Exploiting Complex Fluorophore Interactions to Monitor Virus Capsid Disassembly
Abstract
:1. Introduction
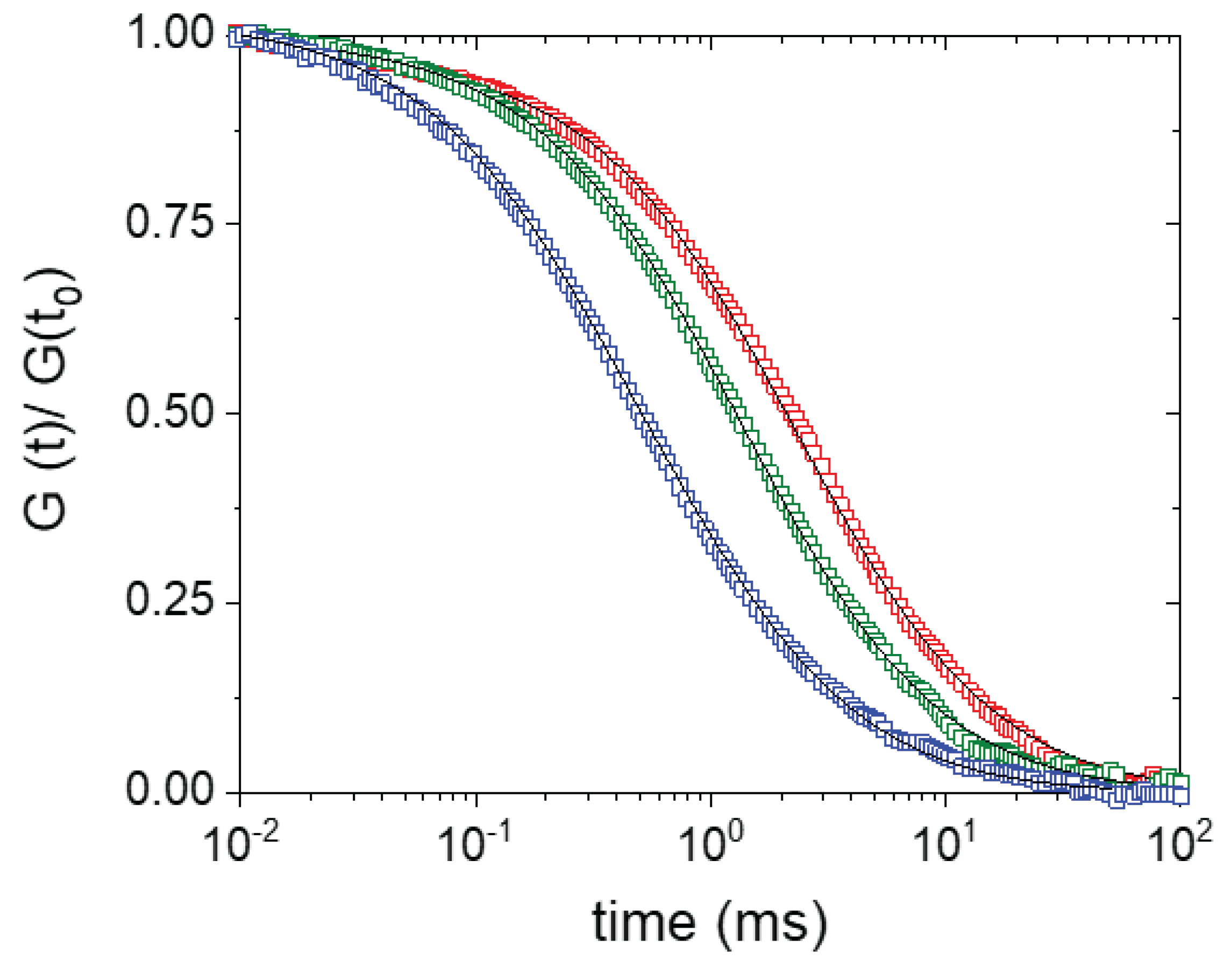
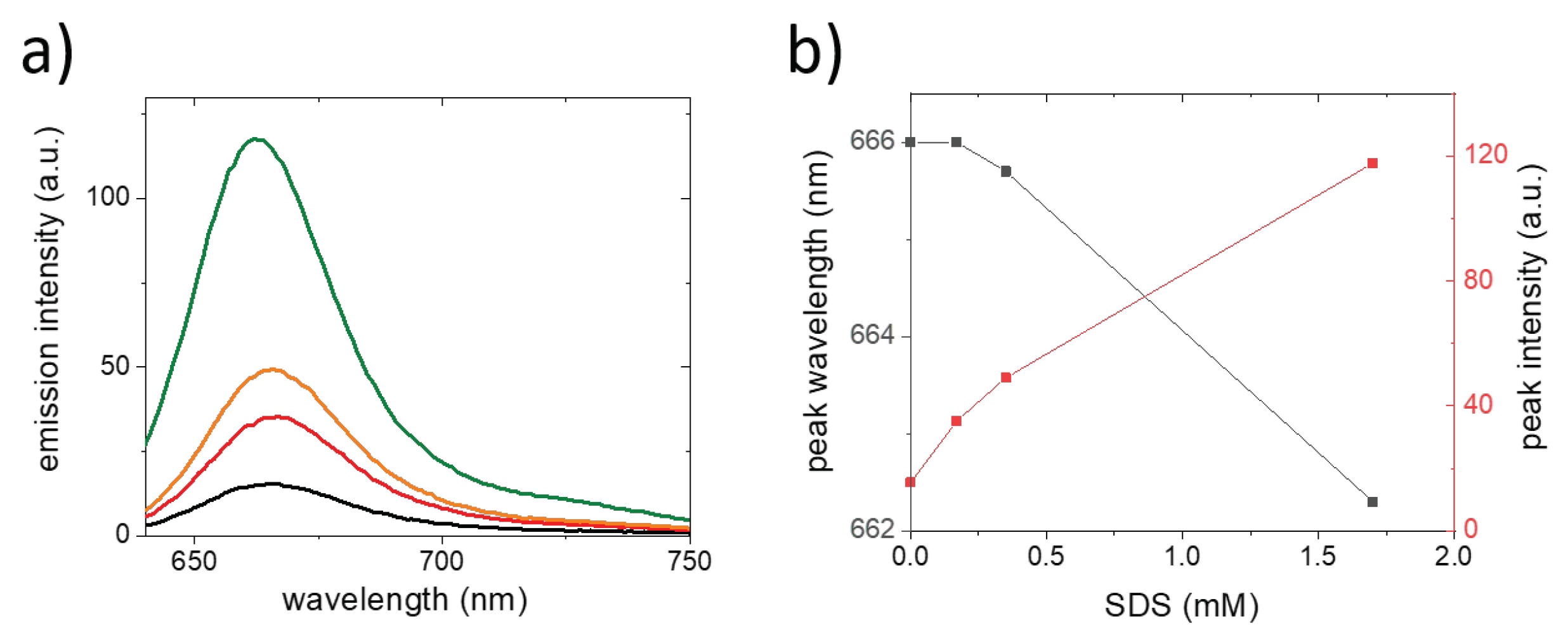
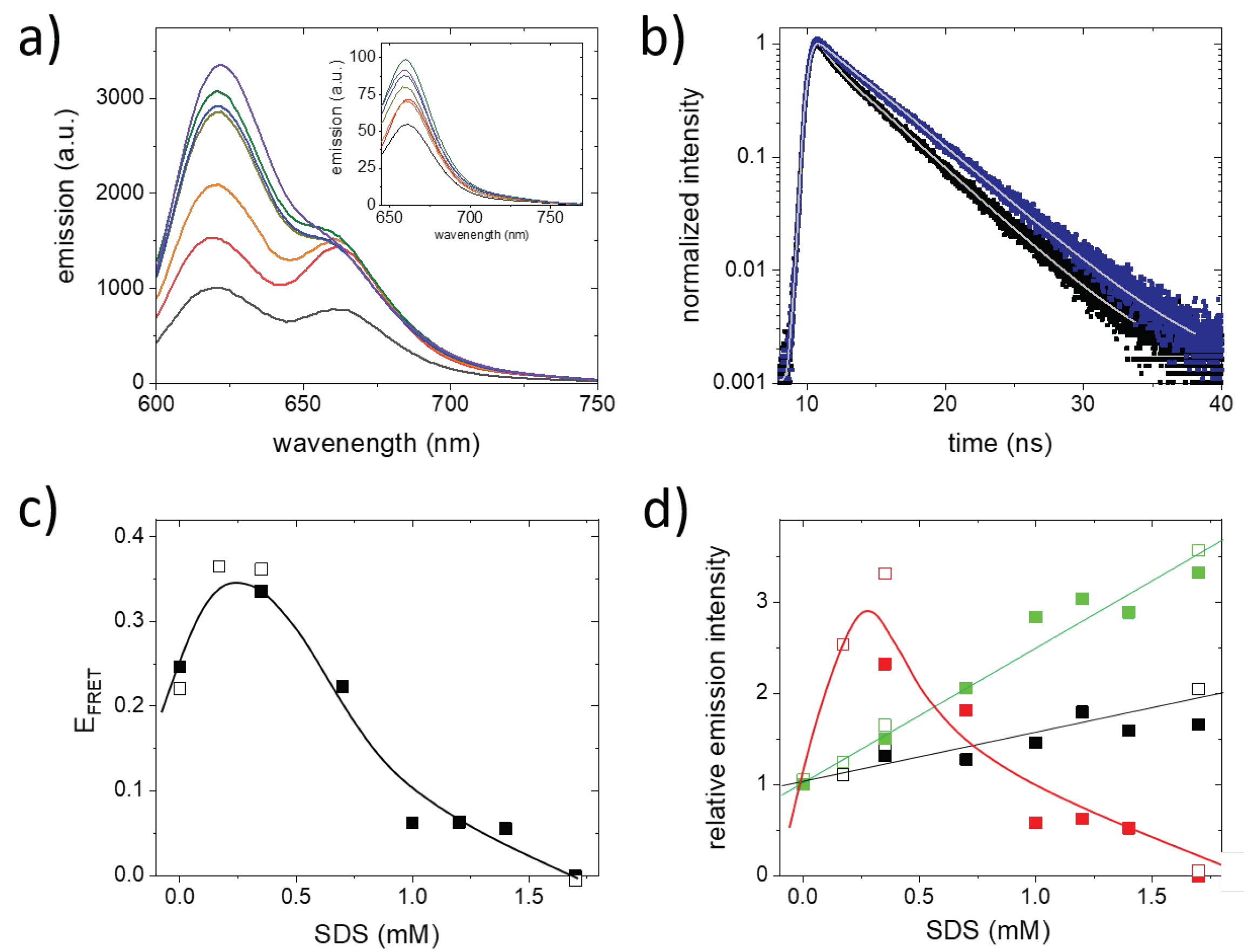
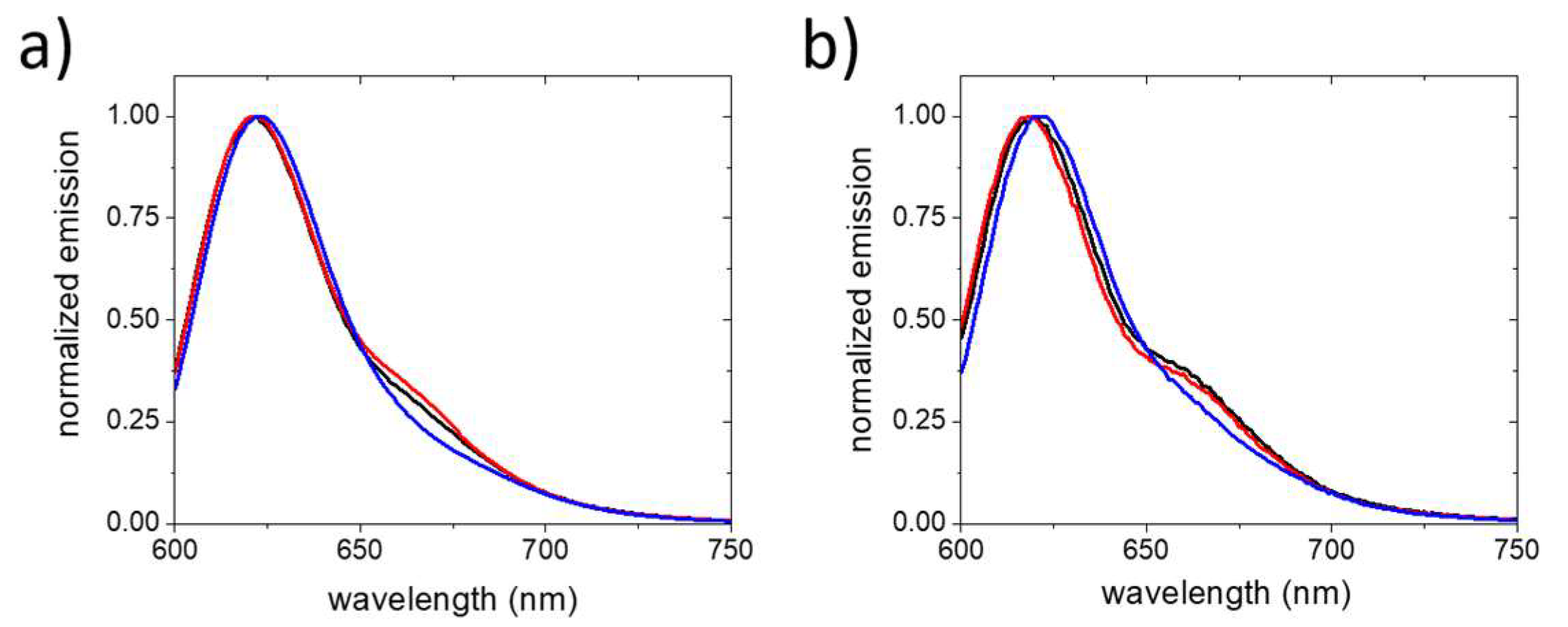
2. Results and Discussion
3. Materials and Methods
3.1. Preparation of Fluorescently Labeled CCMV
3.2. Characterization of Fluorescently Labeled CCMV
3.3. Virus Capsid Disassembly
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Fakhree, M.A.A.; Konings, I.B.M.; Kole, J.; Cambi, A.; Blum, C.; Claessens, M. The Localization of Alpha-synuclein in the Endocytic Pathway. Neuroscience 2021, 457, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Fakhree, M.A.A.; Zijlstra, N.; Raiss, C.C.; Siero, C.J.; Grabmayr, H.; Bausch, A.R.; Blum, C.; Claessens, M. The number of alpha-synuclein proteins per vesicle gives insights into its physiological function. Sci. Rep. 2016, 6, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lippincott-Schwartz, J.; Snapp, E.; Kenworthy, A. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2001, 2, 444–456. [Google Scholar] [CrossRef]
- Marczynski, M.; Rickert, C.A.; Semerdzhiev, S.A.; van Dijk, W.R.; Segers-Nolten, I.M.J.; Claessens, M.; Lieleg, O. alpha-Synuclein Penetrates Mucin Hydrogels Despite Its Mucoadhesive Properties. Biomacromolecules 2019, 20, 4332–4344. [Google Scholar] [CrossRef]
- Sigal, Y.M.; Zhou, R.B.; Zhuang, X.W. Visualizing and discovering cellular structures with super-resolution microscopy. Science 2018, 361, 880–887. [Google Scholar] [CrossRef] [Green Version]
- Weiss, S. Fluorescence spectroscopy of single biomolecules. Science 1999, 283, 1676–1683. [Google Scholar] [CrossRef] [Green Version]
- Gaur, P.; Galkin, M.; Kurochka, A.; Ghosh, S.; Yushchenko, D.A.; Shvadchak, V.V. Fluorescent Probe for Selective Imaging of a-Synuclein Fibrils in Living Cells. ACS Chem. Neurosci. 2021, 12, 1293–1298. [Google Scholar] [CrossRef]
- Klymchenko, A.S. Solvatochromic and Fluorogenic Dyes as Environment-Sensitive Probes: Design and Biological Applications. Acc. Chem. Res. 2017, 50, 366–375. [Google Scholar] [CrossRef] [Green Version]
- Semerdzhiev, S.A.; Shvadchak, V.V.; Subramaniam, V.; Claessens, M. Non-uniform self-assembly: On the anisotropic architecture of alpha-synuclein supra-fibrillar aggregates. Sci. Rep. 2017, 7, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fakhree, M.A.A.; Nolten, I.S.; Blum, C.; Claessens, M. Different Conformational Subensembles of the Intrinsically Disordered Protein alpha-Synuclein in Cells. J. Phys. Chem. Lett. 2018, 9, 1249–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jares-Erijman, E.A.; Jovin, T.M. FRET imaging. Nat. Biotechnol. 2003, 21, 1387–1395. [Google Scholar] [CrossRef]
- Selvin, P.R. The renaissance of fluorescence resonance energy transfer. Nat. Struct. Biol. 2000, 7, 730–734. [Google Scholar] [CrossRef]
- Pieters, B.; van Eldijk, M.B.; Nolte, R.J.M.; Mecinovic, J. Natural supramolecular protein assemblies. Chem. Soc. Rev. 2016, 45, 24–39. [Google Scholar] [CrossRef] [Green Version]
- Garmann, R.F.; Comas-Garcia, M.; Gopal, A.; Knobler, C.M.; Gelbart, W.M. The Assembly Pathway of an Icosahedral Single-Stranded RNA Virus Depends on the Strength of Inter-Subunit Attractions. J. Mol. Biol. 2014, 426, 1050–1060. [Google Scholar] [CrossRef] [Green Version]
- Schröder, T.; Scheible, M.B.; Steiner, F.; Vogelsang, J.; Tinnefeld, P. Interchromophoric Interactions Determine the Maximum Brightness Density in DNA Origami Structures. Nano Lett. 2019, 19, 1275–1281. [Google Scholar] [CrossRef] [Green Version]
- Mateu, M.G. Assembly, stability and dynamics of virus capsids. Arch. Biochem. Biophys. 2013, 531, 65–79. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.J.; Nolte, R.J.M.; Cornelissen, J. Virus-based nanocarriers for drug delivery. Adv. Drug Deliv. Rev. 2012, 64, 811–825. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Hsiao, S.C.; Carrico, Z.M.; Francis, M.B. Genome-Free Viral Capsids as Multivalent Carriers for Taxol Delivery. Angew. Chem.Int. Edit. 2009, 48, 9493–9497. [Google Scholar] [CrossRef]
- Minten, I.J.; Nolte, R.J.M.; Cornelissen, J. Complex Assembly Behavior During the Encapsulation of Green Fluorescent Protein Analogs in Virus Derived Protein Capsules. Macromol. Biosci. 2010, 10, 539–545. [Google Scholar] [CrossRef] [PubMed]
- De Hoog, H.P.M.; Arends, I.; Rowan, A.E.; Cornelissen, J.; Nolte, R.J.M. A hydrogel-based enzyme-loaded polymersome reactor. Nanoscale 2010, 2, 709–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, R.L.C.; Robinson, M.D.M.; Ajabali, A.A.A.; Karunanithy, G.; Lyons, B.; Raj, R.; Raoufmoghaddam, S.; Mohammed, S.; Claridge, T.D.W.; Baldwin, A.J.; et al. Monitoring the Disassembly of Virus-like Particles by F-19-NMR. J. Am. Chem. Soc. 2017, 139, 5277–5280. [Google Scholar] [CrossRef]
- Mertens, B.S.; Velev, O.D. Characterization and control of surfactant-mediated Norovirus interactions. Soft Matter 2015, 11, 8621–8631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wobus, C.E.; Hugle-Dorr, B.; Girod, A.; Petersen, G.; Hallek, M.; Kleinschmidt, J.A. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: Epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J. Virol. 2000, 74, 9281–9293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamza, I.A.; Jurzik, L.; Uberla, K.; Wilhelm, M. Methods to detect infectious human enteric viruses in environmental water samples. Int. J. Hyg. Environ. Health. 2011, 214, 424–436. [Google Scholar] [CrossRef]
- Leisi, R.; Bieri, J.; Roth, N.J.; Ros, C. Determination of parvovirus retention profiles in virus filter membranes using laser scanning microscopy. J. Membr. Sci. 2020, 603, 14. [Google Scholar] [CrossRef]
- Lu, R.Q.; Zhang, C.; Piatkovsky, M.; Ulbricht, M.; Herzberg, M.; Nguyen, T.H. Improvement of virus removal using ultrafiltration membranes modified with grafted zwitterionic polymer hydrogels. Water Res. 2017, 116, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, T.R.; Patil, A.; Raza, B.G.; Reurink, D.; van den Hengel, S.K.; Rutjes, S.A.; Husman, A.M.D.; Roesink, H.D.W.; de Vos, W.M. Cationically modified membranes using covalent layer-by-layer assembly for antiviral applications in drinking water. J. Membr. Sci. 2019, 570, 494–503. [Google Scholar] [CrossRef]
- Asculai, S.S.; Weis, M.T.; Rancourt, M.W.; Kupferberg, A.B. INACTIVATION OF HERPES-SIMPLEX VIRUSES BY NON-IONIC SURFACTANTS. Antimicrob. Agents Chemother. 1978, 13, 686–690. [Google Scholar] [CrossRef] [Green Version]
- Berlier, J.E.; Rothe, A.; Buller, G.; Bradford, J.; Gray, D.R.; Filanoski, B.J.; Telford, W.G.; Yue, S.; Liu, J.X.; Cheung, C.Y.; et al. Quantitative comparison of long-wavelength Alexa Fluor dyes to Cy dyes: Fluorescence of the dyes and their bioconjugates. J. Histochem. Cytochem. 2003, 51, 1699–1712. [Google Scholar] [CrossRef] [Green Version]
- Luchowski, R.; Matveeva, E.G.; Gryczynski, I.; Terpetschnig, E.A.; Patsenker, L.; Laczko, G.; Borejdo, J.; Gryczynski, Z. Single Molecule Studies of Multiple-Fluorophore Labeled Antibodies. Effect of Homo-FRET on the Number of Photons Available Before Photobleaching. Curr. Pharm. Biotechnol. 2008, 9, 411–420. [Google Scholar] [CrossRef] [Green Version]
- Szabo, A.; Szendi-Szatmari, T.; Ujlaky-Nagy, L.; Radi, I.; Vereb, G.; Szollosi, J.; Nagy, P. The Effect of Fluorophore Conjugation on Antibody Affinity and the Photophysical Properties of Dyes. Biophys. J. 2018, 114, 688–700. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.F.; Knutson, J.R. Mechanism of fluorescence concentration quenching of carboxyfluorescein in liposomes: Energy transfer to nonfluorescent dimers. Anal. Biochem. 1988, 172, 61–77. [Google Scholar] [CrossRef]
- Buschmann, V.; Weston, K.D.; Sauer, M. Spectroscopic Study and Evaluation of Red-Absorbing Fluorescent Dyes. Bioconjugate Chem. 2003, 14, 195–204. [Google Scholar] [CrossRef]
- Nicoli, F.; Roos, M.K.; Hemmig, E.A.; Di Antonio, M.; de Vivie-Riedle, R.; Liedl, T. Proximity-Induced H-Aggregation of Cyanine Dyes on DNA-Duplexes. J. Phys. Chem. A 2016, 120, 9941–9947. [Google Scholar] [CrossRef] [PubMed]
- Valdes-Aguilera, O.; Neckers, D.C. Aggregation phenomena in xanthene dyes. Acc. Chem. Res. 1989, 22, 171–177. [Google Scholar] [CrossRef]
- Chatterjee, S.; Molenaar, R.; Tromp, L.; Wagterveld, R.M.; Roesink, H.D.W.; Cornelissen, J.; Claessens, M.; Blum, C. Optimizing fluorophore density for single virus counting: A photophysical approach. Methods Appl. Fluoresc. 2021, 9, 025001. [Google Scholar] [CrossRef]
- Comellas-Aragones, M.; Engelkamp, H.; Claessen, V.I.; Sommerdijk, N.A.J.M.; Rowan, A.E.; Christianen, P.C.M.; Maan, J.C.; Verduin, B.J.M.; Cornelissen, J.J.L.M.; Nolte, R.J.M. A virus-based single-enzyme nanoreactor. Nat. Nanotechnol. 2007, 2, 635–639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verduin, B.J.M. Preparation of Ccmv-Protein in Connection with Its Association into a Spherical-Particle. FEBS Lett. 1974, 45, 50–54. [Google Scholar] [CrossRef] [Green Version]
- Reddy, V.S.; Natarajan, P.; Okerberg, B.; Li, K.; Damodaran, K.V.; Morton, R.T.; Brooks, C.; Johnson, J.E. Virus Particle Explorer (VIPER), a Website for virus capsid structures and their computational analyses. J. Virol. 2001, 75, 11943–11947. [Google Scholar] [CrossRef] [Green Version]
- Speir, J.A.; Munshi, S.; Wang, G.; Baker, T.S.; Johnson, J.E. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure 1995, 3, 63–78. [Google Scholar] [CrossRef] [Green Version]
- Banks, P.R.; Paquette, D.M. Comparison of Three Common Amine Reactive Fluorescent Probes Used for Conjugation to Biomolecules by Capillary Zone Electrophoresis. Bioconjugate Chem. 1995, 6, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Bancroft, J.B.; Hiebert, E.; Rees, M.W.; Markham, R. Properties of cowpea chlorotic mottle virus, its protein and nucleic acid. Virology 1968, 34, 224–239. [Google Scholar] [CrossRef]
- Krüse, J.; Verduin, B.J.M.; Visser, A.J.W.G. Fluorescence of Cowpea-Chlorotic-Mottle Virus Modified with Pyridoxal 5′-Phosphate. Eur. J. Biochem. 1980, 105, 395–401. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chatterjee, S.; Schotpoort, B.A.; Elbert, T.; Cornelissen, J.J.L.M.; Claessens, M.M.A.E.; Blum, C. Exploiting Complex Fluorophore Interactions to Monitor Virus Capsid Disassembly. Molecules 2021, 26, 5750. https://doi.org/10.3390/molecules26195750
Chatterjee S, Schotpoort BA, Elbert T, Cornelissen JJLM, Claessens MMAE, Blum C. Exploiting Complex Fluorophore Interactions to Monitor Virus Capsid Disassembly. Molecules. 2021; 26(19):5750. https://doi.org/10.3390/molecules26195750
Chicago/Turabian StyleChatterjee, Swarupa, Bram A. Schotpoort, Thieme Elbert, Jeroen J. L. M. Cornelissen, Mireille M. A. E. Claessens, and Christian Blum. 2021. "Exploiting Complex Fluorophore Interactions to Monitor Virus Capsid Disassembly" Molecules 26, no. 19: 5750. https://doi.org/10.3390/molecules26195750
APA StyleChatterjee, S., Schotpoort, B. A., Elbert, T., Cornelissen, J. J. L. M., Claessens, M. M. A. E., & Blum, C. (2021). Exploiting Complex Fluorophore Interactions to Monitor Virus Capsid Disassembly. Molecules, 26(19), 5750. https://doi.org/10.3390/molecules26195750






