Analysis and Authentication of Avocado Oil Using High Resolution NMR Spectroscopy
Abstract
1. Introduction
2. Results and Discussion
2.1. NMR Assignment of the Avocado Oil Spectra
2.2. Fatty Acid Profile
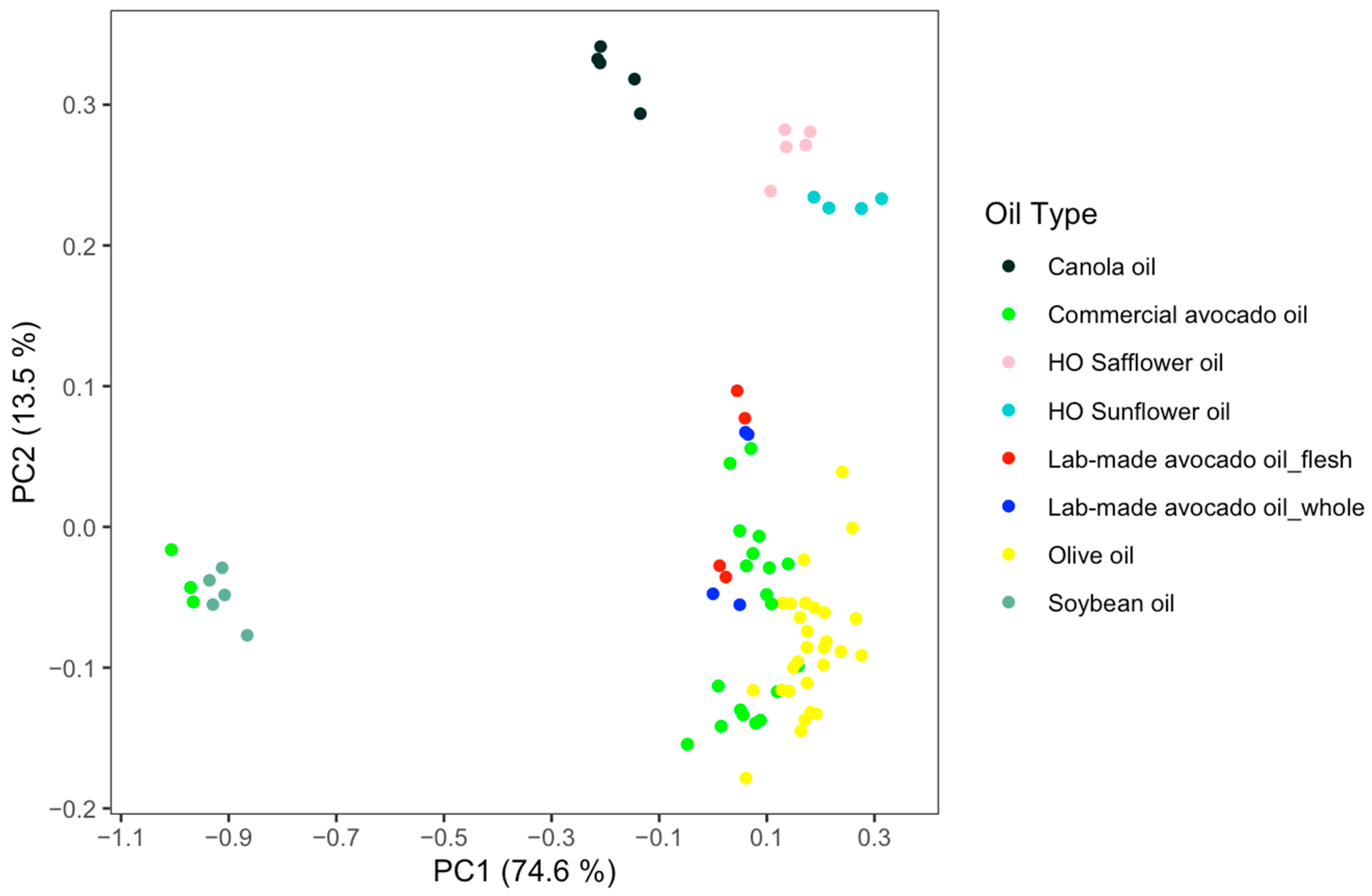
2.3. Spectral Comparison
2.4. Untargeted Analysis
3. Materials and Methods
3.1. Samples and Chemicals
3.2. Sample Preparation for NMR Experiments
3.3. NMR Experiments
3.4. Multivariate Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Werman, M.J.; Neeman, I. Avocado oil production and chemical characteristics. J. Am. Oil Chem. Soc. 1987, 64, 229–232. [Google Scholar] [CrossRef]
- Terés, S.; Barceló-Coblijn, G.; Benet, M.; Álvarez, R.; Bressani, R.; Halver, J.E.; Escribá, P.V. Oleic acid content is responsible for the reduction in blood pressure induced by olive oil. Proc. Natl. Acad. Sci. USA 2008, 105, 13811–13816. [Google Scholar] [CrossRef] [PubMed]
- Altendorf, S. Major Tropical Fruits Market Review 2018; FAO: Rome, Italy, 2019. [Google Scholar]
- Flores, M.; Saravia, C.; Vergara, C.E.; Avila, F.; Valdés, H.; Ortiz-Viedma, J. Avocado Oil: Characteristics, Properties, and Applications. Molecules 2019, 24, 2172. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.O.; Dorantes, L.; Galíndez, J.; Guzmán, R.I. Effect of Different Extraction Methods on Fatty Acids, Volatile Compounds, and Physical and Chemical Properties of Avocado (Persea americana Mill.) Oil. J. Agric. Food Chem. 2003, 51, 2216–2221. [Google Scholar] [CrossRef]
- Corzzini, S.C.S.; Barros, H.D.F.Q.; Grimaldi, R.; Cabral, F.A. Extraction of edible avocado oil using supercritical CO2 and a CO2/ethanol mixture as solvents. J. Food Eng. 2017, 194, 40–45. [Google Scholar] [CrossRef]
- Costagli, G.; Betti, M. Avocado oil extraction processes: Method for cold-pressed high-quality edible oil production versus traditional production. J. Agric. Eng. 2015, 46, 115–122. [Google Scholar] [CrossRef]
- Martínez-Padilla, L.P.; Franke, L.; Xu, X.-Q.; Juliano, P. Improved extraction of avocado oil by application of sono-physical processes. Ultrason. Sonochem. 2018, 40, 720–726. [Google Scholar] [CrossRef]
- Fernandes, G.D.; Gómez-Coca, R.B.; Pérez-Camino, M.C.; Moreda, W.; Barrera-Arellano, D. Chemical characterization of commercial and single-variety avocado oils. Grasas Aceites 2018, 69, e256. [Google Scholar] [CrossRef]
- Tan, C.X.; Tan, S.S.; Tan, S.T. Influence of Geographical Origins on the Physicochemical Properties of Hass Avocado Oil. J. Am. Oil Chem. Soc. 2017, 94, 1431–1437. [Google Scholar] [CrossRef]
- Yanty, N.A.M.; Marikkar, J.M.N.; Long, K. Effect of Varietal Differences on Composition and Thermal Characteristics of Avocado Oil. J. Am. Oil Chem. Soc. 2011, 88, 1997–2003. [Google Scholar] [CrossRef]
- Donetti, M.; Terry, L.A. Biochemical markers defining growing area and ripening stage of imported avocado fruit cv. Hass. J. Food Compos. Anal. 2014, 34, 90–98. [Google Scholar] [CrossRef]
- Green, H.S.; Wang, S.C. First report on quality and purity evaluations of avocado oil sold in the US. Food Control 2020, 116, 107328. [Google Scholar] [CrossRef]
- Sacchi, R.; Medina, I.; Aubourg, S.P.; Addeo, F.; Paolillo, L. Proton nuclear magnetic resonance rapid and structure-specific determination ofω-3 polyunsaturated fatty acids in fish lipids. J. Am. Oil Chem. Soc. 1993, 70, 225–228. [Google Scholar] [CrossRef]
- Igarashi, T.; Aursand, M.; Hirata, Y.; Gribbestad, I.S.; Wada, S.; Nonaka, M. Nondestructive quantitative determination of docosahexaenoic acid and n−3 fatty acids in fish oils by high-resolution 1H nuclear magnetic resonance spectroscopy. J. Am. Oil Chem. Soc. 2000, 77, 737–748. [Google Scholar] [CrossRef]
- Tang, F.; Vasas, M.; Hatzakis, E.; Spyros, A. Magnetic resonance applications in food analysis. Annu. Rep. NMR Spectrosc. 2019, 98, 239–306. [Google Scholar]
- Hatzakis, E. Nuclear Magnetic Resonance (NMR) Spectroscopy in Food Science: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 189–220. [Google Scholar] [CrossRef] [PubMed]
- Vigli, G.; Philippidis, A.; Spyros, A.; Dais, P. Classification of Edible Oils by Employing 31P and 1H NMR Spectroscopy in Combination with Multivariate Statistical Analysis. A Proposal for the Detection of Seed Oil Adulteration in Virgin Olive Oils. J. Agric. Food Chem. 2003, 51, 5715–5722. [Google Scholar] [CrossRef]
- Kupče, Ē.; Claridge, T.D.W. New NOAH Modules for Structure Elucidation at Natural Isotopic Abundance. J. Magn. Reson. 2019, 307, 106568. [Google Scholar] [CrossRef]
- Tang, F.; Hatzakis, E. NMR-Based Analysis of Pomegranate Juice Using Untargeted Metabolomics Coupled with Nested and Quantitative Approaches. Anal. Chem. 2020, 92, 11177–11185. [Google Scholar] [CrossRef]
- Ahmed, R.; Varras, P.C.; Siskos, M.G.; Siddiqui, H.; Choudhary, M.I.; Gerothanassis, I.P. NMR and Computational Studies as Analytical and High-Resolution Structural Tool for Complex Hydroperoxides and Diastereomeric Endo-Hydroperoxides of Fatty Acids in Solution-Exemplified by Methyl Linolenate. Molecules 2020, 25, 4902. [Google Scholar] [CrossRef]
- Prescha, A.; Grajzer, M.; Dedyk, M.; Grajeta, H. The Antioxidant Activity and Oxidative Stability of Cold-Pressed Oils. J. Am. Oil Chem. Soc. 2014, 91, 1291–1301. [Google Scholar] [CrossRef] [PubMed]
- Salvo, A.; Rotondo, A.; Torre, G.L.L.; Cicero, N.; Dugo, G. Determination of 1,2/1,3-diglycerides in Sicilian extra-virgin olive oils by 1H-NMR over a one-year storage period. Nat. Prod. Res. 2017, 31, 822–828. [Google Scholar] [CrossRef] [PubMed]
- Guillén, M.D.; Ruiz, A. Rapid simultaneous determination by proton NMR of unsaturation and composition of acyl groups in vegetable oils. Eur. J. Lipid Sci. Technol. 2003, 105, 688–696. [Google Scholar] [CrossRef]
- Castejón, D.; Fricke, P.; Cambero, M.I.; Herrera, A. Automatic 1H-NMR Screening of Fatty Acid Composition in Edible Oils. Nutrients 2016, 8, 93. [Google Scholar] [CrossRef]
- Williamson, K.; Hatzakis, E. NMR analysis of roasted coffee lipids and development of a spent ground coffee application for the production of bioplastic precursors. Food Res. Int. 2019, 119, 683–692. [Google Scholar] [CrossRef]
- Dais, P.; Misiak, M.; Hatzakis, E. Analysis of marine dietary supplements using NMR spectroscopy. Anal. Methods 2015, 7, 5226–5238. [Google Scholar] [CrossRef]
- Manaf, Y.N.; Rahardjo, A.P.; Yusof, Y.A.; Desa, M.N.; Nusantoro, B.P. Lipid characteristics and tocopherol content of the oils of native avocado cultivars grown in Indonesia. Int. J. Food Prop. 2018, 21, 2758–2771. [Google Scholar] [CrossRef]
- Ruiz-Aracama, A.; Goicoechea, E.; Guillén, M.D. Direct study of minor extra-virgin olive oil components without any sample modification. 1H NMR multisupression experiment: A powerful tool. Food Chem. 2017, 228, 301–314. [Google Scholar] [CrossRef]
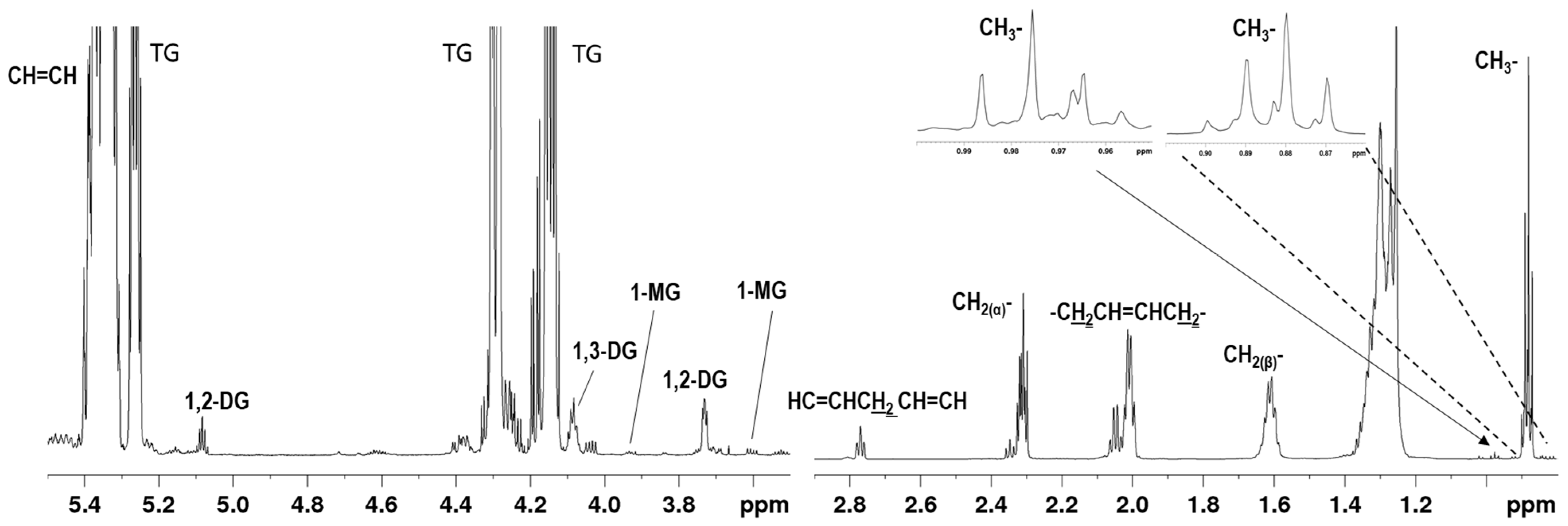
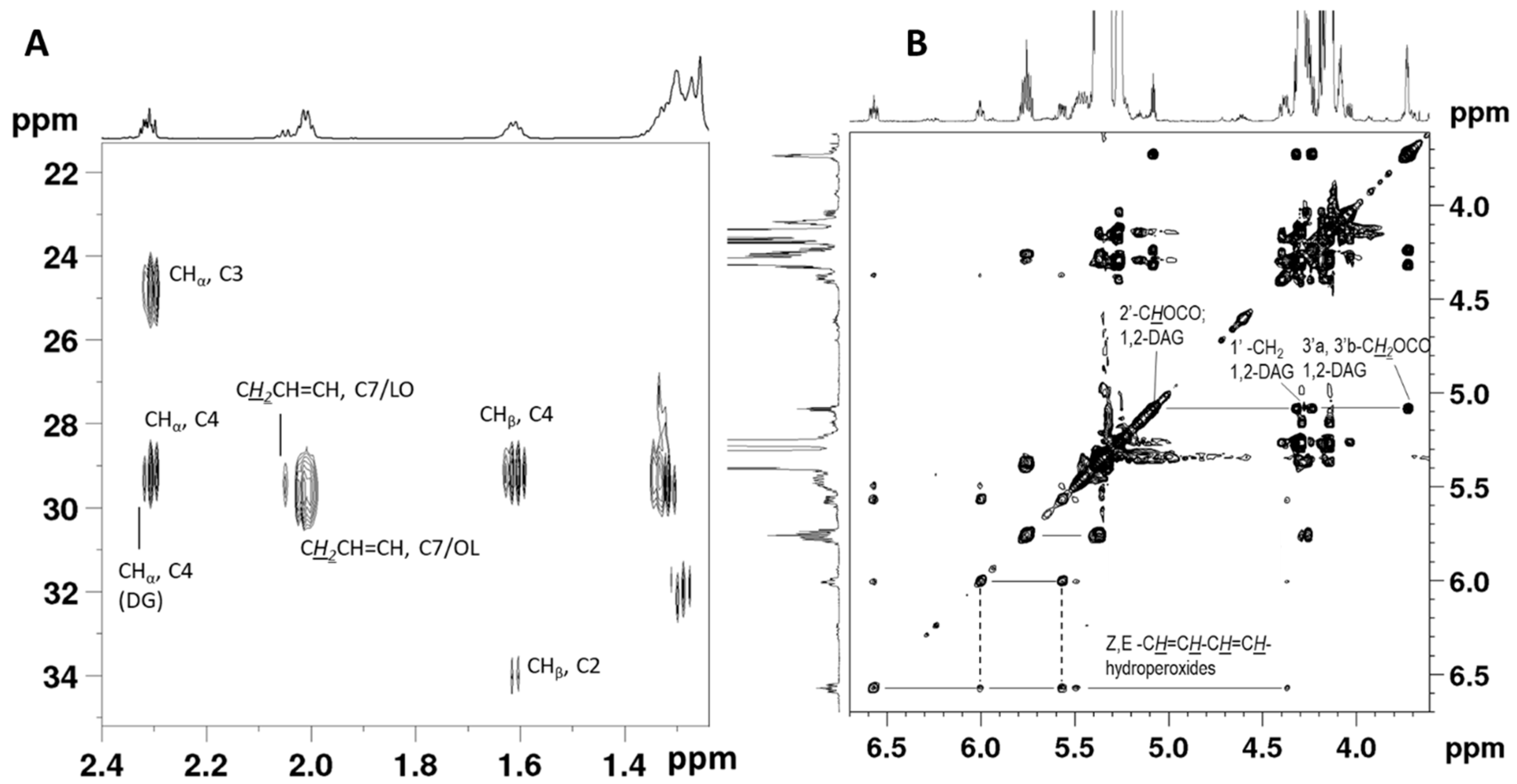
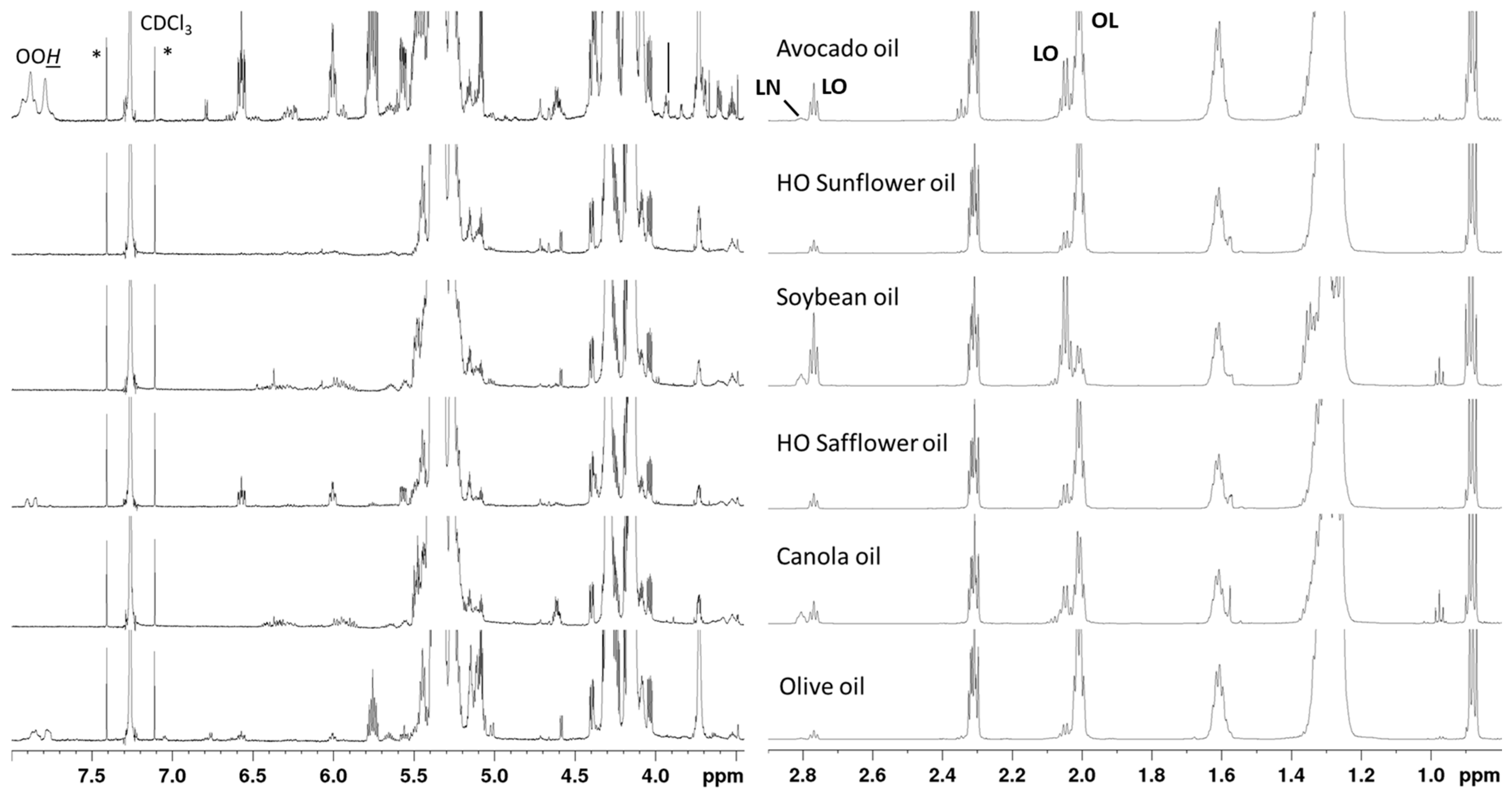

| Signal | δ (ppm) | Functional Group | Component |
|---|---|---|---|
| 1 | 0.880 | CH3CH2 (t), J = 7.1 Hz | SFA, OL |
| 2 | 0.883 | CH3CH2 (t), J = 7.2 Hz | LO |
| 3 | 0.976 | CH3CH2 (t), J = 7.5 Hz | LN |
| 4 | 1.145–1.450 | -(CH2)n- (envelope) | All acyl chains |
| 5 | 1.612 | -CH2(β), m | All acyl chains |
| 6 | 2.009 | CH2CH=CH (m) | OL |
| 7 | 2.048 | CH2CH=CH (m) | LO, LN |
| 8 | 2.308 | -CH2(α) (m) | TG, All acyl chains |
| 9 | 2.347 | -CH2α (t), J = 7.6 Hz | DG, 1-MG, FFA All acyl chains |
| 10 | 2.769 | CH=CHCH2CH=CH (m) | LO |
| 11 | 2.805 | CH=CHCH2CH=CH (br) | LN |
| 12 | 3.602 | 3’a-CH2OCO (dd) | 1-MG |
| 13 | 3.690 | 3’a-CH2OCO (dd) | 1-MG |
| 14 | 3.730 | 3’a, 3’b-CH2OCO | 1,2-DG |
| 15 | 3.839 | 1’a,’b, 3’a,’b HOCH2-CH(OR)-CH2OH (m) | 2-MG |
| 16 | 3.935 | 2’-CHOH | 1-MG |
| 17 | 4.080 | 2’-CHOH (br) | 1,3 DG |
| 18 | 4.144 | CH2OCO (dd) J = 11.96 Hz J = 4.35 Hz | TG |
| 19 | 4.185 | 1’b, 3’b-CH2OCO (dd) J = 11.35 Hz J = 4.35 Hz | 1,3 DG |
| 20 | 4.295 | CH2OCO (dd) J = 11.96 Hz J = 4.35 Hz | TG |
| 21 | 5.083 | 2’-CHOCO (m) | 1,2-DG |
| 22 | 5.264 | 2’-CHOCO (m) | TG |
| 23 | 5.300–5.400 | CH=CH (m) | All acyl chains |
| Component | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| OL/PO | 60.7 | 21.5 | 72.0 | 70.6 | 70.2 | 67.7 | 67.9 | 66.2 | 70.6 | 64.6 |
| LO | 18.9 | 55.1 | 15.7 | 12.8 | 10.6 | 16.8 | 16.3 | 16.8 | 15.3 | 18.7 |
| LN | 0.9 | 6.8 | 0.5 | 0.6 | 0.6 | 0.4 | 1.1 | 0.9 | 1.0 | 1.1 |
| SFA | 19.5 | 16.6 | 11.8 | 16.0 | 18.6 | 15.1 | 12.6 | 16.1 | 13.1 | 15.6 |
| UD | 78.05 | 76.5 | 85.3 | 81.3 | 80.9 | 82.7 | 84.1 | 80.5 | 83.9 | 81.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, F.; Green, H.S.; Wang, S.C.; Hatzakis, E. Analysis and Authentication of Avocado Oil Using High Resolution NMR Spectroscopy. Molecules 2021, 26, 310. https://doi.org/10.3390/molecules26020310
Tang F, Green HS, Wang SC, Hatzakis E. Analysis and Authentication of Avocado Oil Using High Resolution NMR Spectroscopy. Molecules. 2021; 26(2):310. https://doi.org/10.3390/molecules26020310
Chicago/Turabian StyleTang, Fenfen, Hilary S. Green, Selina C. Wang, and Emmanuel Hatzakis. 2021. "Analysis and Authentication of Avocado Oil Using High Resolution NMR Spectroscopy" Molecules 26, no. 2: 310. https://doi.org/10.3390/molecules26020310
APA StyleTang, F., Green, H. S., Wang, S. C., & Hatzakis, E. (2021). Analysis and Authentication of Avocado Oil Using High Resolution NMR Spectroscopy. Molecules, 26(2), 310. https://doi.org/10.3390/molecules26020310