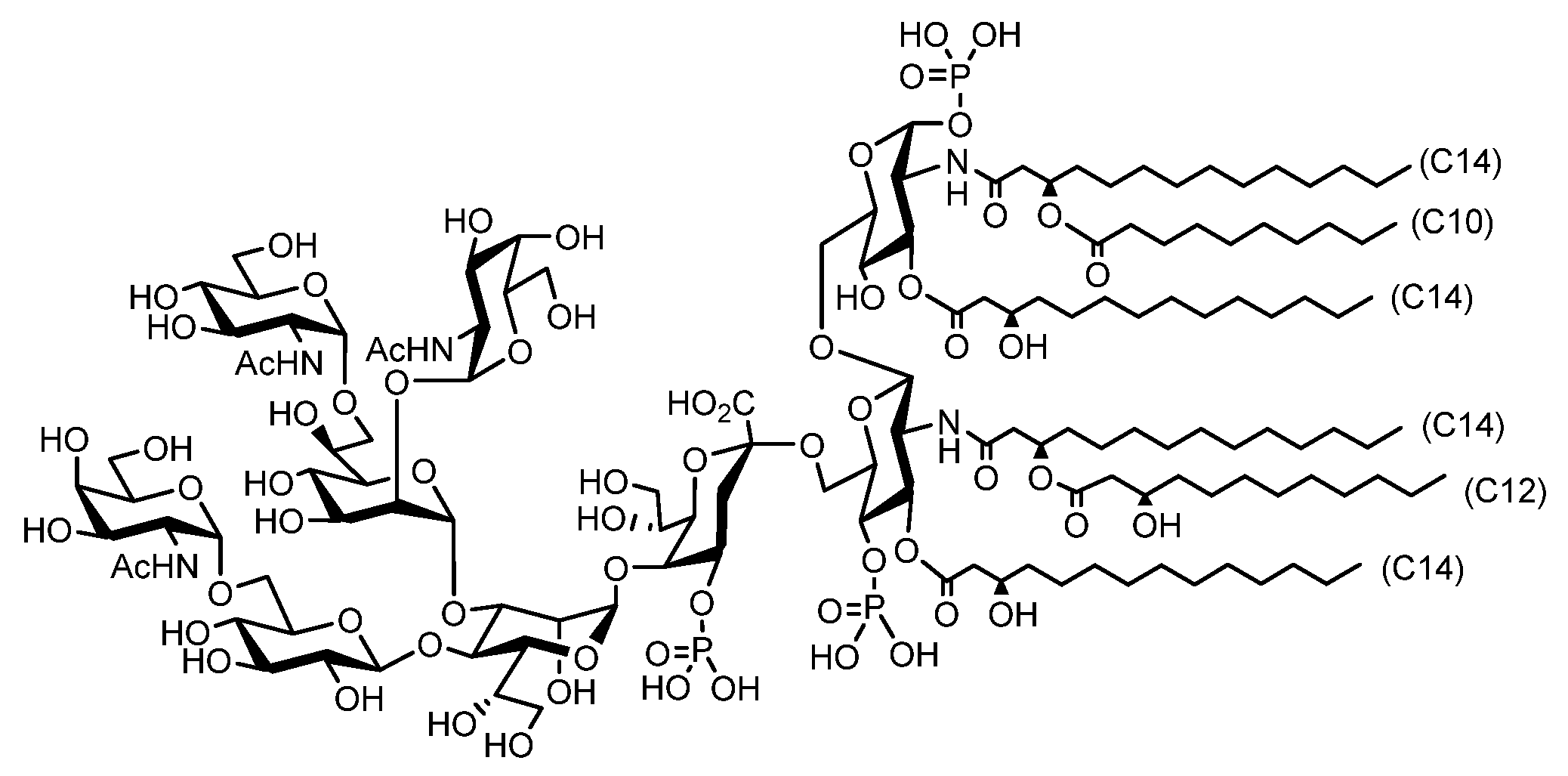
- Shimoyama, A.; Di Lorenzo, F.; Yamaura, H.; Mizote, K.; Palmigiano, A.; Pither, M.D.; Speciale, I.; Uto, T.; Masui, S.; Sturiale, L.; et al. Lipopolysaccharide from Gut-Associated Lymphoid Tissue-Resident Alcaligenes faecalis: Complete Structure Determination and Chemical Synthesis of its Lipid As. Angew. Chem. Int. Ed. Engl. 2021, 60, 10023–10031. [Google Scholar] [CrossRef]
- Rietschel, E.T.; Endotoxin, O.W. Historical Perspectives. In Endotoxin in Health and Disease; Helmut Brade, H., Ed.; CRC Press: Boca Raton, FA, USA, 1999; pp. 16–30. [Google Scholar]
- Imoto, M.; Kusumoto, S.; Shiba, T.; Naoki, H.; Iwashita, T.; Rietschel, E.T.; Wollenweber, H.W.; Galanos, C.; Luderitz, O. Chemical-Structure of Escherichia-Coli Lipid-A—Linkage Site of Acyl-Groups in the Disaccharide Backbone. Tetrahedron Lett. 1983, 24, 4017–4020. [Google Scholar] [CrossRef]
- Imoto, M.; Kusumoto, S.; Shiba, T.; Rietschel, E.T.; Galanos, C.; Luederitz, O. Chemical structure of Escherichia coli lipid A. Tetrahedron Lett. 1985, 26, 907–908. [Google Scholar] [CrossRef]
- Imoto, M.; Yoshimura, H.; Shimamoto, T.; Sakaguchi, N.; Kusumoto, S.; Shiba, T. Total synthesis of Escherichia coli lipid A, the endotoxically active principle of cell-surface lipopolysaccharide. Bull. Chem. Soc. Jpn. 1987, 60, 2205–2214. [Google Scholar] [CrossRef]
- Takayama, K.; Qureshi, N.; Mascagni, P. Complete structure of lipid A obtained from the lipopolysaccharides of the heptoseless mutant of Salmonella typhimurium. J. Biol. Chem. 1983, 258, 12801–12803. [Google Scholar] [CrossRef]
- Flad, H.D.; Loppnow, H.; Feist, W.; Wang, M.H.; Brade, H.; Kusumoto, S.; Rietschel, E.T.; Ulmer, A.J. Interleukin 1 and tumor necrosis factor: Studies on the induction by lipopolysaccharide partial structures. Lymphokine Res. 1989, 8, 235–238. [Google Scholar]
- Wang, M.H.; Feist, W.; Herzbeck, H.; Brade, H.; Kusumoto, S.; Rietschel, E.T.; Flad, H.D.; Ulmer, A.J. Suppressive effect of lipid A partial structures on lipopolysaccharide or lipid A-induced release of interleukin 1 by human monocytes. FEMS Microbiol. Immunol. 1990, 2, 179–185. [Google Scholar]
- Lemaitre, B.; Nicolas, E.; Michaut, L.; Reichhart, J.M.; Hoffmann, J.A. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 1996, 86, 973–983. [Google Scholar] [CrossRef]
- Medzhitov, R.; Preston-Hurlburt, P.; Janeway, C.A., Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 1997, 388, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Poltorak, A.; He, X.; Smirnova, I.; Liu, M.Y.; Van Huffel, C.; Du, X.; Birdwell, D.; Alejos, E.; Silva, M.; Galanos, C.; et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science 1998, 282, 2085–2088. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Shimazu, R.; Akashi, S.; Ogata, H.; Nagai, Y.; Fukudome, K.; Miyake, K.; Kimoto, M. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J. Exp. Med. 1999, 189, 1777–1782. [Google Scholar] [CrossRef]
- Akashi, S.; Saitoh, S.; Wakabayashi, Y.; Kikuchi, T.; Takamura, N.; Nagai, Y.; Kusumoto, Y.; Fukase, K.; Kusumoto, S.; Adachi, Y.; et al. Lipopolysaccharide interaction with cell surface Toll-like receptor 4-MD-2: Higher affinity than that with MD-2 or CD14. J. Exp. Med. 2003, 198, 1035–1042. [Google Scholar] [CrossRef]
- Akashi, S.; Nagai, Y.; Ogata, H.; Oikawa, M.; Fukase, K.; Kusumoto, S.; Kawasaki, K.; Nishijima, M.; Hayashi, S.; Kimoto, M.; et al. Human MD-2 confers on mouse Toll-like receptor 4 species-specific lipopolysaccharide recognition. Int. Immunol. 2001, 13, 1595–1599. [Google Scholar] [CrossRef]
- Ohto, U.; Fukase, K.; Miyake, K.; Satow, Y. Crystal structures of human MD-2 and its complex with antiendotoxic lipid IVa. Science 2007, 316, 1632–1634. [Google Scholar] [CrossRef]
- Kim, H.M.; Park, B.S.; Kim, J.I.; Kim, S.E.; Lee, J.; Oh, S.C.; Enkhbayar, P.; Matsushima, N.; Lee, H.; Yoo, O.J.; et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell 2007, 130, 906–917. [Google Scholar] [CrossRef]
- Park, B.S.; Song, D.H.; Kim, H.M.; Choi, B.S.; Lee, H.; Lee, J.O. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 2009, 458, 1191–1195. [Google Scholar] [CrossRef] [PubMed]
- Ohto, U.; Fukase, K.; Miyake, K.; Shimizu, T. Structural basis of species-specific endotoxin sensing by innate immune receptor TLR4/MD-2. Proc. Natl. Acad. Sci. USA 2012, 109, 7421–7426. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.; Holst, O.; Di Lorenzo, F.; Callaghan, M.; Nurisso, A.; D’Errico, G.; Zamyatina, A.; Peri, F.; Berisio, R.; Jerala, R.; et al. Chemistry of lipid A: At the heart of innate immunity. Chemistry 2015, 21, 500–519. [Google Scholar] [CrossRef]
- Fukase, K.F.; Fujimoto, Y.; Shimoyama, A.; Tanaka, K. Synthesis of Bacterial Glycoconjugates and Their Bio-functional Studies in Innate Immunity. J. Syn. Org. Chem. JPN 2012, 70, 113–130. [Google Scholar] [CrossRef]
- Kusumoto, S.; Fukase, K. Synthesis of endotoxic principle of bacterial lipopolysaccharide and its recognition by innate immune system of hosts. Chem. Rec. 2006, 6, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Brade, L.; Brandenburg, K.; Kuhn, H.M.; Kusumoto, S.; Macher, I.; Rietschel, E.T.; Brade, H. The immunogenicity and antigenicity of lipid A are influenced by its physicochemical state and environment. Infect. Immun. 1987, 55, 2636–2644. [Google Scholar] [CrossRef] [PubMed]
- Tanimura, N.; Saitoh, S.; Ohto, U.; Akashi-Takamura, S.; Fujimoto, Y.; Fukase, K.; Shimizu, T.; Miyake, K. The attenuated inflammation of MPL is due to the lack of CD14-dependent tight dimerization of the TLR4/MD2 complex at the plasma membrane. Int. Immunol. 2014, 26, 307–314. [Google Scholar] [CrossRef]
- Yoshizaki, H.; Fukuda, N.; Sato, K.; Oikawa, M.; Fukase, K.; Suda, Y.; Kusumoto, S. First Total Synthesis of the Re-Type Lipopolysaccharide This work was supported by the Research for the Future Program (No. 97L00502) from the Japan Society for the Promotion of Science. H.Y. is grateful for a JSPS Research Fellowship for Young Scientists (No. 1241) from the Japan Society for the Promotion of Science. The authors are grateful to Mr. Seiji Adachi for his skillful measurement of NMR spectra. Angew. Chem. Int. Ed. Engl. 2001, 40, 1475–1480. [Google Scholar]
- Mata-Haro, V.; Cekic, C.; Martin, M.; Chilton, P.M.; Casella, C.R.; Mitchell, T.C. The vaccine adjuvant monophosphoryl lipid A as a TRIF-biased agonist of TLR4. Science 2007, 316, 1628–1632. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.; Michalek, S.M.; Katz, J. Role of innate immune factors in the adjuvant activity of monophosphoryl lipid A. Infect. Immun. 2003, 71, 2498–24507. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, K.; Tsukano, H.; Watanabe, H.; Lindner, B.; Matsuura, M. Modification of the structure and activity of lipid A in Yersinia pestis lipopolysaccharide by growth temperature. Infect. Immun. 2002, 70, 4092–4098. [Google Scholar] [CrossRef] [PubMed]
- Knirel, Y.A.; Lindner, B.; Vinogradov, E.V.; Kocharova, N.A.; Senchenkova, S.N.; Shaikhutdinova, R.Z.; Dentovskaya, S.V.; Fursova, N.K.; Bakhteeva, I.V.; Titareva, G.M.; et al. Temperature-dependent variations and intraspecies diversity of the structure of the lipopolysaccharide of Yersinia pestis. Biochemistry 2005, 44, 1731–1743. [Google Scholar] [CrossRef]
- Hynes, S.O.; Ferris, J.A.; Szponar, B.; Wadstrom, T.; Fox, J.G.; O’Rourke, J.; Larsson, L.; Yaquian, E.; Ljungh, A.; Clyne, M.; et al. Comparative chemical and biological characterization of the lipopolysaccharides of gastric and enterohepatic helicobacters. Helicobacter 2004, 9, 313–323. [Google Scholar] [CrossRef]
- Nielsen, H.; Birkholz, S.; Andersen, L.P.; Moran, A.P. Neutrophil activation by Helicobacter pylori lipopolysaccharides. J. Infect. Dis. 1994, 170, 135–139. [Google Scholar] [CrossRef]
- Perez-Perez, G.I.; Shepherd, V.L.; Morrow, J.D.; Blaser, M.J. Activation of human THP-1 cells and rat bone marrow-derived macrophages by Helicobacter pylori lipopolysaccharide. Infect. Immun. 1995, 63, 1183–1187. [Google Scholar] [CrossRef]
- Danesh, J.; Wong, Y.; Ward, M.; Muir, J. Chronic infection with Helicobacter pylori, Chlamydia pneumoniae, or cytomegalovirus: Population based study of coronary heart disease. Heart 1999, 81, 245–247. [Google Scholar] [CrossRef]
- Triantafilou, M.; Gamper, F.G.; Lepper, P.M.; Mouratis, M.A.; Schumann, C.; Harokopakis, E.; Schifferle, R.E.; Hajishengallis, G.; Triantafilou, K. Lipopolysaccharides from atherosclerosis-associated bacteria antagonize TLR4, induce formation of TLR2/1/CD36 complexes in lipid rafts and trigger TLR2-induced inflammatory responses in human vascular endothelial cells. Cell Microbiol. 2007, 9, 2030–2039. [Google Scholar] [CrossRef]
- Imamura, M.; Tsutsui, H.; Yasuda, K.; Uchiyama, R.; Yumikura-Futatsugi, S.; Mitani, K.; Hayashi, S.; Akira, S.; Taniguchi, S.; Van Rooijen, N.; et al. Contribution of TIR domain-containing adapter inducing IFN-beta-mediated IL-18 release to LPS-induced liver injury in mice. J. Hepatol. 2009, 51, 333–341. [Google Scholar] [CrossRef]
- Kanneganti, T.D.; Lamkanfi, M.; Kim, Y.G.; Chen, G.; Park, J.H.; Franchi, L.; Vandenabeele, P.; Nunez, G. Pannexin-1-mediated recognition of bacterial molecules activates the cryopyrin inflammasome independent of Toll-like receptor signaling. Immunity 2007, 26, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhao, Y.; Wang, Y.; Gao, W.; Ding, J.; Li, P.; Hu, L.; Shao, F. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 2014, 514, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Vatanen, T.; Kostic, A.D.; d’Hennezel, E.; Siljander, H.; Franzosa, E.A.; Yassour, M.; Kolde, R.; Vlamakis, H.; Arthur, T.D.; Hamalainen, A.M.; et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 2016, 165, 842–853. [Google Scholar] [CrossRef]
- d’Hennezel, E.; Abubucker, S.; Murphy, L.O.; Cullen, T.W. Total Lipopolysaccharide from the Human Gut Microbiome Silences Toll-Like Receptor Signaling. mSystems 2017, 2, e00046_17. [Google Scholar] [CrossRef] [PubMed]
- Naito, T.; Mulet, C.; De Castro, C.; Molinaro, A.; Saffarian, A.; Nigro, G.; Berard, M.; Clerc, M.; Pedersen, A.B.; Sansonetti, P.J.; et al. Lipopolysaccharide from Crypt-Specific Core Microbiota Modulates the Colonic Epithelial Proliferation-to-Differentiation Balance. mBio 2017, 8, e01680-17. [Google Scholar] [CrossRef]
- Erturk-Hasdemir, D.; Oh, S.F.; Okan, N.A.; Stefanetti, G.; Gazzaniga, F.S.; Seeberger, P.H.; Plevy, S.E.; Kasper, D.L. Symbionts exploit complex signaling to educate the immune system. Proc. Natl. Acad. Sci. USA 2019, 116, 26157–26166. [Google Scholar] [CrossRef]
- Steimle, A.; Michaelis, L.; Di Lorenzo, F.; Kliem, T.; Munzner, T.; Maerz, J.K.; Schafer, A.; Lange, A.; Parusel, R.; Gronbach, K.; et al. Weak Agonistic LPS Restores Intestinal Immune Homeostasis. Mol. Ther. 2019, 27, 1974–1991. [Google Scholar] [CrossRef]
- Di Lorenzo, F.; De Castro, C.; Silipo, A.; Molinaro, A. Lipopolysaccharide structures of Gram-negative populations in the gut microbiota and effects on host interactions. Fems Microbiol. Rev. 2019, 43, 257–272. [Google Scholar] [CrossRef]
- Di Lorenzo, F.; Pither, M.D.; Martufi, M.; Scarinci, I.; Guzman-Caldentey, J.; Lakomiec, E.; Jachymek, W.; Bruijns, S.C.M.; Santamaria, S.M.; Frick, J.S.; et al. Pairing Bacteroides vulgatus LPS Structure with Its Immunomodulatory Effects on Human Cellular Models. ACS Cent. Sci. 2020, 6, 1602–1616. [Google Scholar] [CrossRef]
- Zhu, Q.; Shen, Z.; Chiodo, F.; Nicolardi, S.; Molinaro, A.; Silipo, A.; Yu, B. Chemical synthesis of glycans up to a 128-mer relevant to the O-antigen of Bacteroides vulgatus. Nat. Commun. 2020, 11, 4142. [Google Scholar] [CrossRef]
- Obata, T.; Goto, Y.; Kunisawa, J.; Sato, S.; Sakamoto, M.; Setoyama, H.; Matsuki, T.; Nonaka, K.; Shibata, N.; Gohda, M.; et al. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. Proc. Natl. Acad. Sci. USA 2010, 107, 7419–7424. [Google Scholar] [CrossRef]
- Kunisawa, J.; Kiyono, H. Alcaligenes is Commensal Bacteria Habituating in the Gut-Associated Lymphoid Tissue for the Regulation of Intestinal IgA Responses. Front. Immunol. 2012, 3, 65. [Google Scholar] [CrossRef] [PubMed]
- Kunisawa, J.; Nochi, T.; Kiyono, H. Immunological commonalities and distinctions between airway and digestive immunity. Trends Immunol. 2008, 29, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Fagarasan, S.; Kawamoto, S.; Kanagawa, O.; Suzuki, K. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu. Rev. Immunol. 2010, 28, 243–273. [Google Scholar] [CrossRef]
- Fung, T.C.; Bessman, N.J.; Hepworth, M.R.; Kumar, N.; Shibata, N.; Kobuley, D.; Wang, K.; Ziegler, C.G.K.; Goc, J.; Shima, T.; et al. Lymphoid-Tissue-Resident Commensal Bacteria Promote Members of the IL-10 Cytokine Family to Establish Mutualism. Immunity 2016, 44, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Sonnenberg, G.F.; Monticelli, L.A.; Alenghat, T.; Fung, T.C.; Hutnick, N.A.; Kunisawa, J.; Shibata, N.; Grunberg, S.; Sinha, R.; Zahm, A.M.; et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science 2012, 336, 1321–1325. [Google Scholar] [CrossRef]
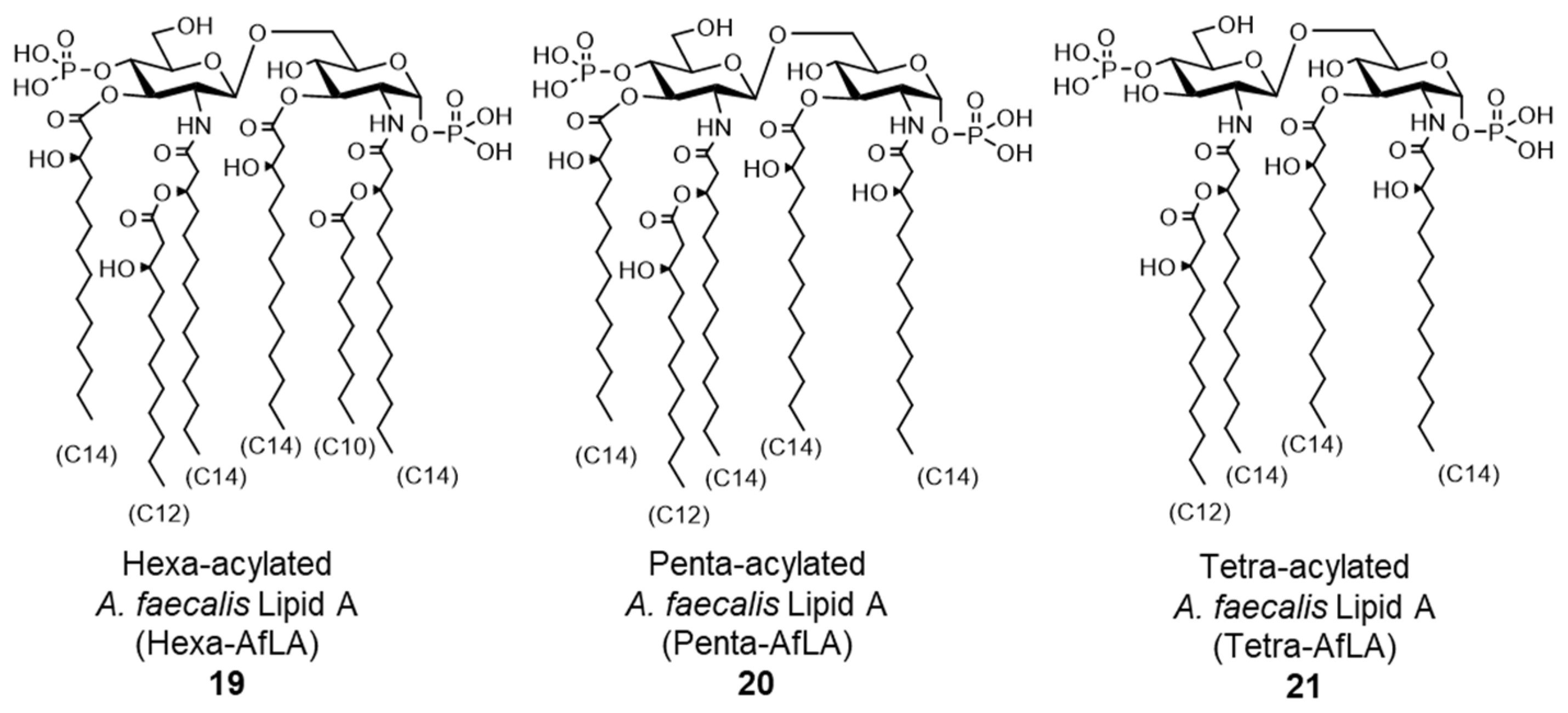
- Shibata, N.; Kunisawa, J.; Hosomi, K.; Fujimoto, Y.; Mizote, K.; Kitayama, N.; Shimoyama, A.; Mimuro, H.; Sato, S.; Kishishita, N.; et al. Lymphoid tissue-resident Alcaligenes LPS induces IgA production without excessive inflammatory responses via weak TLR4 agonist activity. Mucosal Immunol. 2018, 11, 693–702. [Google Scholar] [CrossRef]
- Hosomi, K.; Shibata, N.; Shimoyama, A.; Uto, T.; Nagatake, T.; Tojima, Y.; Nishino, T.; Takeyama, H.; Fukase, K.; Kiyono, H.; et al. Lymphoid Tissue-Resident Alcaligenes Establish an Intracellular Symbiotic Environment by Creating a Unique Energy Shift in Dendritic Cells. Front. Microbiol. 2020, 11, 561005. [Google Scholar] [CrossRef]
- Hexa-Acylated A. faecalis Lipid A (Hexa-AfLA). Available online: https://www.peptide.co.jp/catalog/f-cat?k_code=24018-s (accessed on 13 September 2021).
- Yoshii, K.; Hosomi, K.; Shimoyama, A.; Wang, Y.; Yamaura, H.; Nagatake, T.; Suzuki, H.; Lan, H.; Kiyono, H.; Fukase, K.; et al. Chemically Synthesized Alcaligenes Lipid A Shows a Potent and Safe Nasal Vaccine Adjuvant Activity for the Induction of Streptococcus pneumoniae-Specific IgA and Th17 Mediated Protective Immunity. Microorganisms 2020, 8, 1102. [Google Scholar] [CrossRef]
- Wang, Y.; Hosomi, K.; Shimoyama, A.; Yoshii, K.; Yamaura, H.; Nagatake, T.; Nishino, T.; Kiyono, H.; Fukase, K.; Kunisawa, J. Adjuvant Activity of Synthetic Lipid A of Alcaligenes, a Gut-Associated Lymphoid Tissue-Resident Commensal Bacterium, to Augment Antigen-Specific IgG and Th17 Responses in Systemic Vaccine. Vaccines (Basel) 2020, 8, 395. [Google Scholar] [CrossRef]
- Kariluoto, S.; Aittamaa, M.; Korhola, M.; Salovaara, H.; Vahteristo, L.; Piironen, V. Effects of yeasts and bacteria on the levels of folates in rye sourdoughs. Int. J. Food. Microbiol. 2006, 106, 137–143. [Google Scholar] [CrossRef]
- Dutkiewicz, J.; Mackiewicz, B.; Lemieszek, M.K.; Golec, M.; Milanowski, J. Pantoea agglomerans: A mysterious bacterium of evil and good. Part IV. Beneficial effects. Ann. Agric. Envrion. Med. 2016, 23, 206–222. [Google Scholar] [CrossRef] [PubMed]
- Hebishima, T.; Matsumoto, Y.; Watanabe, G.; Soma, G.; Kohchi, C.; Taya, K.; Hayashi, Y.; Hirota, Y. Oral administration of immunopotentiator from Pantoea agglomerans 1 (IP-PA1) improves the survival of B16 melanoma-inoculated model mice. Exp. Anim. 2011, 60, 101–109. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tsukioka, D.; Nishizawa, T.; Miyase, T.; Achiwa, K.; Suda, T.; Soma, G.; Mizuno, D. Structural characterization of lipid A obtained from Pantoea agglomerans lipopolysaccharide. FEMS Microbiol. Lett. 1997, 149, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Pallach, M.; Di Lorenzo, F.; Facchini, F.A.; Gully, D.; Giraud, E.; Peri, F.; Duda, K.A.; Molinaro, A.; Silipo, A. Structure and inflammatory activity of the LPS isolated from Acetobacter pasteurianus CIP103108. Int. J. Biol. Macromol. 2018, 119, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Ozono, M.; Furuyashiki, M.; Baba, R.; Hashiguchi, S.; Suda, Y.; Fukase, K.; Fujimoto, Y. Characterization of a Novel d-Glycero-d-talo-oct-2-ulosonic acid-substituted Lipid A Moiety in the Lipopolysaccharide Produced by the Acetic Acid Bacterium Acetobacter pasteurianus NBRC 3283. J. Biol. Chem. 2016, 291, 21184–21194. [Google Scholar] [CrossRef]
- Debarry, J.; Hanuszkiewicz, A.; Stein, K.; Holst, O.; Heine, H. The allergy-protective properties of Acinetobacter lwoffii F78 are imparted by its lipopolysaccharide. Allergy 2010, 65, 690–697. [Google Scholar] [CrossRef]
- Ingale, S.; Wolfert, M.A.; Gaekwad, J.; Buskas, T.; Boons, G.-J. Robust immune responses elicited by a fully synthetic three-component vaccine. Nat. Chem. Biol. 2007, 3, 663–667. [Google Scholar] [CrossRef]
- Khan, S.; Weterings, J.J.; Britten, C.M.; de Jong, A.R.; Graafland, D.; Melief, C.J.M.; van der Burg, S.H.; van der Marel, G.; Overkleeft, H.S.; Filippov, D.V.; et al. Chirality of TLR-2 ligand Pam3CysSK4 in fully synthetic peptide conjugates critically influences the induction of specific CD8+ T-cells. Mol. Immunol. 2009, 46, 1084–1091. [Google Scholar] [CrossRef]
- Kaiser, A.; Gaidzik, N.; Becker, T.; Menge, C.; Groh, K.; Cai, H.; Li, Y.-M.; Gerlitzki, B.; Schmitt, E.; Kunz, H. Fully Synthetic Vaccines Consisting of Tumor-Associated MUC1 Glycopeptides and a Lipopeptide Ligand of the Toll-like Receptor 2. Angew. Chem. Int. Ed. 2010, 49, 3688–3692. [Google Scholar] [CrossRef]
- Wilkinson, B.L.; Day, S.; Malins, L.R.; Apostolopoulos, V.; Payne, R.J. Self-Adjuvanting Multicomponent Cancer Vaccine Candidates Combining Per-Glycosylated MUC1 Glycopeptides and the Toll-like Receptor 2 Agonist Pam3CysSer. Angew. Chem. Int. Ed. 2011, 50, 1635–1639. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, B.L.; Day, S.; Chapman, R.; Perrier, S.; Apostolopoulos, V.; Payne, R.J. Synthesis and Immunological Evaluation of Self-Assembling and Self-Adjuvanting Tricomponent Glycopeptide Cancer-Vaccine Candidates. Chem.—A Eur. J. 2012, 18, 16540–16548. [Google Scholar] [CrossRef]
- Lakshminarayanan, V.; Thompson, P.; Wolfert, M.A.; Buskas, T.; Bradley, J.M.; Pathangey, L.B.; Madsen, C.S.; Cohen, P.A.; Gendler, S.J.; Boons, G.J. Immune recognition of tumor-associated mucin MUC1 is achieved by a fully synthetic aberrantly glycosylated MUC1 tripartite vaccine. Proc. Natl. Acad. Sci. USA 2012, 109, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Chen, M.-S.; Sun, Z.-Y.; Zhao, Y.-F.; Kunz, H.; Li, Y.-M. Self-Adjuvanting Synthetic Antitumor Vaccines from MUC1 Glycopeptides Conjugated to T-Cell Epitopes from Tetanus Toxoid. Angew. Chem. Int. Ed. 2013, 52, 6106–6110. [Google Scholar] [CrossRef]
- Palitzsch, B.; Hartmann, S.; Stergiou, N.; Glaffig, M.; Schmitt, E.; Kunz, H. A Fully Synthetic Four-Component Antitumor Vaccine Consisting of a Mucin Glycopeptide Antigen Combined with Three Different T-Helper-Cell Epitopes. Angew. Chem. Int. Ed. 2014, 53, 14245–14249. [Google Scholar] [CrossRef]
- Thompson, P.; Lakshminarayanan, V.; Supekar, N.T.; Bradley, J.M.; Cohen, P.A.; Wolfert, M.A.; Gendler, S.J.; Boons, G.-J. Linear synthesis and immunological properties of a fully synthetic vaccine candidate containing a sialylated MUC1 glycopeptide. Chem. Commun. 2015, 51, 10214–10217. [Google Scholar] [CrossRef]
- Ingale, S.; Wolfert, M.A.; Buskas, T.; Boons, G.J. Increasing the antigenicity of synthetic tumor-associated carbohydrate antigens by targeting Toll-like receptors. Chembiochem 2009, 10, 455–463. [Google Scholar] [CrossRef]
- Aiga, T.; Manabe, Y.; Ito, K.; Chang, T.C.; Kabayama, K.; Ohshima, S.; Kametani, Y.; Miura, A.; Furukawa, H.; Inaba, H.; et al. Immunological Evaluation of Co-Assembling a Lipidated Peptide Antigen and Lipophilic Adjuvants: Self-Adjuvanting Anti-Breast-Cancer Vaccine Candidates. Angew. Chem. Int. Ed. Engl. 2020, 59, 17705–17711. [Google Scholar] [CrossRef]
- Skwarczynski, M.; Zhao, G.; Boer, J.C.; Ozberk, V.; Azuar, A.; Cruz, J.G.; Giddam, A.K.; Khalil, Z.G.; Pandey, M.; Shibu, M.A.; et al. Poly(amino acids) as a potent self-adjuvanting delivery system for peptide-based nanovaccines. Sci. Adv. 2020, 6, eaax2285. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhou, Z.; Tang, S.; Guo, Z. Carbohydrate-monophosphoryl lipid a conjugates are fully synthetic self-adjuvanting cancer vaccines eliciting robust immune responses in the mouse. ACS Chem. Biol. 2012, 7, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.; Zhou, Z.; Suryawanshi, S.; Mondal, M.A.; Guo, Z. Fully Synthetic Self-Adjuvanting alpha-2,9-Oligosialic Acid Based Conjugate Vaccines against Group C Meningitis. ACS Cent. Sci. 2016, 2, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Lewicky, J.D.; Ulanova, M.; Jiang, Z.H. Synthesis of a TLR4 Agonist-Carbohydrate Antigen Conjugate As A Self-Adjuvanting Cancer Vaccine. ChemistrySelect 2016, 5, 906–910. [Google Scholar] [CrossRef]
- Reintjens, N.R.M.; Tondini, E.; de Jong, A.R.; Meeuwenoord, N.J.; Chiodo, F.; Peterse, E.; Overkleeft, H.S.; Filippov, D.V.; van der Marel, G.A.; Ossendorp, F.; et al. Self-Adjuvanting Cancer Vaccines from Conjugation-Ready Lipid A Analogues and Synthetic Long Peptides. J. Med. Chem. 2020, 63, 11691–11706. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shimoyama, A.; Fukase, K. Lipid A-Mediated Bacterial–Host Chemical Ecology: Synthetic Research of Bacterial Lipid As and Their Development as Adjuvants. Molecules 2021, 26, 6294. https://doi.org/10.3390/molecules26206294
Shimoyama A, Fukase K. Lipid A-Mediated Bacterial–Host Chemical Ecology: Synthetic Research of Bacterial Lipid As and Their Development as Adjuvants. Molecules. 2021; 26(20):6294. https://doi.org/10.3390/molecules26206294
Chicago/Turabian StyleShimoyama, Atsushi, and Koichi Fukase. 2021. "Lipid A-Mediated Bacterial–Host Chemical Ecology: Synthetic Research of Bacterial Lipid As and Their Development as Adjuvants" Molecules 26, no. 20: 6294. https://doi.org/10.3390/molecules26206294
APA StyleShimoyama, A., & Fukase, K. (2021). Lipid A-Mediated Bacterial–Host Chemical Ecology: Synthetic Research of Bacterial Lipid As and Their Development as Adjuvants. Molecules, 26(20), 6294. https://doi.org/10.3390/molecules26206294





