Epithelial-to-Mesenchymal Transition Is Not a Major Modulating Factor in the Cytotoxic Response to Natural Products in Cancer Cell Lines
Abstract
1. Introduction
2. Results
2.1. Screening of Natural Products in Various Breast, Colon, and Pancreatic Cancer Cell Lines Reveals Notable Anti-Cancer Activity
2.2. Natural Products Induce Mesenchymal-to-Epithelial Transition Selectively in Cancer Cell Lines
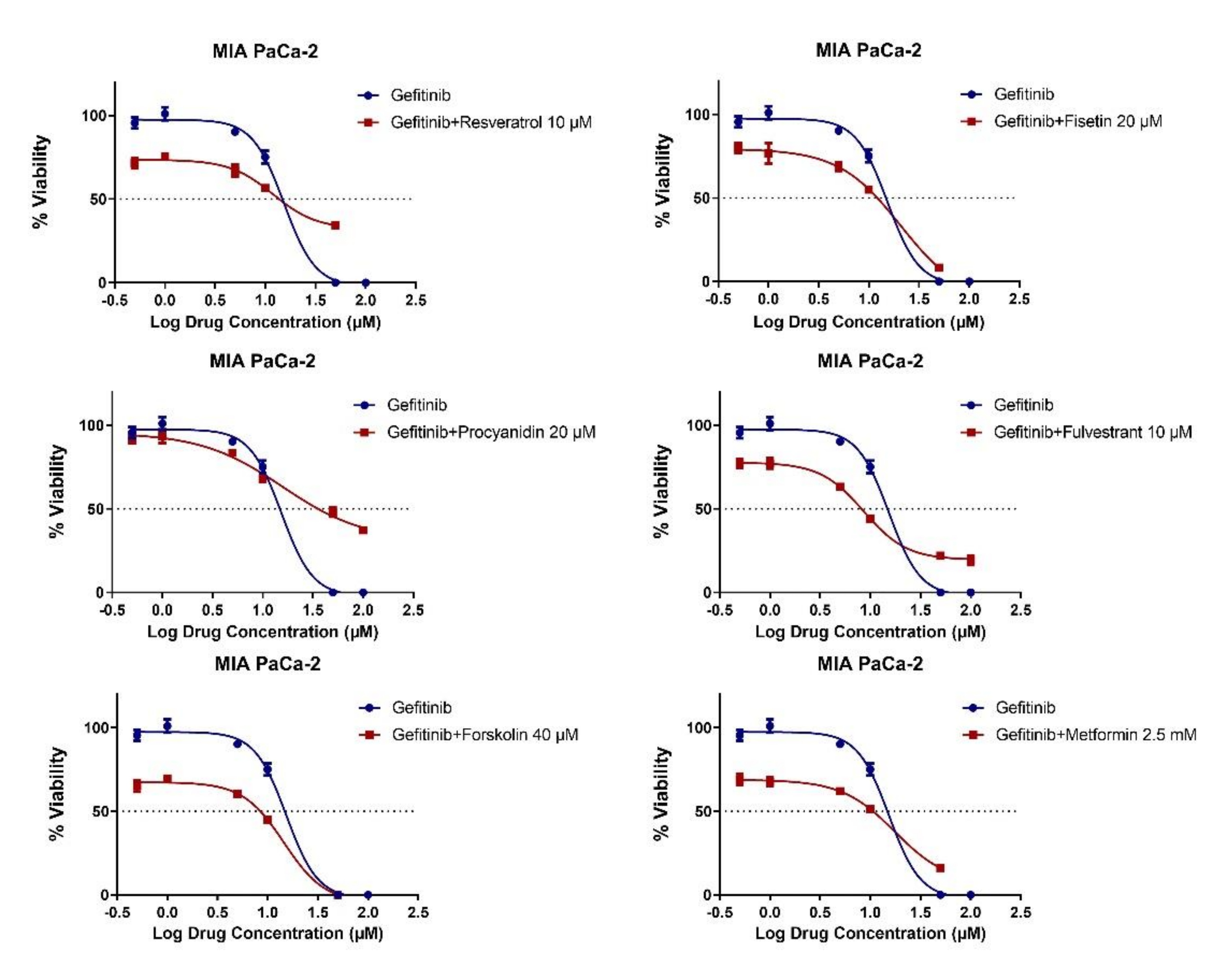
2.3. MET Induction by Natural Compounds Is Not Uniformly Related to Increased Sensitivity to Chemotherapeutics but Can Result in Occasional Synergistic or Additive Effects
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culture Conditions
4.2. Chemicals
4.3. Cell Viability Assay
4.4. Combination Index Analysis
4.5. Quantitative Real Time PCR Analysis
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Sample Availability
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Fischer, K.R.; Durrans, A.; Lee, S.; Sheng, J.; Li, F.; Wong, S.T.; Choi, H.; El Rayes, T.; Ryu, S.; Troeger, J.; et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015, 527, 472–476. [Google Scholar] [CrossRef]
- Gasiule, S.; Dreize, N.; Kaupinis, A.; Razanskas, R.; Ciupas, L.; Stankevicius, V.; Kapustina, Z.; Laurinavicius, A.; Valius, M.; Vilkaitis, G. Molecular Insights into miRNA-Driven Resistance to 5-Fluorouracil and Oxaliplatin Chemotherapy: miR-23b Modulates the Epithelial-Mesenchymal Transition of Colorectal Cancer Cells. J. Clin. Med. 2019, 8, 2115. [Google Scholar] [CrossRef]
- Song, K.A.; Niederst, M.J.; Lochmann, T.L.; Hata, A.N.; Kitai, H.; Ham, J.; Floros, K.V.; Hicks, M.A.; Hu, H.; Mulvey, H.E.; et al. Epithelial-to-Mesenchymal Transition Antagonizes Response to Targeted Therapies in Lung Cancer by Suppressing BIM. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Carstens, J.L.; Kim, J.; Scheible, M.; Kaye, J.; Sugimoto, H.; Wu, C.C.; LeBleu, V.S.; Kalluri, R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015, 527, 525–530. [Google Scholar] [CrossRef]
- Jonckheere, S.; Adams, J.; De Groote, D.; Campbell, K.; Berx, G.; Goossens, S. Epithelial-Mesenchymal Transition (EMT) as a Therapeutic Target. Cells Tissues Organs 2021, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, S.; Meng, X.; Gan, R.Y.; Zhang, J.J.; Li, H.B. Dietary Natural Products for Prevention and Treatment of Breast Cancer. Nutrients 2017, 9, 728. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Kim, B. Anti-Cancer Natural Products and Their Bioactive Compounds Inducing ER Stress-Mediated Apoptosis: A Review. Nutrients 2018, 10, 1021. [Google Scholar] [CrossRef] [PubMed]
- Fabiani, R. Antitumoral Properties of Natural Products. Molecules 2020, 25, 650. [Google Scholar] [CrossRef] [PubMed]
- Guilford, J.M.; Pezzuto, J.M. Natural products as inhibitors of carcinogenesis. Expert Opin. Investig. Drugs 2008, 17, 1341–1352. [Google Scholar] [CrossRef]
- Subramaniam, S.; Selvaduray, K.R.; Radhakrishnan, A.K. Bioactive Compounds: Natural Defense Against Cancer? Biomolecules 2019, 9, 758. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, X.; Lai, M. Modulation of epithelial-to-mesenchymal cancerous transition by natural products. Fitoterapia 2015, 106, 247–255. [Google Scholar] [CrossRef]
- Avila-Carrasco, L.; Majano, P.; Sanchez-Tomero, J.A.; Selgas, R.; Lopez-Cabrera, M.; Aguilera, A.; Gonzalez Mateo, G. Natural Plants Compounds as Modulators of Epithelial-to-Mesenchymal Transition. Front. Pharmacol. 2019, 10, 715. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, Y.; Wang, H.; Li, G.; Yan, H.; Fei, Z.; Xu, Y.; Li, W. Curcumin reverses irinotecan resistance in colon cancer cell by regulation of epithelial-mesenchymal transition. Anti-Cancer Drugs 2018, 29, 334–340. [Google Scholar] [CrossRef]
- Jin, X.; Wei, Y.; Liu, Y.; Lu, X.; Ding, F.; Wang, J.; Yang, S. Resveratrol promotes sensitization to Doxorubicin by inhibiting epithelial-mesenchymal transition and modulating SIRT1/beta-catenin signaling pathway in breast cancer. Cancer Med. 2019, 8, 1246–1257. [Google Scholar] [CrossRef]
- Bahrami, A.; Majeed, M.; Sahebkar, A. Curcumin: A potent agent to reverse epithelial-to-mesenchymal transition. Cell. Oncol. 2019, 42, 405–421. [Google Scholar] [CrossRef] [PubMed]
- Toden, S.; Okugawa, Y.; Jascur, T.; Wodarz, D.; Komarova, N.L.; Buhrmann, C.; Shakibaei, M.; Boland, C.R.; Goel, A. Curcumin mediates chemosensitization to 5-fluorouracil through miRNA-induced suppression of epithelial-to-mesenchymal transition in chemoresistant colorectal cancer. Carcinogenesis 2015, 36, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Cai, J.; Liu, J.; Han, B.; Gao, F.; Gao, W.; Zhang, Y.; Zhang, J.; Zhao, Z.; Jiang, C. Curcumin increases efficiency of gamma-irradiation in gliomas by inhibiting Hedgehog signaling pathway. Cell Cycle 2017, 16, 1181–1192. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhang, R.; Zhang, X.; Zhang, B.; Yao, Q. Curcumin may reverse 5-fluorouracil resistance on colonic cancer cells by regulating TET1-NKD-Wnt signal pathway to inhibit the EMT progress. Biomed. Pharmacother. 2020, 129, 110381. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.; Yokota, T.; Ashihara, H.; Lean, M.E.; Crozier, A. Plant foods and herbal sources of resveratrol. J. Agric. Food Chem. 2002, 50, 3337–3340. [Google Scholar] [CrossRef] [PubMed]
- Yar Saglam, A.S.; Kayhan, H.; Alp, E.; Onen, H.I. Resveratrol enhances the sensitivity of FL118 in triple-negative breast cancer cell lines via suppressing epithelial to mesenchymal transition. Mol. Biol. Rep. 2021, 48, 475–489. [Google Scholar] [CrossRef]
- Xu, J.; Liu, D.; Niu, H.; Zhu, G.; Xu, Y.; Ye, D.; Li, J.; Zhang, Q. Resveratrol reverses Doxorubicin resistance by inhibiting epithelial-mesenchymal transition (EMT) through modulating PTEN/Akt signaling pathway in gastric cancer. J. Exp. Clin. Cancer Res. CR 2017, 36, 19. [Google Scholar] [CrossRef] [PubMed]
- Baribeau, S.; Chaudhry, P.; Parent, S.; Asselin, E. Resveratrol inhibits cisplatin-induced epithelial-to-mesenchymal transition in ovarian cancer cell lines. PLoS ONE 2014, 9, e86987. [Google Scholar] [CrossRef]
- Tong, J.; Shen, Y.; Zhang, Z.; Hu, Y.; Zhang, X.; Han, L. Apigenin inhibits epithelial-mesenchymal transition of human colon cancer cells through NF-kappaB/Snail signaling pathway. Biosci. Rep. 2019, 39, BSR20190452. [Google Scholar] [CrossRef]
- Qin, Y.; Zhao, D.; Zhou, H.G.; Wang, X.H.; Zhong, W.L.; Chen, S.; Gu, W.G.; Wang, W.; Zhang, C.H.; Liu, Y.R.; et al. Apigenin inhibits NF-kappaB and snail signaling, EMT and metastasis in human hepatocellular carcinoma. Oncotarget 2016, 7, 41421–41431. [Google Scholar] [CrossRef]
- Lee, H.H.; Jung, J.; Moon, A.; Kang, H.; Cho, H. Antitumor and Anti-Invasive Effect of Apigenin on Human Breast Carcinoma through Suppression of IL-6 Expression. Int. J. Mol. Sci. 2019, 20, 3143. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.N.; Fu, J.; Shankar, S.; Srivastava, R.K. EGCG enhances the therapeutic potential of gemcitabine and CP690550 by inhibiting STAT3 signaling pathway in human pancreatic cancer. PLoS ONE 2012, 7, e31067. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gong, X.; Jiang, R.; Lin, D.; Zhou, T.; Zhang, A.; Li, H.; Zhang, X.; Wan, J.; Kuang, G.; et al. Fisetin Inhibited Growth and Metastasis of Triple-Negative Breast Cancer by Reversing Epithelial-to-Mesenchymal Transition via PTEN/Akt/GSK3beta Signal Pathway. Front. Pharmacol. 2018, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Pattabiraman, D.R.; Bierie, B.; Kober, K.I.; Thiru, P.; Krall, J.A.; Zill, C.; Reinhardt, F.; Tam, W.L.; Weinberg, R.A. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science 2016, 351, aad3680. [Google Scholar] [CrossRef]
- Li, D.; Zhao, T.; Meng, J.; Jing, Y.; Jia, F.; He, P. Procyanidin B2 inhibits high glucoseinduced epithelialmesenchymal transition in HK2 human renal proximal tubular epithelial cells. Mol. Med. Rep. 2015, 12, 8148–8154. [Google Scholar] [CrossRef]
- Zhao, W.; Shi, F.; Guo, Z.; Zhao, J.; Song, X.; Yang, H. Metabolite of ellagitannins, urolithin A induces autophagy and inhibits metastasis in human sw620 colorectal cancer cells. Mol. Carcinog. 2018, 57, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Sarrias, A.; Tome-Carneiro, J.; Bellesia, A.; Tomas-Barberan, F.A.; Espin, J.C. The ellagic acid-derived gut microbiota metabolite, urolithin A, potentiates the anticancer effects of 5-fluorouracil chemotherapy on human colon cancer cells. Food Funct. 2015, 6, 1460–1469. [Google Scholar] [CrossRef]
- Cheng, F.; Dou, J.; Zhang, Y.; Wang, X.; Wei, H.; Zhang, Z.; Cao, Y.; Wu, Z. Urolithin A Inhibits Epithelial-Mesenchymal Transition in Lung Cancer Cells via P53-Mdm2-Snail Pathway. Onco Targets Ther. 2021, 14, 3199–3208. [Google Scholar] [CrossRef]
- Qu, C.; Zhang, W.; Zheng, G.; Zhang, Z.; Yin, J.; He, Z. Metformin reverses multidrug resistance and epithelial-mesenchymal transition (EMT) via activating AMP-activated protein kinase (AMPK) in human breast cancer cells. Mol. Cell. Biochem. 2014, 386, 63–71. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Z.; Hu, L. The regulatory effects of metformin on the [SNAIL/miR-34]:[ZEB/miR-200] system in the epithelial-mesenchymal transition (EMT) for colorectal cancer (CRC). Eur. J. Pharmacol. 2018, 834, 45–53. [Google Scholar] [CrossRef]
- de Anda-Jauregui, G.; Hernandez-Lemus, E. Computational Oncology in the Multi-Omics Era: State of the Art. Front. Oncol. 2020, 10, 423. [Google Scholar] [CrossRef]
- Di Matteo, S.; Nevi, L.; Overi, D.; Landolina, N.; Faccioli, J.; Giulitti, F.; Napoletano, C.; Oddi, A.; Marziani, A.M.; Costantini, D.; et al. Metformin exerts anti-cancerogenic effects and reverses epithelial-to-mesenchymal transition trait in primary human intrahepatic cholangiocarcinoma cells. Sci. Rep. 2021, 11, 2557. [Google Scholar] [CrossRef]
- Adachi, S.I.; Sasaki, K.; Kondo, S.; Komatsu, W.; Yoshizawa, F.; Isoda, H.; Yagasaki, K. Antihyperuricemic Effect of Urolithin A in Cultured Hepatocytes and Model Mice. Molecules 2020, 25, 5136. [Google Scholar] [CrossRef]
- El-Wetidy, M.S.; Ahmad, R.; Rady, I.; Helal, H.; Rady, M.I.; Vaali-Mohammed, M.A.; Al-Khayal, K.; Traiki, T.B.; Abdulla, M.H. Urolithin A induces cell cycle arrest and apoptosis by inhibiting Bcl-2, increasing p53-p21 proteins and reactive oxygen species production in colorectal cancer cells. Cell Stress Chaperones 2021, 26, 473–493. [Google Scholar] [CrossRef] [PubMed]
- Follin-Arbelet, V.; Misund, K.; Naderi, E.H.; Ugland, H.; Sundan, A.; Blomhoff, H.K. The natural compound forskolin synergizes with dexamethasone to induce cell death in myeloma cells via BIM. Sci. Rep. 2015, 5, 13001. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, G.; Jin, G.; Yao, K.; Zhao, Z.; Bie, L.; Guo, Y.; Li, N.; Deng, W.; Chen, X.; et al. Resveratrol suppresses colon cancer growth by targeting the AKT/STAT3 signaling pathway. Int. J. Mol. Med. 2019, 43, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Scholz, C.; Zang, C.; Schefe, J.H.; Habbel, P.; Regierer, A.C.; Schulz, C.O.; Possinger, K.; Eucker, J. Metformin and the mTOR inhibitor everolimus (RAD001) sensitize breast cancer cells to the cytotoxic effect of chemotherapeutic drugs in vitro. Anticancer Res. 2012, 32, 1627–1637. [Google Scholar]
- Liu, Y.S.; Chang, Y.C.; Kuo, W.W.; Chen, M.C.; Hsu, H.H.; Tu, C.C.; Yeh, Y.L.; Viswanadha, V.P.; Liao, P.H.; Huang, C.Y. Inhibition of protein phosphatase 1 stimulates noncanonical ER stress eIF2alpha activation to enhance fisetin-induced chemosensitivity in HDAC inhibitor-resistant hepatocellular carcinoma cells. Cancers 2019, 11, 918. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Guo, X.; Wang, H.; Zhou, T.; Wang, X. Differences in the Effects of EGCG on Chromosomal Stability and Cell Growth between Normal and Colon Cancer Cells. Molecules 2018, 23, 788. [Google Scholar] [CrossRef]
- Park, J.B. The effects of fulvestrant, an estrogen receptor antagonist, on the proliferation, differentiation and mineralization of osteoprecursor cells. Mol. Med. Rep. 2013, 7, 555–558. [Google Scholar] [CrossRef]
- Safari, Z.; Safaralizadeh, R.; Seyedzadeh, M.H.; Valinezad Orang, A.; Zare, A.; Hosseinpour Feizi, M.A.; Kardar, G.A. The Induction of Metformin Inhibitory Effects on Tumor Cell Growth in Hypoxic Condition. Iran. J. Allergy Asthma Immunol. 2015, 14, 605–614. [Google Scholar]
- Shilpi, A.; Parbin, S.; Sengupta, D.; Kar, S.; Deb, M.; Rath, S.K.; Pradhan, N.; Rakshit, M.; Patra, S.K. Mechanisms of DNA methyltransferase-inhibitor interactions: Procyanidin B2 shows new promise for therapeutic intervention of cancer. Chem. Biol. Interact. 2015, 233, 122–138. [Google Scholar] [CrossRef]
- Smith, M.L.; Murphy, K.; Doucette, C.D.; Greenshields, A.L.; Hoskin, D.W. The Dietary Flavonoid Fisetin Causes Cell Cycle Arrest, Caspase-Dependent Apoptosis, and Enhanced Cytotoxicity of Chemotherapeutic Drugs in Triple-Negative Breast Cancer Cells. J. Cell Biochem. 2016, 117, 1913–1925. [Google Scholar] [CrossRef]
- Sutcliffe, T.C.; Winter, A.N.; Punessen, N.C.; Linseman, D.A. Procyanidin B2 Protects Neurons from Oxidative, Nitrosative, and Excitotoxic Stress. Antioxidants 2017, 6, 77. [Google Scholar] [CrossRef]
- Syng-Ai, C.; Kumari, A.L.; Khar, A. Effect of curcumin on normal and tumor cells: Role of glutathione and bcl-2. Mol. Cancer Ther. 2004, 3, 1101–1108. [Google Scholar] [PubMed]
- van Ginkel, P.R.; Sareen, D.; Subramanian, L.; Walker, Q.; Darjatmoko, S.R.; Lindstrom, M.J.; Kulkarni, A.; Albert, D.M.; Polans, A.S. Resveratrol inhibits tumor growth of human neuroblastoma and mediates apoptosis by directly targeting mitochondria. Clin. Cancer Res. 2007, 13, 5162–5169. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Qi, M.; Li, P.; Zhan, Y.; Shao, H. Apigenin in cancer therapy: Anti-cancer effects and mechanisms of action. Cell Biosci. 2017, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Sun, H.; Zha, W.; Cui, W.; Xu, L.; Min, Q.; Wu, J. Apigenin Attenuates Adriamycin-Induced Cardiomyocyte Apoptosis via the PI3K/AKT/mTOR Pathway. Evid.-Based Complement. Alternat. Med. 2017, 2017, 2590676. [Google Scholar] [CrossRef] [PubMed]
- Costea, T.; Hudita, A.; Ciolac, O.A.; Galateanu, B.; Ginghina, O.; Costache, M.; Ganea, C.; Mocanu, M.M. Chemoprevention of Colorectal Cancer by Dietary Compounds. Int. J. Mol. Sci. 2018, 19, 3787. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. [Google Scholar] [CrossRef]
- Lohse, I.; Wildermuth, E.; Brothers, S.P. Naturally occurring compounds as pancreatic cancer therapeutics. Oncotarget 2018, 9, 35448–35457. [Google Scholar] [CrossRef][Green Version]
- Yue, Q.; Gao, G.; Zou, G.; Yu, H.; Zheng, X. Natural Products as Adjunctive Treatment for Pancreatic Cancer: Recent Trends and Advancements. BioMed Res. Int. 2017, 2017, 8412508. [Google Scholar] [CrossRef]
- Yilmaz-Ozcan, S.; Sade, A.; Kucukkaraduman, B.; Kaygusuz, Y.; Senses, K.M.; Banerjee, S.; Gure, A.O. Epigenetic mechanisms underlying the dynamic expression of cancer-testis genes, PAGE2-2B and SPANX-B, during mesenchymal-to-epithelial transition. PLoS ONE 2014, 9, e107905. [Google Scholar] [CrossRef]
- Akbar, M.W.; Isbilen, M.; Belder, N.; Canli, S.D.; Kucukkaraduman, B.; Turk, C.; Sahin, O.; Gure, A.O. A Stemness and EMT Based Gene Expression Signature Identifies Phenotypic Plasticity and is A Predictive but Not Prognostic Biomarker for Breast Cancer. J. Cancer 2020, 11, 949–961. [Google Scholar] [CrossRef]
- Goodwin, R.A.; Asmis, T.R. Overview of systemic therapy for colorectal cancer. Clin. Colon Rectal Surg. 2009, 22, 251–256. [Google Scholar] [CrossRef]
- Guglielmi, A.P.; Sobrero, A.F. Second-line therapy for advanced colorectal cancer. Gastrointest. Cancer Res. GCR 2007, 1, 57–63. [Google Scholar]
- Mohammed, S.; Van Buren, G., 2nd; Fisher, W.E. Pancreatic cancer: Advances in treatment. World J. Gastroenterol. 2014, 20, 9354–9360. [Google Scholar] [CrossRef]
- Binenbaum, Y.; Na’ara, S.; Gil, Z. Gemcitabine resistance in pancreatic ductal adenocarcinoma. Drug Resist. Updates Rev. Comment. Antimicrob. Anticancer. Chemother. 2015, 23, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.; Janakiram, N.B.; Li, Q.; Madka, V.; Ely, M.; Lightfoot, S.; Crawford, H.; Steele, V.E.; Rao, C.V. The epidermal growth factor receptor inhibitor gefitinib prevents the progression of pancreatic lesions to carcinoma in a conditional LSL-KrasG12D/+ transgenic mouse model. Cancer Prev. Res. 2010, 3, 1417–1426. [Google Scholar] [CrossRef] [PubMed]
- Ramos, P.; Bentires-Alj, M. Mechanism-based cancer therapy: Resistance to therapy, therapy for resistance. Oncogene 2015, 34, 3617–3626. [Google Scholar] [CrossRef] [PubMed]
- Boumahdi, S.; de Sauvage, F.J. The great escape: Tumour cell plasticity in resistance to targeted therapy. Nat. Rev. Drug Discov. 2020, 19, 39–56. [Google Scholar] [CrossRef]
- Bukowski, K.; Kciuk, M.; Kontek, R. Mechanisms of Multidrug Resistance in Cancer Chemotherapy. Int. J. Mol. Sci. 2020, 21, 3233. [Google Scholar] [CrossRef]
- Smith, B.N.; Bhowmick, N.A. Role of EMT in Metastasis and Therapy Resistance. J. Clin. Med. 2016, 5, 17. [Google Scholar] [CrossRef]
- de Oliveira Junior, R.G.; Christiane Adrielly, A.F.; da Silva Almeida, J.R.G.; Grougnet, R.; Thiery, V.; Picot, L. Sensitization of tumor cells to chemotherapy by natural products: A systematic review of preclinical data and molecular mechanisms. Fitoterapia 2018, 129, 383–400. [Google Scholar] [CrossRef]
- Britton, R.G.; Kovoor, C.; Brown, K. Direct molecular targets of resveratrol: Identifying key interactions to unlock complex mechanisms. Ann. N. Y. Acad. Sci. 2015, 1348, 124–133. [Google Scholar] [CrossRef]
- Byers, L.A.; Diao, L.; Wang, J.; Saintigny, P.; Girard, L.; Peyton, M.; Shen, L.; Fan, Y.; Giri, U.; Tumula, P.K.; et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin. Cancer Res. 2013, 19, 279–290. [Google Scholar] [CrossRef]
- George, J.T.; Jolly, M.K.; Xu, S.; Somarelli, J.A.; Levine, H. Survival Outcomes in Cancer Patients Predicted by a Partial EMT Gene Expression Scoring Metric. Cancer Res. 2017, 77, 6415–6428. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.Z.; Miow, Q.H.; Miki, Y.; Noda, T.; Mori, S.; Huang, R.Y.; Thiery, J.P. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol. Med. 2014, 6, 1279–1293. [Google Scholar] [CrossRef]
- Chakraborty, P.; George, J.T.; Tripathi, S.; Levine, H.; Jolly, M.K. Comparative Study of Transcriptomics-Based Scoring Metrics for the Epithelial-Hybrid-Mesenchymal Spectrum. Front. Bioeng. Biotechnol. 2020, 8, 220. [Google Scholar] [CrossRef] [PubMed]
- Park, S.M.; Gaur, A.B.; Lengyel, E.; Peter, M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008, 22, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Lyu, J.; Dunlap, M.; Harvey, S.E.; Cheng, C. A combinatorially regulated RNA splicing signature predicts breast cancer EMT states and patient survival. RNA 2020, 26, 1257–1267. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
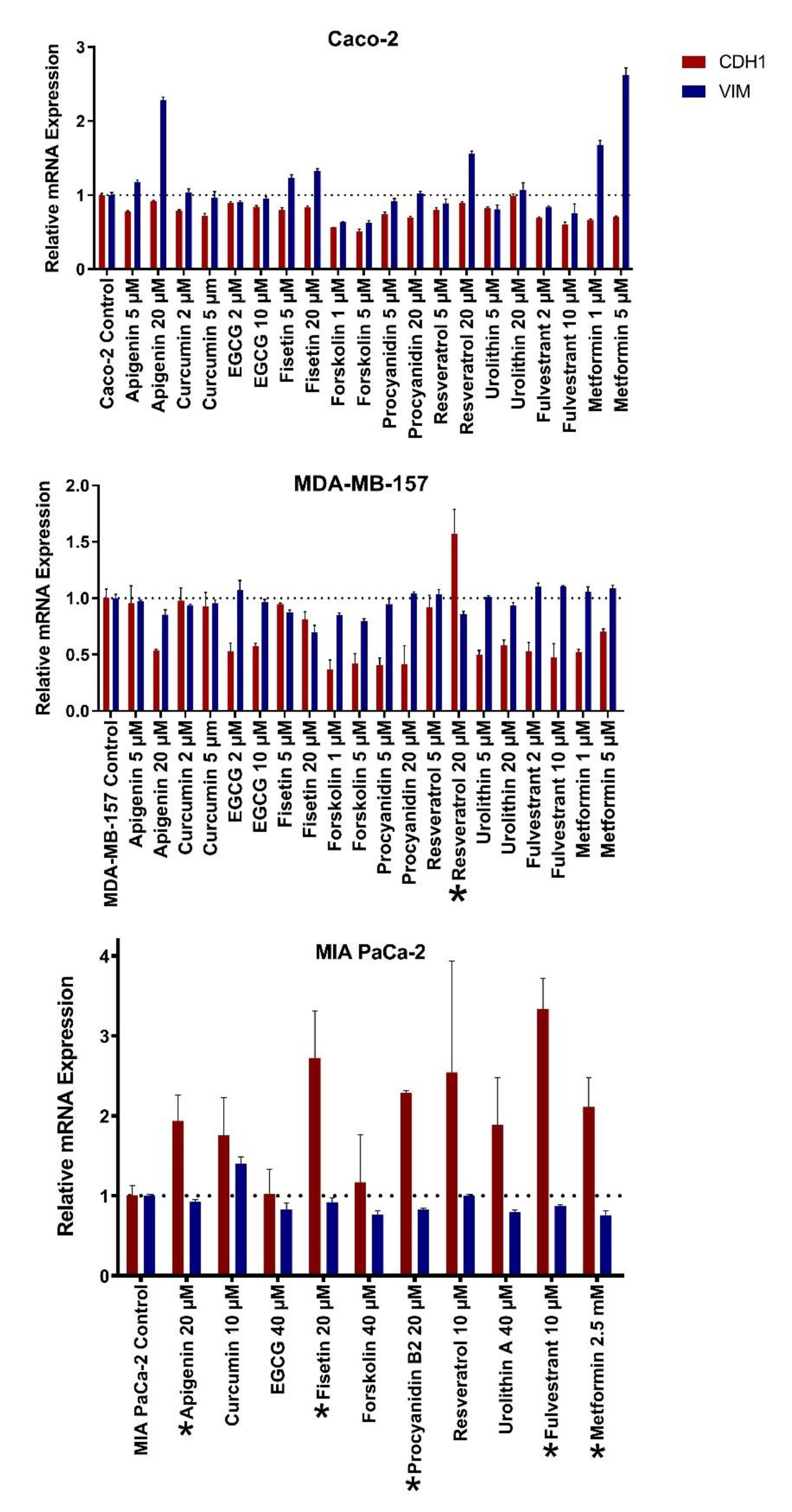
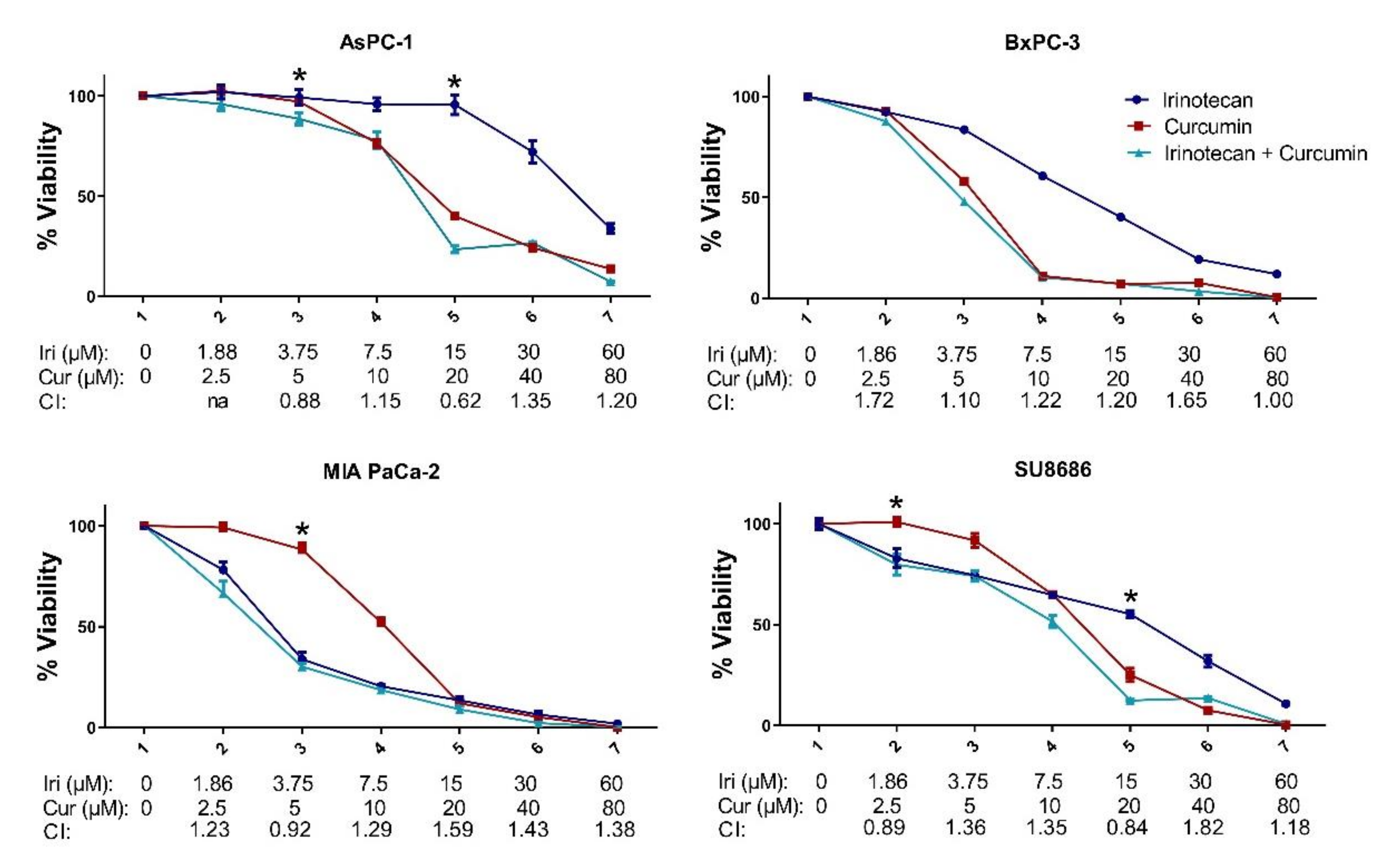
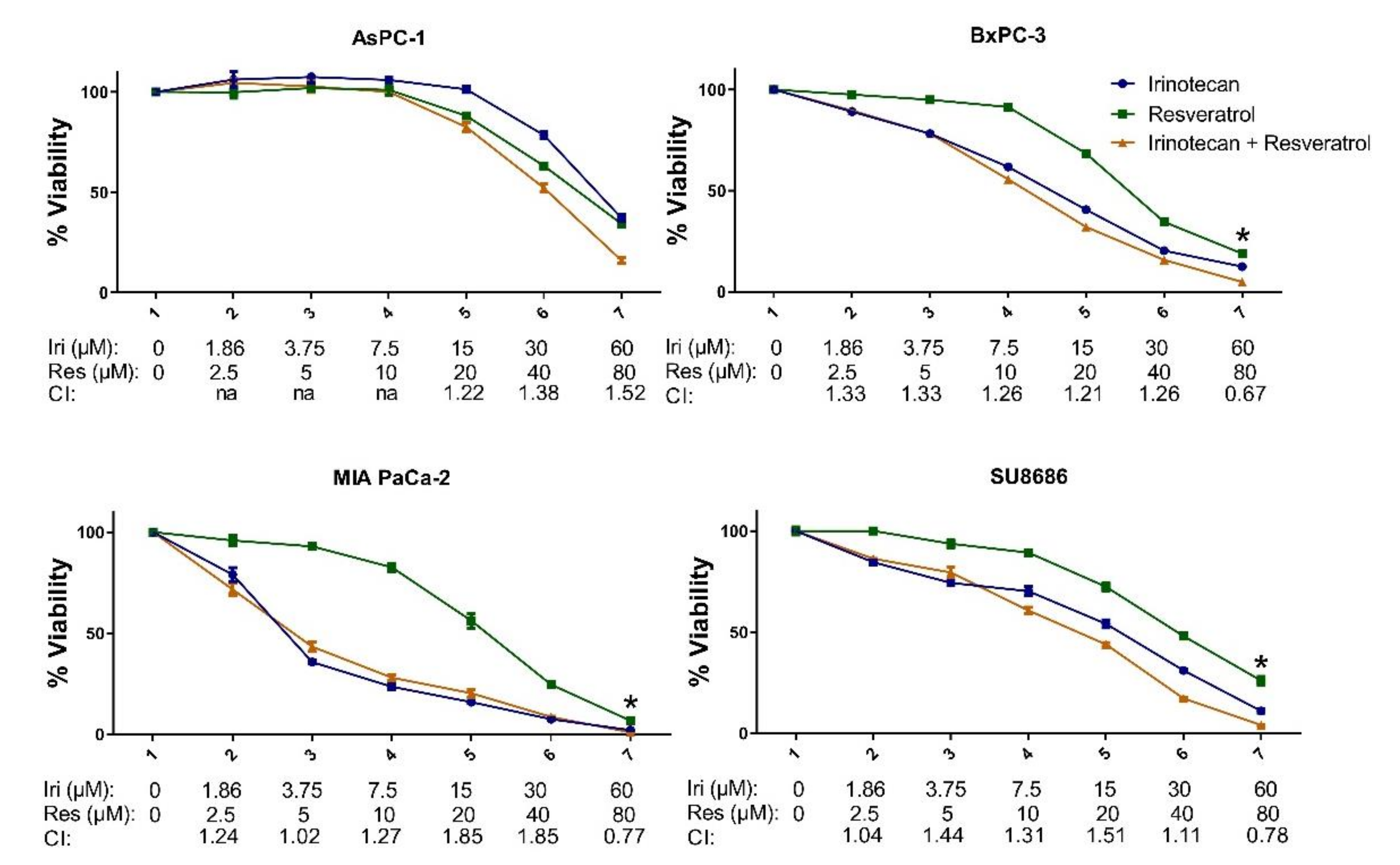
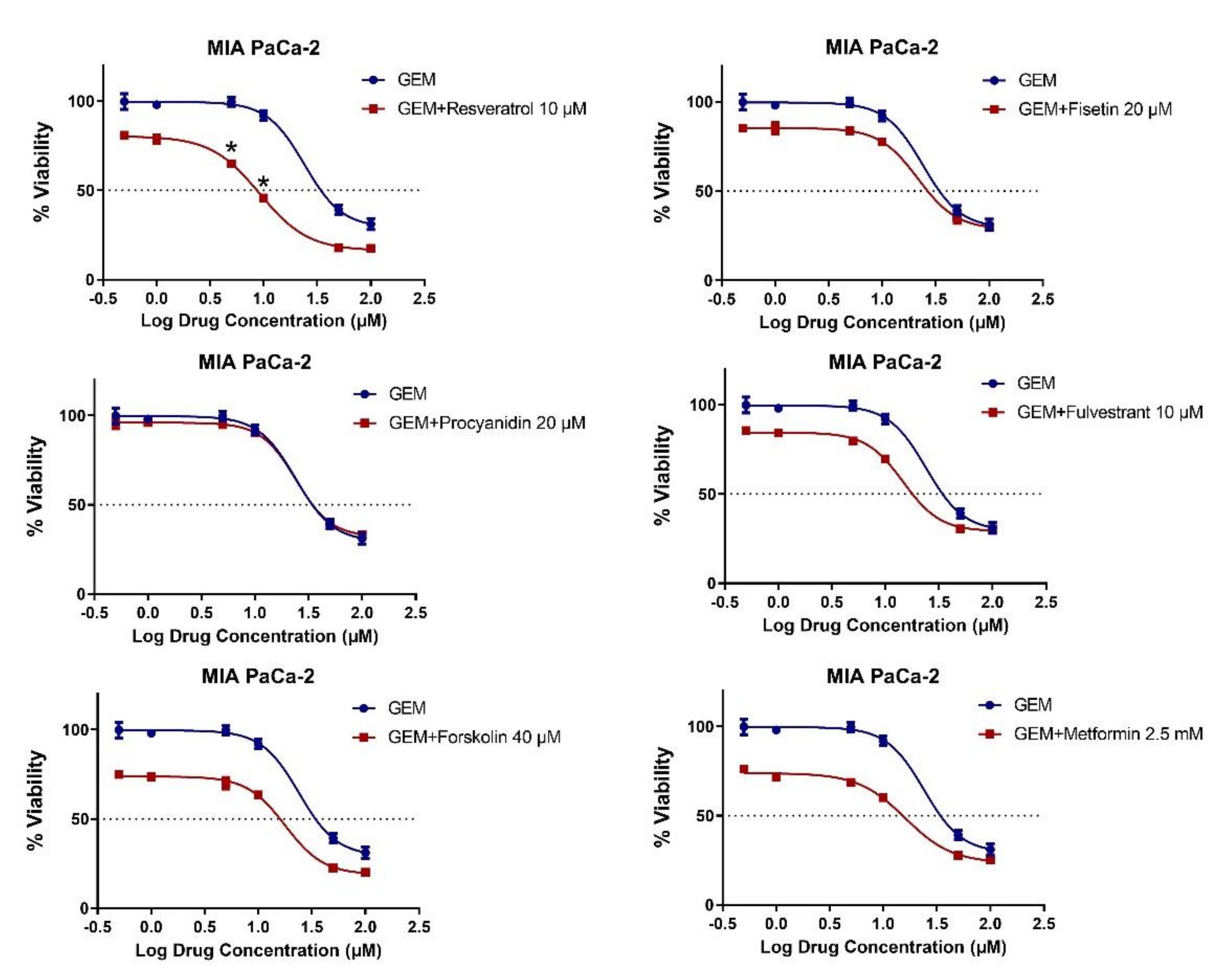
| CAL-51 | MDA-MB-157 | MDA-MB-231 | MDA-MB-436 | MDA-MB-453 | MDA-MB-468 | |
|---|---|---|---|---|---|---|
| Apigenin | 34 ± 2.9 * | 78 ± 4.6 | >100 | 52 ± 4.2 | 36 ± 5.8 | 43 ± 13 |
| Curcumin | 24 ± 3.4 | 23 ± 1.2 | 40 ± 2.7 | 12 ± 2.2 | 14 ± 3.2 | 30 ± 3.8 |
| EGCG | 80 ± 3.2 | >100 | 79 ± 3.6 | >100 | 85 ± 3.8 | 83 ± 3.1 |
| Fisetin | 41 ± 7.6 | 39 ± 2.1 | >100 | 86 ± 9.3 | 72 ± 5.1 | >100 |
| Forskolin | 48 ± 3.3 | >100 | >100 | >100 | 38 ± 5.3 | >100 |
| Procyanidin B2 | >100 | >100 | >100 | >100 | >100 | >100 |
| Resveratrol | 59 ± 4.4 | 65 ± 5.6 | 33 ± 2.6 | 54 ± 5.7 | 64 ± 7.8 | 30 ± 5.7 |
| Urolithin A | >100 | >100 | >100 | >100 | 57 ± 3.3 | >100 |
| Fulvestrant | >100 | >100 | >100 | >100 | 29 ± 7.5 | >100 |
| Metformin(μM) | >100 | >100 | >100 | >100 | >100 | >100 |
| HCT8 | KM12 | LS513 | LoVo | SW837 | WiDr | |
|---|---|---|---|---|---|---|
| Apigenin | 19 ± 2.9 * | 26 ± 2.3 | 12 ± 3.9 | 28 ± 6.8 | 50 ± 3.6 | 40 ± 8 |
| Curcumin | 12 ± 1.1 | 10 ± 1.9 | 6 ± 0.6 | 9 ± 0.6 | 15 ± 1.6 | 8 ± 0.5 |
| EGCG | 14 ± 2.6 | 17 ± 1.7 | 17 ± 1.9 | 23 ± 2.9 | 15 ± 2.1 | 20 ± 2.9 |
| Fisetin | 35 ± 6.9 | 16 ± 3.1 | 26 ± 3.3 | 24 ± 3.1 | 38 ± 4.3 | 66 ± 7.9 |
| Forskolin | >100 | 4 ± 0.3 | >100 | >100 | >100 | >100 |
| Procyanidin B2 | >100 | >100 | >100 | >100 | >100 | >100 |
| Resveratrol | 35 ± 5.1 | 47 ± 4.6 | 24 ± 1.9 | 27 ± 3.3 | 25 ± 1.8 | 39 ± 2.7 |
| Urolithin A | 40 ± 3.3 | >100 | 30 ± 3.1 | 28 ± 2.7 | 40 ± 2.2 | 52 ± 3 |
| Fulvestrant | >100 | >100 | >100 | >100 | >100 | >100 |
| Metformin(mM) | 2 ± 1.2 | 13 ± 4.1 | 2 ± 2.1 | 10 ± 2.5 | 23 ± 2.7 | 10 ± 3.3 |
| IC50 (μM) | AsPC-1 | BxPC-3 | MIA PaCa-2 | SU8686 |
|---|---|---|---|---|
| Apigenin | 34 ± 1.3 * | 18 ± 2.3 | 40 ± 5.4 | 21 ± 0.9 |
| Curcumin | 18 ± 2.2 | 7 ± 0.4 | 11 ± 0.8 | 17 ± 1.7 |
| EGCG | 35 ± 7.6 | 26 ± 3.7 | 87 ± 10.2 | 22 ± 2.4 |
| Fisetin | 90 ± 7.7 | 25 ± 1.1 | 38 ± 4.7 | 45 ± 11.9 |
| Forskolin | >100 | 45 ± 3.1 | >100 | >100 |
| Procyanidin B2 | >100 | >100 | >100 | >100 |
| Resveratrol | 65 ± 4.4 | 32 ± 1.7 | 23 ± 1.8 | 40 ± 4.1 |
| Urolithin A | >100 | 92 ± 4.8 | 96 ± 1.8 | >100 |
| Fulvestrant | >100 | >100 | >100 | >100 |
| Metformin(mM) | 40 ± 2.1 | 7 ± 1.2 | 27 ± 3.7 | 4 ± 5.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kucukkaraduman, B.; Cicek, E.G.; Akbar, M.W.; Demirkol Canli, S.; Vural, B.; Gure, A.O. Epithelial-to-Mesenchymal Transition Is Not a Major Modulating Factor in the Cytotoxic Response to Natural Products in Cancer Cell Lines. Molecules 2021, 26, 5858. https://doi.org/10.3390/molecules26195858
Kucukkaraduman B, Cicek EG, Akbar MW, Demirkol Canli S, Vural B, Gure AO. Epithelial-to-Mesenchymal Transition Is Not a Major Modulating Factor in the Cytotoxic Response to Natural Products in Cancer Cell Lines. Molecules. 2021; 26(19):5858. https://doi.org/10.3390/molecules26195858
Chicago/Turabian StyleKucukkaraduman, Baris, Ekin Gokce Cicek, Muhammad Waqas Akbar, Secil Demirkol Canli, Burcak Vural, and Ali Osmay Gure. 2021. "Epithelial-to-Mesenchymal Transition Is Not a Major Modulating Factor in the Cytotoxic Response to Natural Products in Cancer Cell Lines" Molecules 26, no. 19: 5858. https://doi.org/10.3390/molecules26195858
APA StyleKucukkaraduman, B., Cicek, E. G., Akbar, M. W., Demirkol Canli, S., Vural, B., & Gure, A. O. (2021). Epithelial-to-Mesenchymal Transition Is Not a Major Modulating Factor in the Cytotoxic Response to Natural Products in Cancer Cell Lines. Molecules, 26(19), 5858. https://doi.org/10.3390/molecules26195858






