Cannabis-Derived Compounds Cannabichromene and Δ9-Tetrahydrocannabinol Interact and Exhibit Cytotoxic Activity against Urothelial Cell Carcinoma Correlated with Inhibition of Cell Migration and Cytoskeleton Organization
Abstract
1. Introduction
2. Results
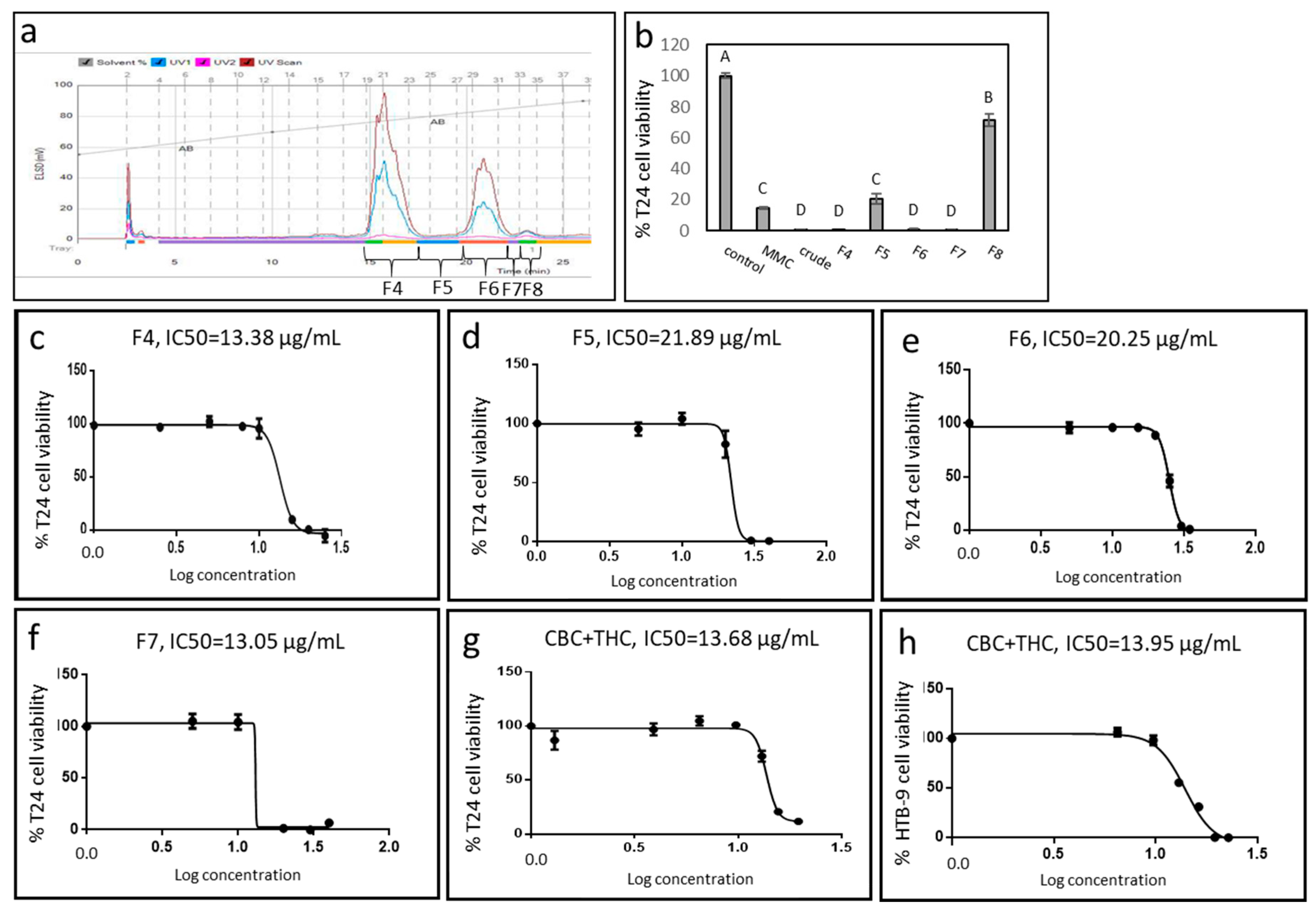
2.1. Cytotoxic Activity of IGB Cannabis Strain Extracts and Fractions Against Urothelial Cell Carcinoma
2.2. Active Fraction Chemical Composition and Determination of Activity
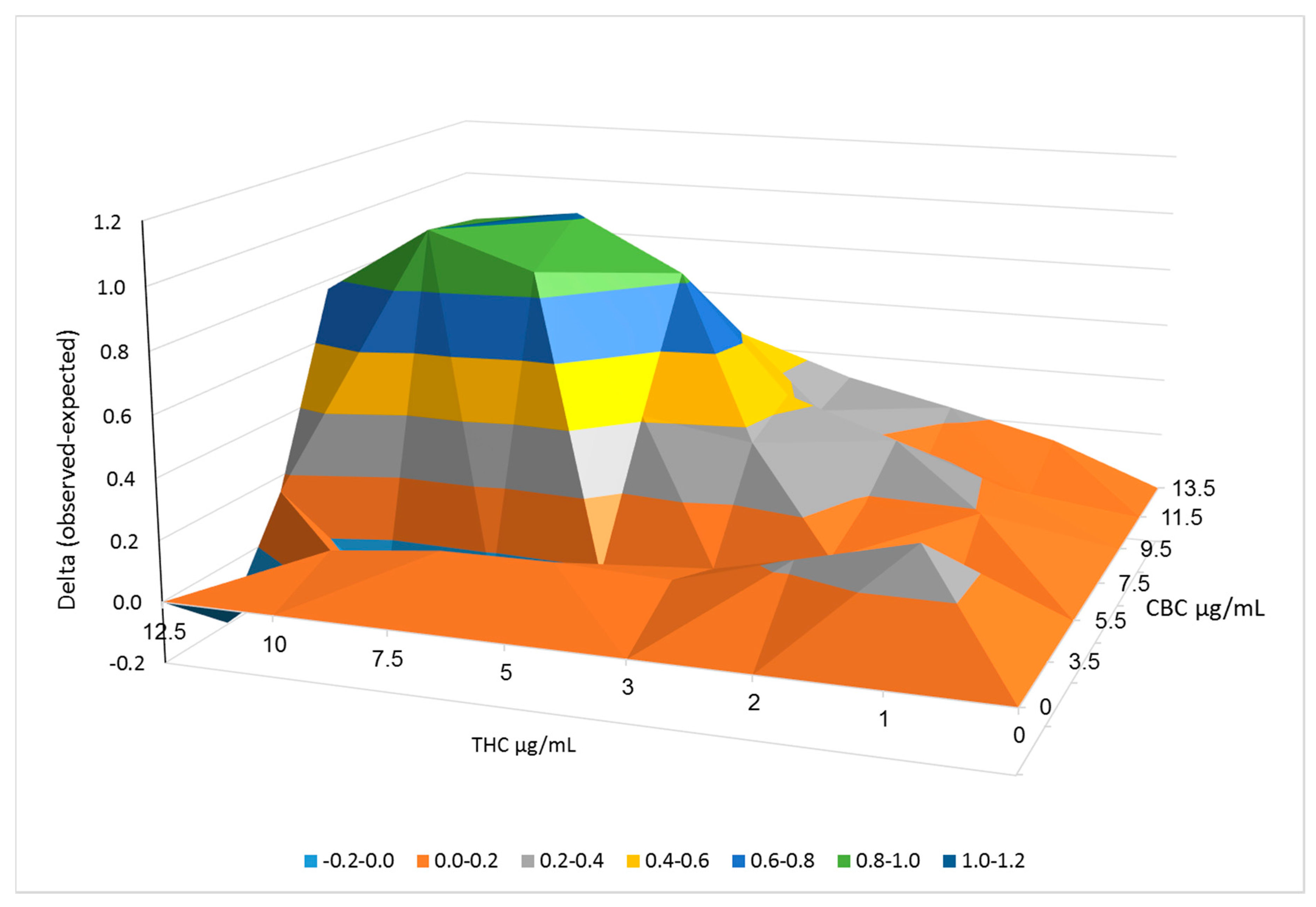
2.3. Synergistic Interactions between CBC and THC
2.4. Determination of CB1, CB2, TRPA1, TRPV1 and TRPV2 Receptor Involvement in Cytotoxic Activity
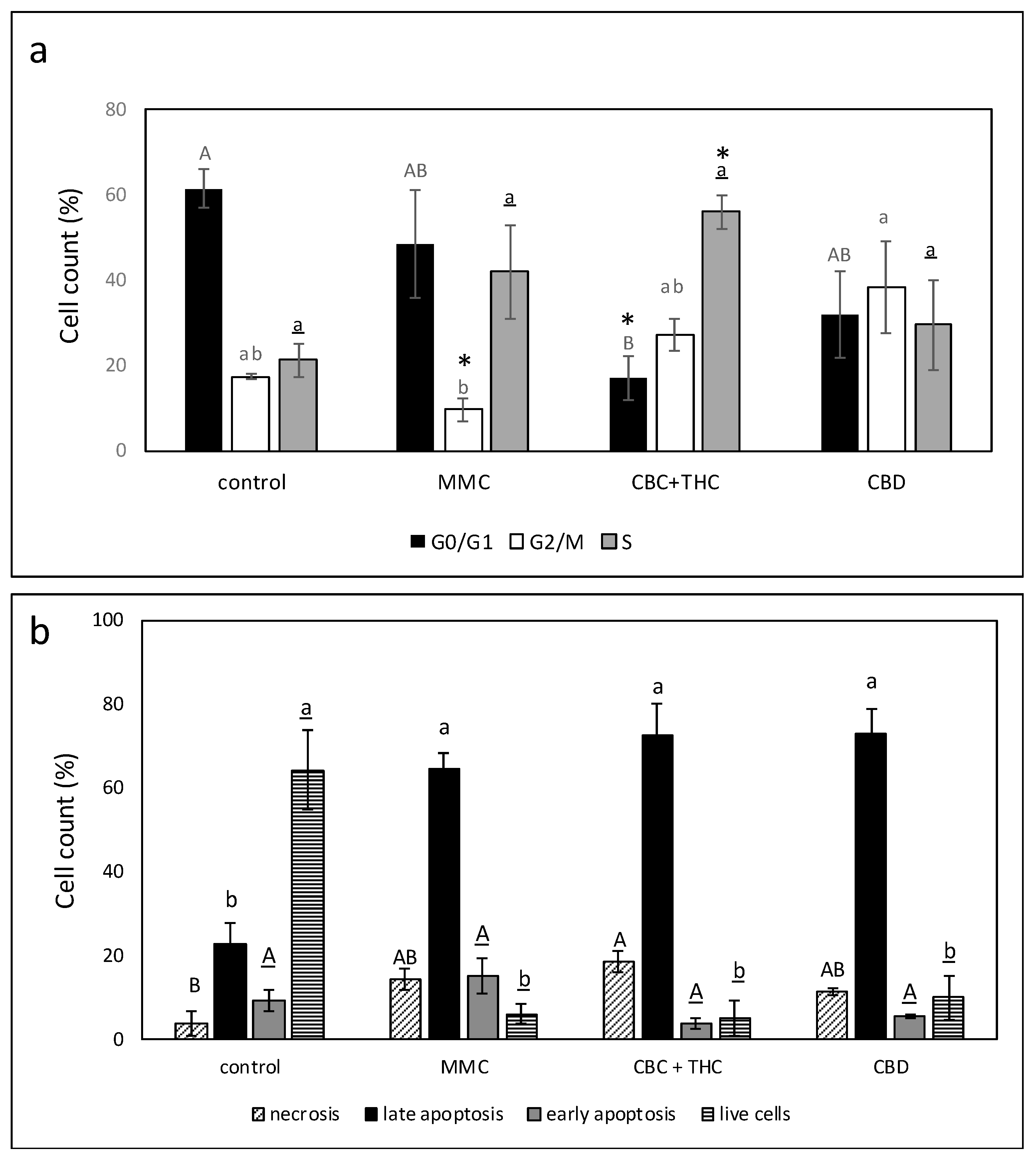
2.5. Determination of the Effect of CBC + THC or CBD Treatments on Cell Cycle Arrest and Apoptosis
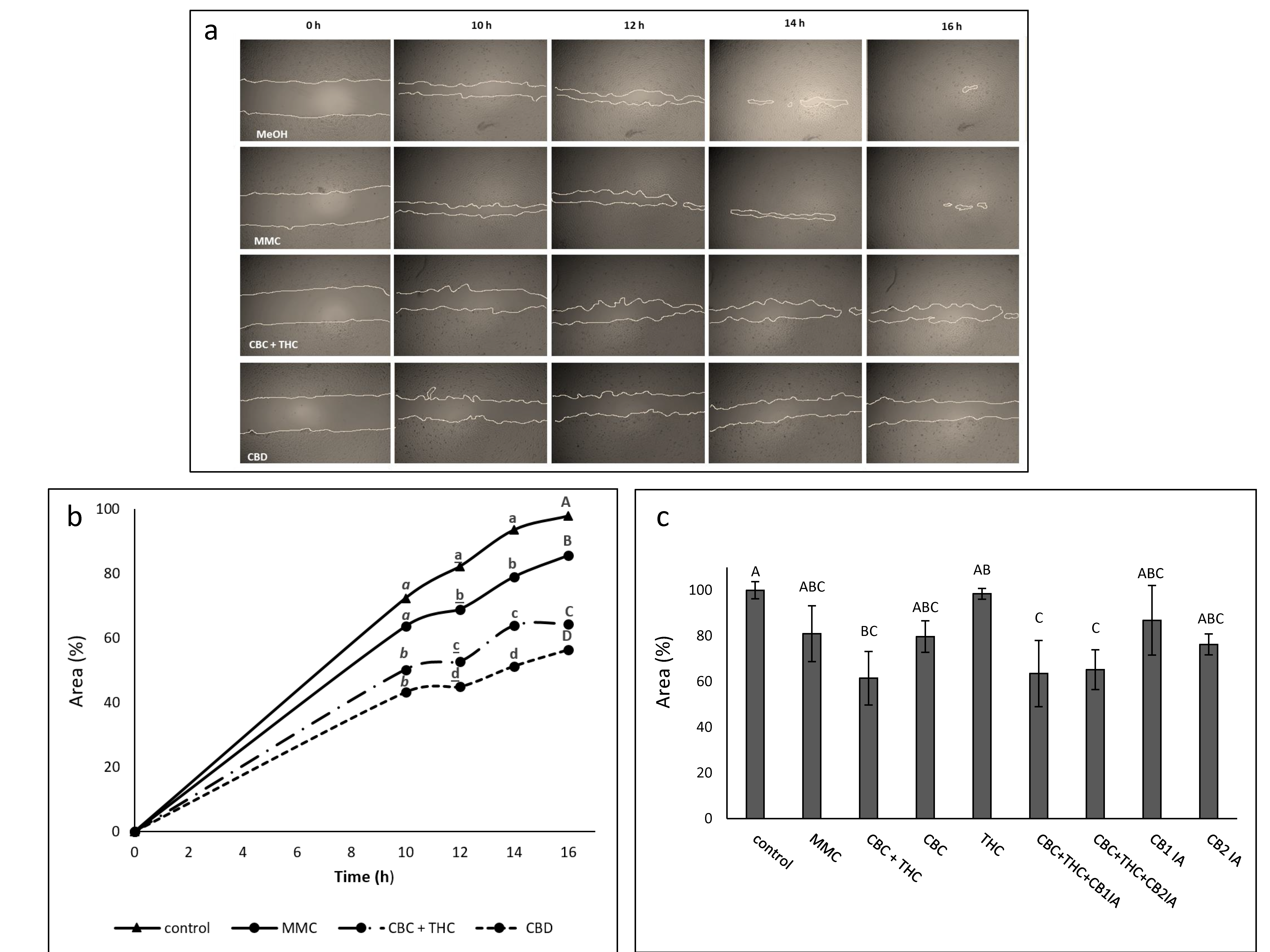
2.6. Determination of the Effect of CBC + THC and CBD Treatments on Cell Motility
2.7. Determination of the Effect of CBC + THC and CBD Treatments on Cell Invasion
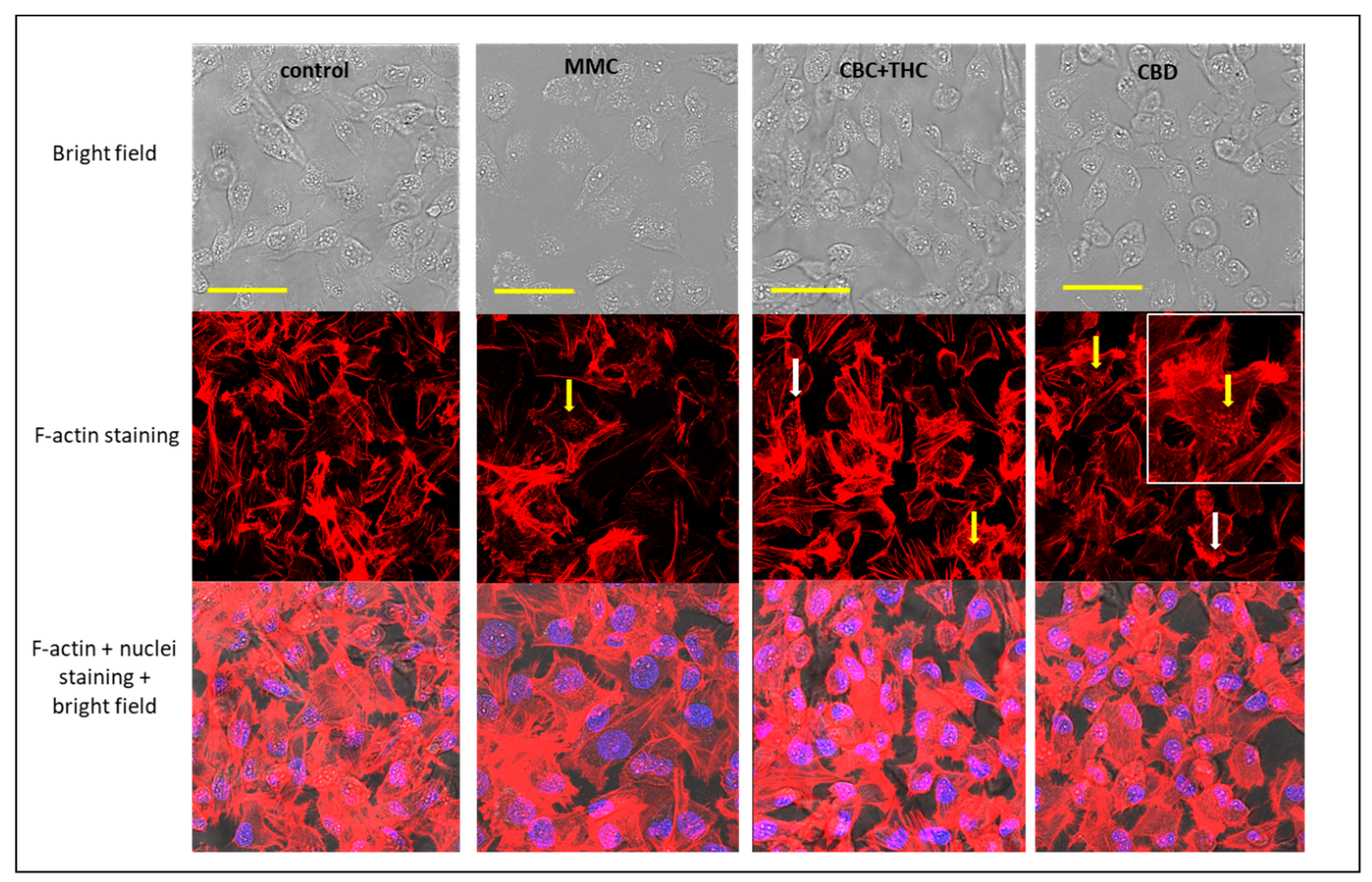
2.8. Determination of the Effect of CBC + THC and CBD Treatments on F-actin Filaments
3. Discussion
4. Materials and Methods
4.1. Plant Growth and Extract Preparation
4.2. Extract Fractionation
4.3. Standard/Material Preparation
4.4. Chemical Analysis
4.5. Cell Cultures
4.6. Cell Proliferation Assay
4.7. Analysis of Combined Drug Effects
4.8. Apoptosis Assay and Cell Cycle Analysis
4.9. Cell Migration and Cell Invasion Assays
4.10. Cytoskeleton Staining
4.11. Quantitative Real-Time (qRT) PCR
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Thomas, A.A.; Wallner, L.P.; Quinn, V.P.; Slezak, J.; Van Den Eeden, S.K.; Chien, G.W.; Jacobsen, S.J. Association between cannabis use and the risk of bladder cancer: Results from the California Men’s Health Study. Urology 2015, 85, 388–393. [Google Scholar] [CrossRef]
- Hanuš, L.O.; Meyer, S.M.; Muñoz, E.; Taglialatela-Scafati, O.; Appendino, G. Phytocannabinoids: A unified critical inventory. Nat. Prod. Rep. 2016, 33, 1357–1392. [Google Scholar] [CrossRef]
- Berman, P.; Futoran, K.; Lewitus, G.M.; Mukha, D.; Benami, M.; Shlomi, T.; Meiri, D. A new ESI-LC/MS approach for comprehensive metabolic profiling of phytocannabinoids in Cannabis. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef]
- Aizpurua-Olaizola, O.; Soydaner, U.; Öztürk, E.; Schibano, D.; Simsir, Y.; Navarro, P.; Etxebarria, N.; Usobiaga, A. Evolution of the cannabinoid and terpene content during the growth of Cannabis sativa plants from different chemotypes. J. Nat. Prod. 2016, 79, 324–331. [Google Scholar] [CrossRef]
- Ramer, R.; Hinz, B. Cannabinoids as anticancer drugs. In Advances in Pharmacology, 80th ed.; Kendall, D., Alexander, S.P.H., Eds.; Elsevier Academic Press: Cambridge, MA, USA, 2017; pp. 397–436. [Google Scholar]
- Baram, L.; Peled, E.; Berman, P.; Yellin, B.; Besser, E.; Benami, M.; Louria-Hayon, I.; Lewitus, G.M.; Meiri, D. The heterogeneity and complexity of Cannabis extracts as antitumor agents. Oncotarget 2019, 10, 4091. [Google Scholar] [CrossRef] [PubMed]
- Ramer, R.; Schwarz, R.; Hinz, B. Modulation of the endocannabinoid system as a potential anticancer strategy. Front. Pharmacol. 2019, 10, 430. [Google Scholar] [CrossRef] [PubMed]
- Velasco, G.; Sánchez, C.; Guzmán, M. Towards the use of cannabinoids as antitumour agents. Nat. Rev. Cancer 2012, 12, 436–444. [Google Scholar] [CrossRef]
- Cianchi, F.; Papucci, L.; Schiavone, N.; Lulli, M.; Magnelli, L.; Vinci, M.C.; Messerini, L.; Manera, C.; Ronconi, E.; Romagnani, P. Cannabinoid receptor activation induces apoptosis through tumor necrosis factor α–mediated ceramide de novo synthesis in colon cancer cells. Clin. Cancer Res. 2008, 14, 7691–7700. [Google Scholar] [CrossRef] [PubMed]
- Galve-Roperh, I.; Sánchez, C.; Cortés, M.L.; del Pulgar, T.G.; Izquierdo, M.; Guzmán, M. Anti-tumoral action of cannabinoids: Involvement of sustained ceramide accumulation and extracellular signal-regulated kinase activation. Nat. Med. 2000, 6, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, V.; Philips, B.J.; Su, R.; Smaldone, M.C.; Erickson, V.L.; Chancellor, M.B.; Yoshimura, N.; Tyagi, P. Differential expression of functional cannabinoid receptors in human bladder detrusor and urothelium. J. Urol. 2009, 181, 1932–1938. [Google Scholar] [CrossRef]
- Bettiga, A.; Aureli, M.; Colciago, G.; Murdica, V.; Moschini, M.; Lucianò, R.; Canals, D.; Hannun, Y.; Hedlund, P.; Lavorgna, G. Bladder cancer cell growth and motility implicate cannabinoid 2 receptor-mediated modifications of sphingolipids metabolism. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Russo, E.B. Taming THC: Potential cannabis synergy and phytocannabinoid-terpenoid entourage effects. Br. J. Pharmacol. 2011, 163, 1344–1364. [Google Scholar] [CrossRef] [PubMed]
- Russo, E.B. The case for the entourage effect and conventional breeding of clinical cannabis: No “strain,” no gain. Front. Plant. Sci. 2018, 9, 1969. [Google Scholar] [CrossRef] [PubMed]
- Koltai, H.; Namdar, D. Cannabis phytomolecule ‘entourage’: From domestication to medical use. Trends Plant. Sci. 2020, 25, 976–984. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Jo, M.J.; Yun, H.K.; Kim, D.Y.; Kim, B.R.; Kim, J.L.; Park, S.H.; Na, Y.J.; Jeong, Y.A.; Kim, B.G. Cannabidiol promotes apoptosis via regulation of XIAP/Smac in gastric cancer. Cell Death Dis. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Olson, M.F.; Sahai, E. The actin cytoskeleton in cancer cell motility. Clin. Exp. Metastasis 2009, 26, 273. [Google Scholar] [CrossRef] [PubMed]
- Velasco, G.; Hernández-Tiedra, S.; Dávila, D.; Lorente, M. The use of cannabinoids as anticancer agents. Prog. Neuropsychopharmacol. Biol. Psychiatry 2016, 64, 259–266. [Google Scholar] [CrossRef]
- Velasco, G.; Sánchez, C.; Guzmán, M. Anticancer mechanisms of cannabinoids. Curr. Oncol. 2016, 23, S23. [Google Scholar] [CrossRef]
- De Petrocellis, L.; Ligresti, A.; Schiano Moriello, A.; Iappelli, M.; Verde, R.; Stott, C.G.; Cristino, L.; Orlando, P.; Di Marzo, V. Non-THC cannabinoids inhibit prostate carcinoma growth in vitro and in vivo: Pro-apoptotic effects and underlying mechanisms. Br. J. Pharmacol. 2013, 168, 79–102. [Google Scholar] [CrossRef]
- Pollastro, F.; Caprioglio, D.; Del Prete, D.; Rogati, F.; Minassi, A.; Taglialatela-Scafati, O.; Munoz, E.; Appendino, G. Cannabichromene. Nat. Prod. Commun. 2018, 13, 1934578X1801300922. [Google Scholar] [CrossRef]
- Onaivi, E.S.; Ishiguro, H.; Gong, J.P.; Patel, S.; Meozzi, P.A.; Myers, L.; Perchuk, A.; Mora, Z.; Tagliaferro, P.A.; Gardner, E.; et al. Functional expression of brain neuronal CB2 cannabinoid receptors are involved in the effects of drugs of abuse and in depression. Ann. N.Y. Acad. Sci. 2008, 1139, 434. [Google Scholar] [CrossRef] [PubMed]
- La Porta, C.; Bura, S.A.; Aracil-Fernández, A.; Manzanares, J.; Maldonado, R. Role of CB1 and CB2 cannabinoid receptors in the development of joint pain induced by monosodium iodoacetate. Pain 2013, 154, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Pertwee, R.G.; Howlett, A.; Abood, M.E.; Alexander, S.; Di Marzo, V.; Elphick, M.; Greasley, P.; Hansen, H.S.; Kunos, G.; Mackie, K. International Union of Basic and Clinical Pharmacology. LXXIX. Cannabinoid receptors and their ligands: Beyond CB1 and CB2. Pharmacol. Rev. 2010, 62, 588–631. [Google Scholar] [CrossRef] [PubMed]
- Laprairie, R.; Bagher, A.; Kelly, M.; Denovan-Wright, E. Cannabidiol is a negative allosteric modulator of the cannabinoid CB1 receptor. Br. J. Pharmacol. 2015, 172, 4790–4805. [Google Scholar] [CrossRef] [PubMed]
- Izzo, A.A.; Borrelli, F.; Capasso, R.; Di Marzo, V.; Mechoulam, R. Non-psychotropic plant cannabinoids: New therapeutic opportunities from an ancient herb. Trends Pharmacol. Sci. 2009, 30, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Liu, Y.; Huang, S.; Liu, G.; Xie, C.; Zhou, J.; Fan, W.; Li, Q.; Wang, Q.; Zhong, D.; et al. Overexpression of cannabinoid receptors CB1 and CB2 correlates with improved prognosis of patients with hepatocellular carcinoma. Cancer Genet. Cytogenet. 2006, 171, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Andrade, E.L.; Forner, S.; Bento, A.F.; Leite, D.F.P.; Dias, M.A.; Leal, P.C.; Koepp, J.; Calixto, J.B. TRPA1 receptor modulation attenuates bladder overactivity induced by spinal cord injury. Am. J. Physiol. Renal Physiol. 2011, 300, F1223–F1234. [Google Scholar] [CrossRef]
- Lazzeri, M.; Vannucchi, M.G.; Spinelli, M.; Bizzoco, E.; Beneforti, P.; Turini, D.; Faussone-Pellegrini, M.S. Transient receptor potential vanilloid type 1 (TRPV1) expression changes from normal urothelium to transitional cell carcinoma of human bladder. Eur. Urol. 2005, 48, 691–698. [Google Scholar] [CrossRef]
- Yamada, T.; Ueda, T.; Shibata, Y.; Ikegami, Y.; Saito, M.; Ishida, Y.; Ugawa, S.; Kohri, K.; Shimada, S. TRPV2 activation induces apoptotic cell death in human T24 bladder cancer cells: A potential therapeutic target for bladder cancer. Urology 2010, 76, e1–e509. [Google Scholar] [CrossRef]
- Muller, C.; Morales, P.; Reggio, P.H. Cannabinoid ligands targeting TRP channels. Front. Mol. Neurosci. 2019, 11, 487. [Google Scholar] [CrossRef]
- Houtgraaf, J.H.; Versmissen, J.; van der Giessen, W.J. A concise review of DNA damage checkpoints and repair in mammalian cells. Cardiovasc. Revasc. Med. 2006, 7, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.G.; Chung, H.; Yoo, Y.D.; Lee, J.G.; Choi, Y.I.; Yu, Y.S. Mechanism of growth inhibitory effect of Mitomycin-C on cultured human retinal pigment epithelial cells: Apoptosis and cell cycle arrest. Curr. Eye Res. 2001, 22, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, X.; Liu, Q.; Leung, A.W.; Wang, P.; Xu, C. Sinoporphyrin sodium mediated photodynamic therapy inhibits the migration associated with collapse of F-actin filaments cytoskeleton in MDA-MB-231 cells. Photodiagnosis Photodyn. Ther. 2016, 13, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Pawlik, A.; Nowak, J.M.; Grzanka, D.; Gackowska, L.; Michalkiewicz, J.; Grzanka, A. Hyperthermia induces cytoskeletal alterations and mitotic catastrophe in p53-deficient H1299 lung cancer cells. Acta Histochem. 2013, 115, 8–15. [Google Scholar] [CrossRef]
- Takahashi, K.; Suzuki, K. WAVE2, N-WASP, and Mena facilitate cell invasion via phosphatidylinositol 3-kinase-dependent local accumulation of actin filaments. J. Cell. Biochem. 2011, 112, 3421–3429. [Google Scholar] [CrossRef]
- O’Neill, P.R.; Castillo-Badillo, J.A.; Meshik, X.; Kalyanaraman, V.; Melgarejo, K.; Gautam, N. Membrane flow drives an adhesion-independent amoeboid cell migration mode. Dev. Cell 2018, 46, 9–22. [Google Scholar] [CrossRef]
- Laviña, B.; Castro, M.; Niaudet, C.; Cruys, B.; Álvarez-Aznar, A.; Carmeliet, P.; Bentley, K.; Brakebusch, C.; Betsholtz, C.; Gaengel, K. Defective endothelial cell migration in the absence of Cdc42 leads to capillary-venous malformations. Development 2018, 145, dev161182. [Google Scholar] [CrossRef]
- Mazuz, M.; Moyal, L.; Hodak, E.; Nadarajan, S.; Vinayaka, A.C.; Gorovitz-Haris, B.; Lubin, I.; Drori, A.; Drori, G.; Van Cauwenberghe, O.; et al. Synergistic cytotoxic activity of cannabinoids from Cannabis sativa against cutaneous T-cell lymphoma (CTCL) in-vitro and ex-vivo. Oncotarget 2020, 11, 1141. [Google Scholar] [CrossRef]
- Shiozaki, A.; Kudou, M.; Ichikawa, D.; Fujiwara, H.; Shimizu, H.; Ishimoto, T.; Arita, T.; Kosuga, T.; Konishi, H.; Komatsu, S. Esophageal cancer stem cells are suppressed by tranilast, a TRPV2 channel inhibitor. J. Gastroenterol. 2018, 53, 197–207. [Google Scholar] [CrossRef]
- Available online: http://rsb.info.nih.gov/ij/download.html (accessed on 1 December 2020).
- Ye, Z.; Liang, Z.; Mi, Q.; Guo, Y. Limonene terpenoid obstructs human bladder cancer cell (T24 cell line) growth by inducing cellular apoptosis, caspase activation, G2/M phase cell cycle arrest and stops cancer metastasis. Balkan Union Oncol. 2020, 25, 280–285. [Google Scholar]
- Available online: https://www.jmp.com/en_us/home.html (accessed on 1 December 2020).

| Treatment | Cell Invasion (%) Relative to Vehicle Control |
|---|---|
| MMC 4 µg/mL | 43.7 ± 4.8 |
| CBC+THC (8.6 + 1.4 µg/mL) | 90.5 ± 4.1 |
| CBD (10 µg/mL) | 89.7 ± 6.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anis, O.; Vinayaka, A.C.; Shalev, N.; Namdar, D.; Nadarajan, S.; Anil, S.M.; Cohen, O.; Belausov, E.; Ramon, J.; Mayzlish Gati, E.; et al. Cannabis-Derived Compounds Cannabichromene and Δ9-Tetrahydrocannabinol Interact and Exhibit Cytotoxic Activity against Urothelial Cell Carcinoma Correlated with Inhibition of Cell Migration and Cytoskeleton Organization. Molecules 2021, 26, 465. https://doi.org/10.3390/molecules26020465
Anis O, Vinayaka AC, Shalev N, Namdar D, Nadarajan S, Anil SM, Cohen O, Belausov E, Ramon J, Mayzlish Gati E, et al. Cannabis-Derived Compounds Cannabichromene and Δ9-Tetrahydrocannabinol Interact and Exhibit Cytotoxic Activity against Urothelial Cell Carcinoma Correlated with Inhibition of Cell Migration and Cytoskeleton Organization. Molecules. 2021; 26(2):465. https://doi.org/10.3390/molecules26020465
Chicago/Turabian StyleAnis, Omer, Ajjampura C. Vinayaka, Nurit Shalev, Dvora Namdar, Stalin Nadarajan, Seegehalli M. Anil, Ofer Cohen, Eduard Belausov, Jacob Ramon, Einav Mayzlish Gati, and et al. 2021. "Cannabis-Derived Compounds Cannabichromene and Δ9-Tetrahydrocannabinol Interact and Exhibit Cytotoxic Activity against Urothelial Cell Carcinoma Correlated with Inhibition of Cell Migration and Cytoskeleton Organization" Molecules 26, no. 2: 465. https://doi.org/10.3390/molecules26020465
APA StyleAnis, O., Vinayaka, A. C., Shalev, N., Namdar, D., Nadarajan, S., Anil, S. M., Cohen, O., Belausov, E., Ramon, J., Mayzlish Gati, E., & Koltai, H. (2021). Cannabis-Derived Compounds Cannabichromene and Δ9-Tetrahydrocannabinol Interact and Exhibit Cytotoxic Activity against Urothelial Cell Carcinoma Correlated with Inhibition of Cell Migration and Cytoskeleton Organization. Molecules, 26(2), 465. https://doi.org/10.3390/molecules26020465







