Tissue mRNA for S100A4, S100A6, S100A8, S100A9, S100A11 and S100P Proteins in Colorectal Neoplasia: A Pilot Study
Abstract
1. Introduction
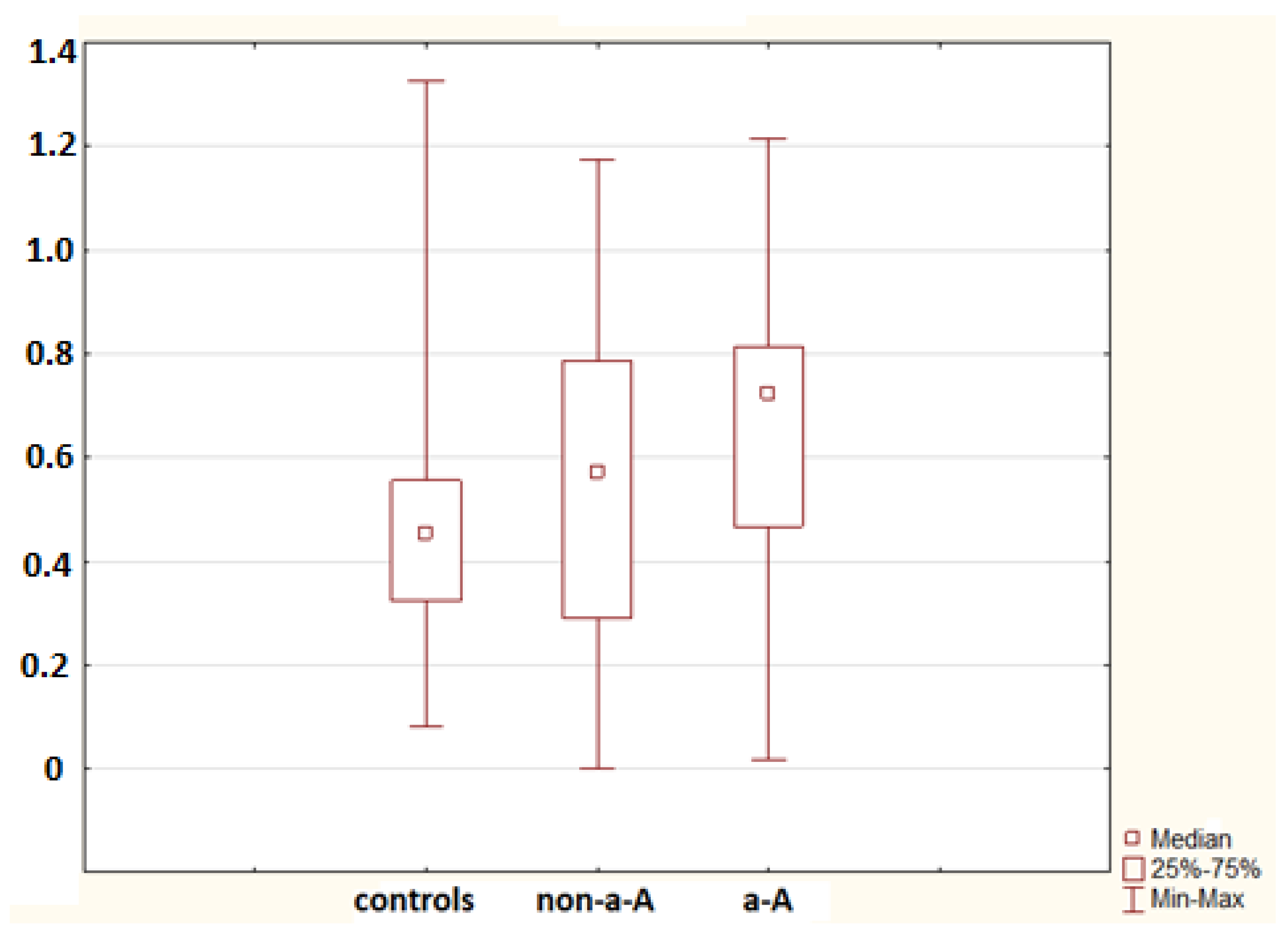
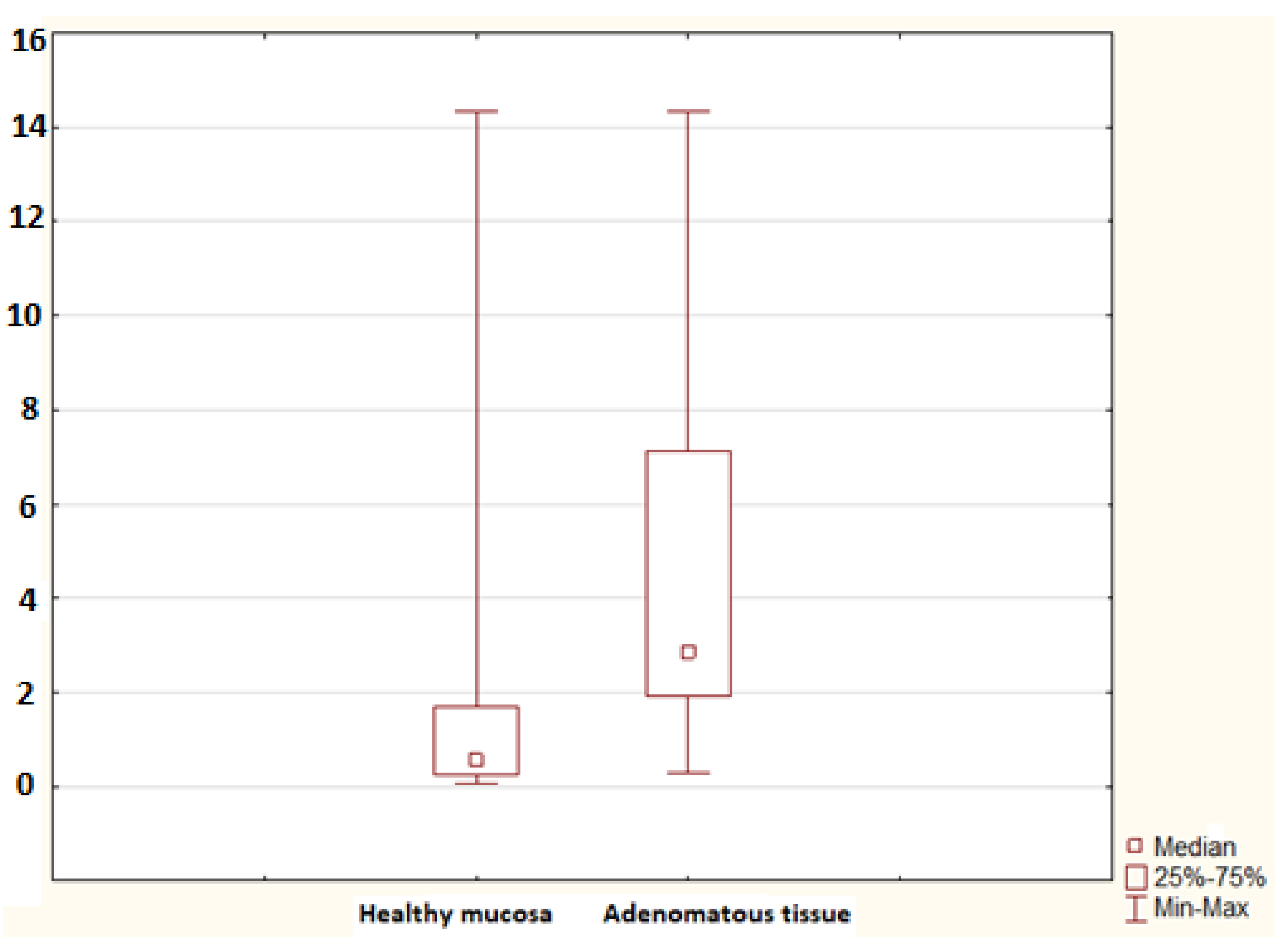
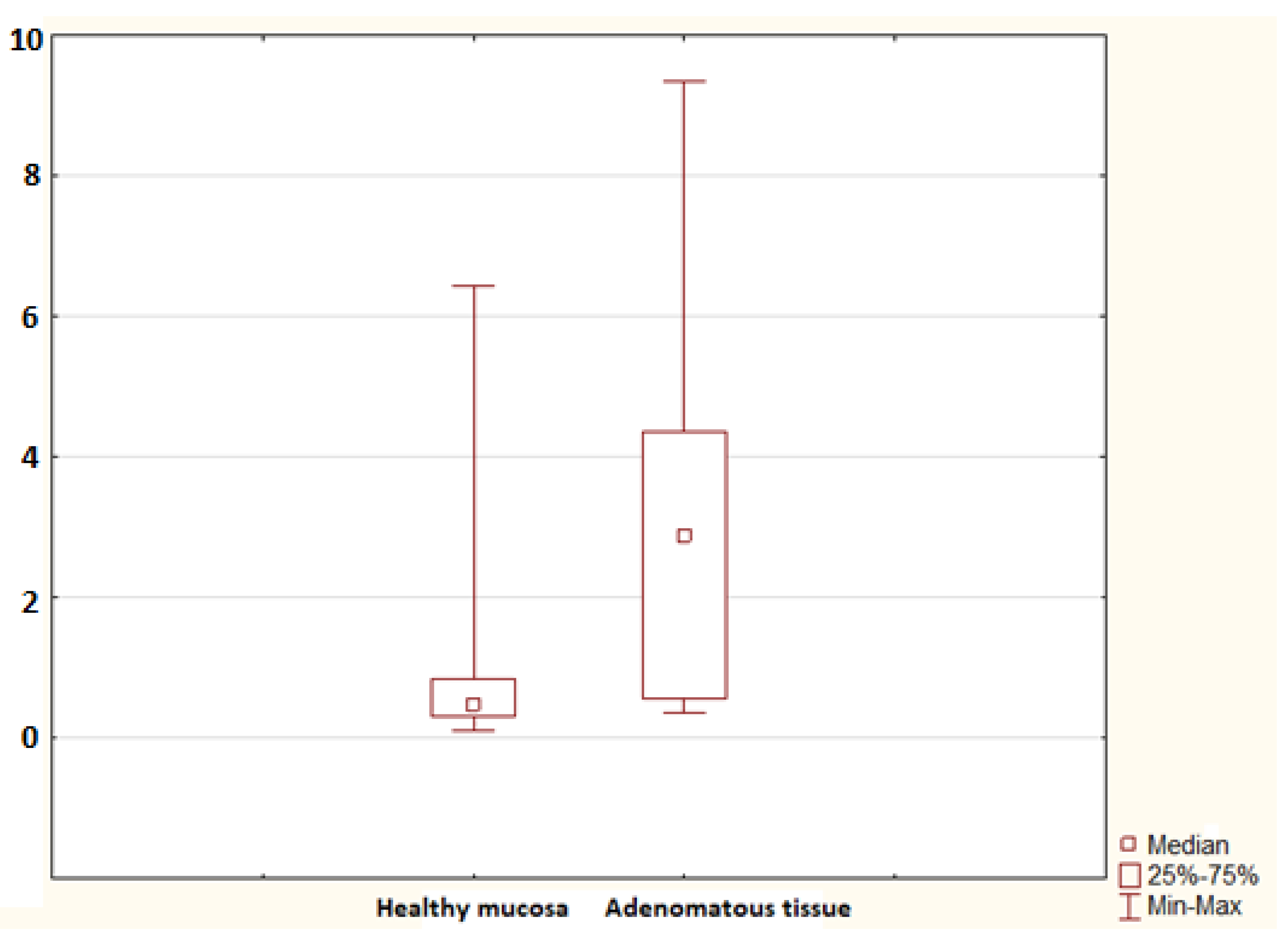
2. Results
3. Discussion
4. Methods
4.1. Subjects
4.2. Endoscopic Procedure
4.3. RNA Isolation qPCR
4.4. Statistical Analysis
4.5. Ethical Issues
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Haigis, K.M. (Ed.) Molecular Pathogenesis of Colorectal Cancer; Springer: Berlin/Heidelberg, Germany, 2013; ISBN 978-1-4614-8412-7. [Google Scholar]
- Kohoutova, D.; Pejchal, J.; Bures, J. Mitotic and apoptotic activity in colorectal neoplasia. BMC Gastroenterol. 2018, 18, 65. [Google Scholar] [CrossRef] [PubMed]
- Kohoutova, D.; Forstlova, M.; Moravkova, P.; Cyrany, J.; Bosak, J.; Smajs, D.; Rejchrt, S.; Bures, J. Bacteriocin production by mucosal bacteria in current and previous colorectal neoplasia. BMC Cancer 2020, 20, 39. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.H.; Goel, A.; Chung, D.C. Pathways of colorectal carcinogenesis. Gastroenterology 2020, 158, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Bresalier, R.S. Colorectal cancer. In Sleisenger and Fordtran’s Gastrointestinal and Liver Disease, 11th ed.; Feldman, M., Friedman, L.S., Brandt, L.J., Eds.; Elsevier: Philadelphia, PA, USA, 2021; pp. 2108–2152. ISBN 978-0-323-60962-3. [Google Scholar]
- Stulík, J.; Osterreicher, J.; Koupilová, K.; Knízek, J.; Bures, J.; Jandík, P.; Langr, F.; Dedic, K.; Schäfer, B.W.; Heizmann, C.W. Differential expression of the Ca2+ binding S100A6 protein in normal, preneoplastic and neoplastic colon mucosa. Eur. J. Cancer 2000, 36, 1050–1059. [Google Scholar] [CrossRef]
- Wang, W.; Yu, S.; Huang, S.; Deng, R.; Ding, Y.; Wu, Y.; Li, X.; Wang, A.; Wang, S.; Chen, W.; et al. A complex role for calcium signaling in colorectal cancer development and progression. Mol. Cancer Res. 2019, 17, 2145–2153. [Google Scholar] [CrossRef]
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 proteins. Curr. Mol. Med. 2013, 13, 1, 24–57. [Google Scholar] [CrossRef]
- Chen, H.; Xu, C.; Jin, Q.; Liu, Z. S100 protein family in human cancer. Am. J. Cancer Res. 2014, 4, 89–115. [Google Scholar]
- Moravkova, P.; Kohoutova, D.; Rejchrt, S.; Cyrany, J.; Bures, J. Role of S100 proteins in colorectal carcinogenesis. Gastroenterol. Res. Pract. 2016, 2016, 2632703. [Google Scholar] [CrossRef]
- Moravkova, P.; Kohoutova, D.; Vavrova, J.; Bures, J. Serum S100A6, S100A8, S100A9 and S100A11 proteins in colorectal neoplasia: Results of a single centre prospective study. Scand. J. Clin. Lab. Investig. 2020, 80, 173–178. [Google Scholar] [CrossRef]
- Stulík, J.; Kovárová, H.; Macela, A.; Bures, J.; Jandík, P.; Langr, F.; Otto, A.; Thiede, B.; Jungblut, P. Overexpression of calcium-binding protein calgranulin B in colonic mucosal diseases. Clin. Chim. Acta 1997, 265, 41–55. [Google Scholar] [CrossRef]
- Stulík, J.; Osterreicher, J.; Koupilová, K.; Knízek, J.; Macela, A.; Bures, J.; Jandík, P.; Langr, F.; Dedic, K.; Jungblut, P.R. The analysis of S100A9 and S100A8 expression in matched sets of macroscopically normal colon mucosa and colorectal carcinoma: The S100A9 and S100A8 positive cells underlie and invade tumor mass. Electrophoresis 1999, 20, 1047–1054. [Google Scholar] [CrossRef]
- Stulík, J.; Koupilova, K.; Osterreicher, J.; Knízek, J.; Macela, A.; Bures, J.; Jandík, P.; Langr, F.; Dedic, K.; Jungblut, P.R. Protein abundance alterations in matched sets of macroscopically normal colon mucosa and colorectal carcinoma. Electrophoresis 1999, 20, 3638–3646. [Google Scholar]
- Kang, Y.G.; Jung, C.K.; Lee, A.; Kang, W.K.; Oh, S.T.; Kang, C.S. Prognostic significance of S100A4 mRNA and protein expression in colorectal cancer. J. Surg. Oncol. 2012, 105, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Ishikawa, T.; Mogushi, K.; Okazaki, S.; Ishiguro, M.; Iida, S.; Mizushima, H.; Tanaka, H.; Uetake, H.; Sugihara, K. Overexpression of the S100A2 protein as a prognostic marker for patients with stage II and III colorectal cancer. Int. J. Oncol. 2016, 48, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Goh, J.Y.; Feng, M.; Wang, W.; Oguz, G.; Yatim, S.M.J.M.; Lee, P.L.; Bao, Y.; Lim, T.H.; Wang, P.; Tam, W.L.; et al. Chromosome 1q21.3 amplification is a trackable biomarker and actionable target for breast cancer recurrence. Nat. Med. 2017, 23, 1319–1330. [Google Scholar] [CrossRef]
- Schäfer, B.W.; Wicki, R.; Engelkamp, D.; Mattei, M.G.; Heizmann, C.W. Isolation of a YAC clone covering a cluster of nine S100 genes on human chromosome 1q21: Rationale for a new nomenclature of the S100 calcium-binding protein family. Genomics 1995, 25, 638–643. [Google Scholar] [CrossRef]
- Orlando, G.; Law, P.J.; Cornish, A.J.; Dobbins, S.E.; Chubb, D.; Broderick, P.; Litchfield, K.; Hariri, F.; Pastinen, T.; Osborne, C.S.; et al. Promoter capture Hi-C-based identification of recurrent noncoding mutations in colorectal cancer. Nat. Genet. 2018, 50, 1375–1380. [Google Scholar] [CrossRef]
- Gibadulinova, A.; Oveckova, I.; Parkkila, S.; Pastorekova, S.; Pastorek, J. Key promoter elements involved in transcriptional activation of the cancer-related gene coding for S100P calcium-binding protein. Oncol. Rep. 2008, 20, 391–396. [Google Scholar]
- Nakamura, N.; Takenaga, K. Hypomethylation of the metastasis-associated S100A4 gene correlates with gene activation in human colon adenocarcinoma cell lines. Clin. Exp. Metastasis 1998, 16, 471–479. [Google Scholar] [CrossRef]
- Wang, X.H.; Zhang, L.H.; Zhong, X.Y.; Xing, X.F.; Liu, Y.Q.; Niu, Z.J.; Peng, Y.; Du, H.; Zhang, G.G.; Hu, Y.; et al. S100A6 overexpression is associated with poor prognosis and is epigenetically up-regulated in gastric cancer. Am. J. Pathol. 2010, 177, 586–597. [Google Scholar] [CrossRef]
- Liu, K.; Zhang, Y.; Zhang, C.; Zhang, Q.; Li, J.; Xiao, F.; Li, Y.; Zhang, R.; Dou, D.; Liang, J.; et al. Methylation of S100A8 is a promising diagnosis and prognostic marker in hepatocellular carcinoma. Oncotarget 2016, 7, 56798–56810. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fang, L.; Zang, Y.; Xu, Z. Identification of Core Genes and Key Pathways via Integrated Analysis of Gene Expression and DNA Methylation Profiles in Bladder Cancer. Med. Sci. Monit. 2018, 24, 3024–3033. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kim, C.N.; Kim, S.Y.; Lee, J.S.; Cho, D.; Kim, J.W.; Yoon, S.Y. Enhanced S100A4 protein expression is clinicopathologically significant to metastatic potential and p53 dysfunction in colorectal cancer. Oncol. Rep. 2009, 22, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Sorci, G.; Riuzzi, F.; Arcuri, C.; Bianchi, R.; Brozzi, F.; Tubaro, C.; Giambanco, I. S100B’s double life: Intracellular regulator and extracellular signal. Biochim. Biophys. Acta 2009, 1793, 1008–1022. [Google Scholar] [CrossRef]
- Schmidt-Hansen, B.; Ornås, D.; Grigorian, M.; Klingelhöfer, J.; Tulchinsky, E.; Lukanidin, E.; Ambartsumian, N. Extracellular S100A4(mts1) stimulates invasive growth of mouse endothelial cells and modulates MMP-13 matrix metalloproteinase activity. Oncogene 2004, 23, 5487–5495. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Xu, Y.; Cai, G.; Guan, Z.; Cai, S. Downregulation of S100A4 expression by RNA interference suppresses cell growth and invasion in human colorectal cancer cells. Oncol. Rep. 2012, 27, 917–922. [Google Scholar] [CrossRef]
- Marenholz, I.; Heizmann, C.W.; Fritz, G. S100 proteins in mouse and man: From evolution to function and pathology (including an update of the nomenclature). Biochem. Biophys. Res. Commun. 2004, 322, 1111–1122. [Google Scholar] [CrossRef]
- Donato, R.; Sorci, G.; Giambanco, I. S100A6 protein: Functional roles. Cell Mol. Life Sci. 2017, 74, 2749–2760. [Google Scholar] [CrossRef]
- Feng, S.; Zhou, Q.; Yang, B.; Li, Q.; Liu, A.; Zhao, Y.; Qiu, C.; Ge, J.; Zhai, H. The effect of S100A6 on nuclear translocation of CacyBP/SIP in colon cancer cells. PLoS ONE 2018, 13, e0192208. [Google Scholar] [CrossRef]
- Kilańczyk, E.; Graczyk, A.; Ostrowska, H.; Kasacka, I.; Leśniak, W.; Filipek, A. S100A6 is transcriptionally regulated by β-catenin and interacts with a novel target, lamin A/C, in colorectal cancer cells. Cell Calcium 2012, 51, 470–477. [Google Scholar] [CrossRef]
- Króliczak, W.; Pietrzak, M.; Puzianowska-Kuznicka, M. P53-dependent suppression of the human calcyclin gene (S100A6): The role of Sp1 and of NFkappaB. Acta Biochim. Pol. 2008, 55, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, K.; Andoh, A.; Ishiguro, S.; Suzuki, N.; Hunai, H.; Kobune-Fujiwara, Y.; Kameyama, M.; Miyoshi, J.; Akedo, H.; Nakamura, H. Increased expression of S100A6 (Calcyclin), a calcium-binding protein of the S100 family, in human colorectal adenocarcinomas. Clin. Cancer Res. 2000, 6, 172–177. [Google Scholar] [PubMed]
- Gebhardt, C.; Németh, J.; Angel, P.; Hess, J. S100A8 and S100A9 in inflammation and cancer. Biochem. Pharmacol. 2006, 72, 1622–1631. [Google Scholar] [CrossRef] [PubMed]
- Ehrchen, J.M.; Sunderkötter, C.; Foell, D.; Vogl, T.; Roth, J. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J. Leukoc. Biol. 2009, 86, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Wu, R.; Chen, L.; Peng, Q.; Li, S.; Zhang, Y.; Zhou, L.; Duan, L. S100A9 regulates MDSCs-mediated immune suppression via the RAGE and TLR4 signaling pathways in colorectal carcinoma. Front. Immunol. 2019, 10, 2243. [Google Scholar] [CrossRef]
- Ghavami, S.; Kerkhoff, C.; Los, M.; Hashemi, M.; Sorg, C.; Karami-Tehrani, F. Mechanism of apoptosis induced by S100A8/A9 in colon cancer cell lines: The role of ROS and the effect of metal ions. J. Leukoc. Biol. 2004, 76, 169–175. [Google Scholar] [CrossRef]
- Ghavami, S.; Rashedi, I.; Dattilo, B.M.; Eshraghi, M.; Chazin, W.J.; Hashemi, M.; Wesselborg, S.; Kerkhoff, C.; Los, M. S100A8/A9 at low concentration promotes tumor cell growth via RAGE ligation and MAP kinase-dependent pathway. J. Leukoc. Biol. 2008, 83, 1484–1492. [Google Scholar] [CrossRef]
- Luley, K.; Noack, F.; Lehnert, H.; Homann, N. Local calprotectin production in colorectal cancer and polyps: Active neutrophil recruitment in carcinogenesis. Int. J. Color. Dis. 2011, 26, 603–607. [Google Scholar] [CrossRef]
- Moris, D.; Spartalis, E.; Angelou, A.; Margonis, G.A.; Papalambros, A.; Petrou, A.; Athanasiou, A.; Schizas, D.; Dimitroulis, D.; Felekouras, E. The value of calprotectin S100A8/A9 complex as a biomarker in colorectal cancer: A systematic review. J. BUON 2016, 21, 859–866. [Google Scholar]
- Turvill, J.; Aghahoseini, A.; Sivarajasingham, N.; Abbas, K.; Choudhry, M.; Polyzois, K.; Lasithiotakis, K.; Volanaki, D.; Kim, B.; Langlands, F.; et al. Faecal calprotectin in patients with suspected colorectal cancer: A diagnostic accuracy study. Br. J. Gen. Pract. 2016, 66, e499–e506. [Google Scholar] [CrossRef]
- Andrés Cerezo, L.; Šumová, B.; Prajzlerová, K.; Veigl, D.; Damgaard, D.; Nielsen, C.H.; Pavelka, K.; Vencovský, J.; Šenolt, L. Calgizzarin (S100A11): A novel inflammatory mediator associated with disease activity of rheumatoid arthritis. Arthritis Res. Ther. 2017, 19, 79. [Google Scholar] [CrossRef] [PubMed]
- Anania, M.C.; Miranda, C.; Vizioli, M.G.; Mazzoni, M.; Cleris, L.; Pagliardini, S.; Manenti, G.; Borrello, M.G.; Pierotti, M.A.; Greco, A. S100A11 overexpression contributes to the malignant phenotype of papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2013, 98, 1591–1600. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J. Expression of S100A11 is a prognostic factor for disease-free survival and overall survival in patients with high-grade serous ovarian vancer. Appl. Immunohistochem. Mol. Morphol. 2017, 25, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zheng, S.; Jing, C.; Zhang, J.; Shen, H.; Xu, X.; Lin, J.; Zhang, B. S100A11 promotes TGF-β1-induced epithelial-mesenchymal transition through SMAD2/3 signaling pathway in intrahepatic cholangiocarcinoma. Future Oncol. 2018, 14, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.F.; Li, T.; Jiang, F.; Ni, W.K.; Guan, C.Q.; Liu, Z.X.; Lu, C.H.; Ni, R.Z.; Wu, W.; Xiao, M.B. Correlation between S100A11 and the TGF-β1/SMAD4 pathway and its effects on the proliferation and apoptosis of pancreatic cancer cell line PANC-1. Mol. Cell. Biochem. 2019, 450, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Li, T.; Ji, Y.; Jiang, F.; Ni, W.; Zhu, J.; Bao, B.; Lu, C.; Ni, R. S100A11 promotes human pancreatic cancer PANC-1 cell proliferation and is involved in the PI3K/AKT signaling pathway. Oncol. Lett. 2018, 15, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Koh, S.A.; Lee, K.H. HGF-mediated S100A11 overexpression enhances proliferation and invasion of gastric cancer. Am. J. Transl. Res. 2018, 10, 3385–3394. [Google Scholar]
- Takamatsu, H.; Yamamoto, K.I.; Tomonobu, N.; Murata, H.; Inoue, Y.; Yamauchi, A.; Sumardika, I.W.; Chen, Y.; Kinoshita, R.; Yamamura, M.; et al. Extracellular S100A11 plays a critical role in spread of the fibroblast population in pancreatic cancers. Oncol. Res. 2019, 27, 713–727. [Google Scholar] [CrossRef]
- Melle, C.; Ernst, G.; Schimmel, B.; Bleul, A.; Mothes, H.; Kaufmann, R.; Settmacher, U.; Von Eggeling, F. Different expression of calgizzarin (S100A11) in normal colonic epithelium, adenoma and colorectal carcinoma. Int. J. Oncol. 2006, 28, 195–200. [Google Scholar] [CrossRef][Green Version]
- Chiang, J.M.; Tan, R.; Wang, J.Y.; Chen, J.S.; Lee, Y.S.; Hsieh, P.S.; Changchien, C.R.; Chen, J.R. S100P, a calcium-binding protein, is preferentially associated with the growth of polypoid tumors in colorectal cancer. Int. J. Mol. Med. 2015, 35, 675–683. [Google Scholar] [CrossRef][Green Version]
- Zuo, Z.; Zhang, P.; Lin, F.; Shang, W.; Bi, R.; Lu, F.; Wu, J.; Jiang, L. Interplay between Trx-1 and S100P promotes colorectal cancer cell epithelial-mesenchymal transition by up-regulating S100A4 through AKT activation. J. Cell. Mol. Med. 2018, 22, 2430–2441. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Y.N.; Lin, G.L.; Qiu, H.Z.; Wu, B.; Wu, H.Y.; Zhao, Y.; Chen, Y.J.; Lu, C.M. S100P, a potential novel prognostic marker in colorectal cancer. Oncol. Rep. 2012, 28, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Lam, F.F.; Jankova, L.; Dent, O.F.; Molloy, M.P.; Kwun, S.Y.; Clarke, C.; Chapuis, P.; Robertson, G.; Beale, P.; Clarke, S.; et al. Identification of distinctive protein expression patterns in colorectal adenoma. Proteom. Clin. Appl. 2010, 4, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, D.; Downs-Kelly, E.; Liu, X.; Pai, R.K.; Patil, D.T.; Rybicki, L.; Bennett, A.E.; Plesec, T.; Cummings, O.; Rex, D.; et al. Reproducibility of the villous component and high-grade dysplasia in colorectal adenomas <1 cm: Implications for endoscopic surveillance. Am. J. Surg. Pathol. 2013, 37, 427–433. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Bustin, S.A.; Mueller, R. Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin. Sci. (Lond.) 2005, 109, 365–379. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peterova, E.; Bures, J.; Moravkova, P.; Kohoutova, D. Tissue mRNA for S100A4, S100A6, S100A8, S100A9, S100A11 and S100P Proteins in Colorectal Neoplasia: A Pilot Study. Molecules 2021, 26, 402. https://doi.org/10.3390/molecules26020402
Peterova E, Bures J, Moravkova P, Kohoutova D. Tissue mRNA for S100A4, S100A6, S100A8, S100A9, S100A11 and S100P Proteins in Colorectal Neoplasia: A Pilot Study. Molecules. 2021; 26(2):402. https://doi.org/10.3390/molecules26020402
Chicago/Turabian StylePeterova, Eva, Jan Bures, Paula Moravkova, and Darina Kohoutova. 2021. "Tissue mRNA for S100A4, S100A6, S100A8, S100A9, S100A11 and S100P Proteins in Colorectal Neoplasia: A Pilot Study" Molecules 26, no. 2: 402. https://doi.org/10.3390/molecules26020402
APA StylePeterova, E., Bures, J., Moravkova, P., & Kohoutova, D. (2021). Tissue mRNA for S100A4, S100A6, S100A8, S100A9, S100A11 and S100P Proteins in Colorectal Neoplasia: A Pilot Study. Molecules, 26(2), 402. https://doi.org/10.3390/molecules26020402





