π–π Noncovalent Interaction Involving 1,2,4- and 1,3,4-Oxadiazole Systems: The Combined Experimental, Theoretical, and Database Study
Abstract
:1. Introduction
2. Results and Discussion
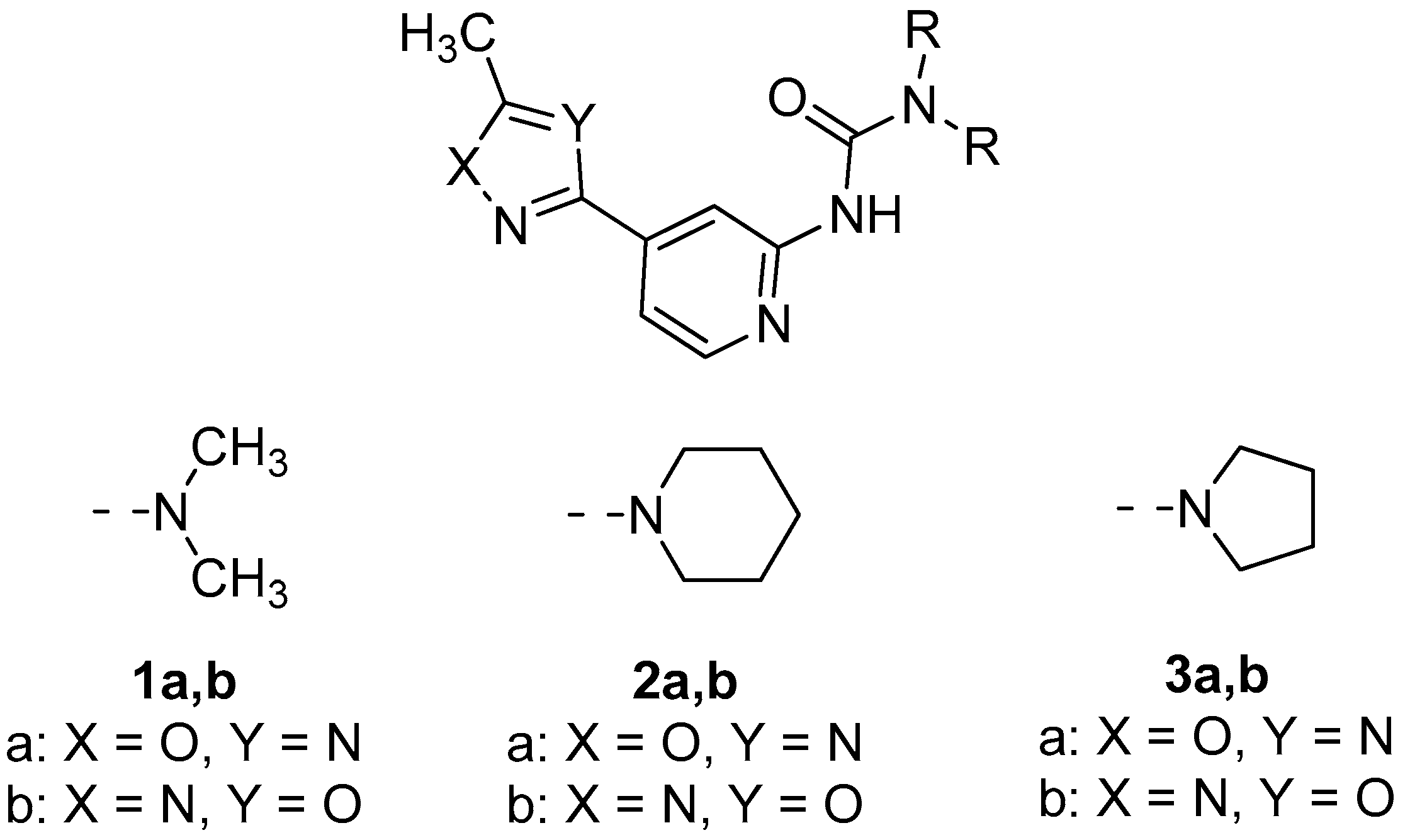
2.1. General Consideration of XRD Structures of (1–3)a,b
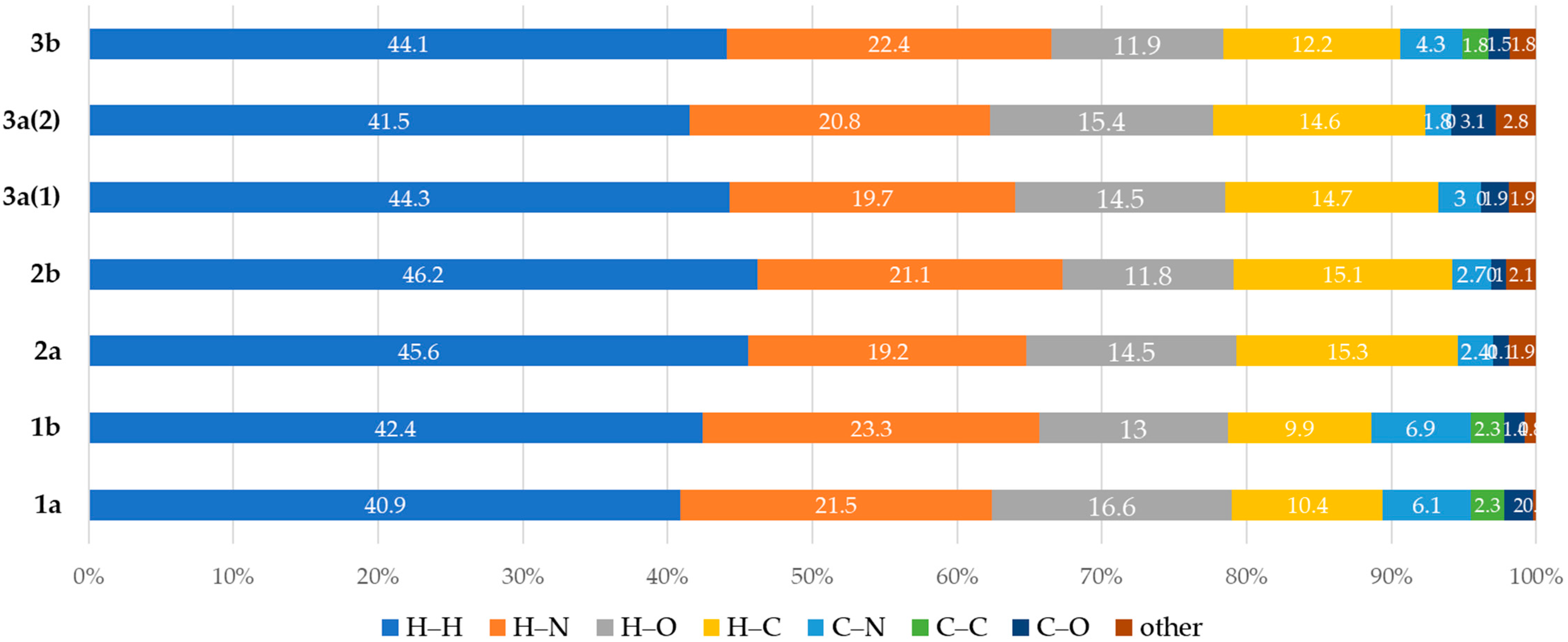
2.2. Hirshfeld Surface Analyses of (1–3)a,b
2.3. Noncovalent Bonding Patterns
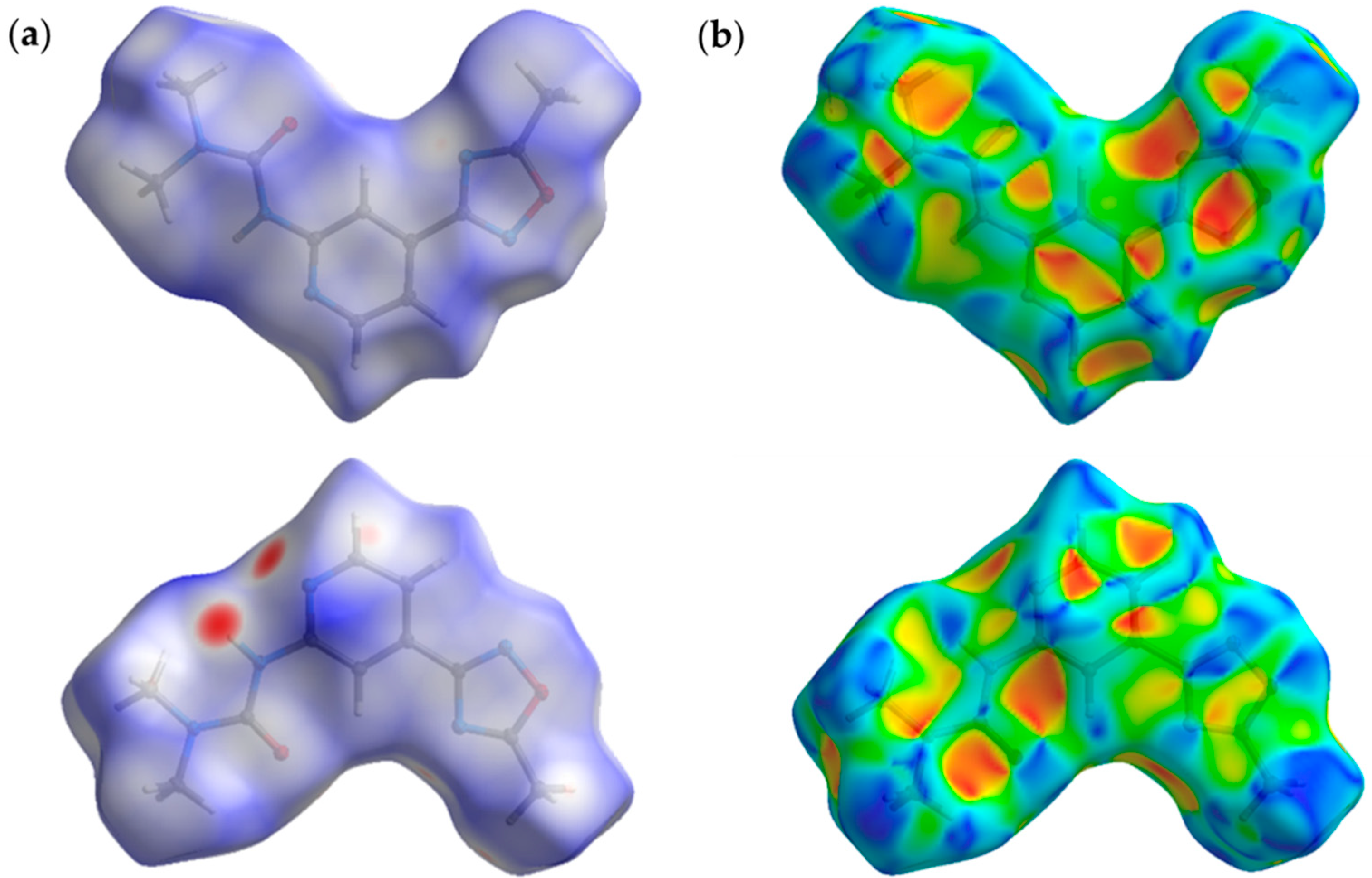
2.4. Theoretical Study of Noncovalent Interactions in (1–3)a,b
2.5. Supramolecular Association in Solution
2.6. CSD and PDB Search of (Oxadiazole)···π Interaction
3. Material and Methods
3.1. General
3.2. Oxadiazoles Preparation and Characterization
- 1,1-Dimethyl-3-(4-(5-methyl-1,3,4-oxadiazol-2-yl)pyridin-2-yl)urea 1b
- N-(4-(5-Methyl-1,2,4-oxadiazol-3-yl)pyridin-2-yl)pyrrolidine-1-carboxamide 3a
- N-(4-(5-Methyl-1,3,4-oxadiazol-2-yl)pyridin-2-yl)pyrrolidine-1-carboxamide 3b
3.3. Crystallography
3.4. Hirshfeld Surface Analysis
3.5. Computational Study
3.6. Databases Search Processing
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Burley, S.; Petsko, G. Aromatic-aromatic interaction: A mechanism of protein structure stabilization. Science 1985, 229, 23–28. [Google Scholar] [CrossRef]
- Chelli, R.; Gervasio, F.L.; Procacci, P.; Schettino, V. Stacking and T-shape Competition in Aromatic−Aromatic Amino Acid Interactions. J. Am. Chem. Soc. 2002, 124, 6133–6143. [Google Scholar] [CrossRef] [PubMed]
- Schneider, H.-J. Binding Mechanisms in Supramolecular Complexes. Angew. Chem. Int. Ed. 2009, 48, 3924–3977. [Google Scholar] [CrossRef] [PubMed]
- Meyer, E.A.; Castellano, R.K.; Diederich, F. Interactions with Aromatic Rings in Chemical and Biological Recognition. Angew. Chem. Int. Ed. 2003, 42, 1210–1250. [Google Scholar] [CrossRef]
- Sutton, C.; Risko, C.; Brédas, J.-L. Noncovalent Intermolecular Interactions in Organic Electronic Materials: Implications for the Molecular Packing vs Electronic Properties of Acenes. Chem. Mater. 2016, 28, 3–16. [Google Scholar] [CrossRef]
- Bu, R.; Xiong, Y.; Zhang, C. π–π Stacking Contributing to the Low or Reduced Impact Sensitivity of Energetic Materials. Cryst. Growth Des. 2020, 20, 2824–2841. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Q.; Vo, T.T.; Parrish, D.A.; Shreeve, J.M. Energetic Salts with π-Stacking and Hydrogen-Bonding Interactions Lead the Way to Future Energetic Materials. J. Am. Chem. Soc. 2015, 137, 1697–1704. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, S.E.; Seguin, T.J.; Guan, Y.; Doney, A.C. Noncovalent Interactions in Organocatalysis and the Prospect of Computational Catalyst Design. Acc. Chem. Res. 2016, 49, 1061–1069. [Google Scholar] [CrossRef]
- Neel, A.J.; Hilton, M.J.; Sigman, M.S.; Toste, F.D. Exploiting non-covalent π interactions for catalyst design. Nature 2017, 543, 637–646. [Google Scholar] [CrossRef]
- Salonen, L.M.; Ellermann, M.; Diederich, F. Aromatic Rings in Chemical and Biological Recognition: Energetics and Structures. Angew. Chem. Int. Ed. 2011, 50, 4808–4842. [Google Scholar] [CrossRef]
- Juhás, M.; Zitko, J. Molecular Interactions of Pyrazine-Based Compounds to Proteins. J. Med. Chem. 2020, 63, 8901–8916. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.; Lone, M.; Al-Othman, Z.; Al-Warthan, A.; Sanagi, M. Heterocyclic Scaffolds: Centrality in Anticancer Drug Development. Curr. Drug Targets 2015, 16, 711–734. [Google Scholar] [CrossRef]
- Heravi, M.M.; Zadsirjan, V. Prescribed drugs containing nitrogen heterocycles: An overview. RSC Adv. 2020, 10, 44247–44311. [Google Scholar] [CrossRef]
- Kerru, N.; Gummidi, L.; Maddila, S.; Gangu, K.K.; Jonnalagadda, S.B. A Review on Recent Advances in Nitrogen-Containing Molecules and Their Biological Applications. Molecules 2020, 25, 1909. [Google Scholar] [CrossRef] [PubMed]
- Gomtsyan, A. Heterocycles in drugs and drug discovery. Chem. Heterocycl. Compd. 2012, 48, 7–10. [Google Scholar] [CrossRef]
- Bhutani, P.; Joshi, G.; Raja, N.; Bachhav, N.; Rajanna, P.K.; Bhutani, H.; Paul, A.T.; Kumar, R.U.S. FDA Approved Drugs from 2015–June 2020: A Perspective. J. Med. Chem. 2021, 64, 2339–2381. [Google Scholar] [CrossRef]
- Delost, M.D.; Smith, D.T.; Anderson, B.J.; Njardarson, J.T. From Oxiranes to Oligomers: Architectures of U.S. FDA Approved Pharmaceuticals Containing Oxygen Heterocycles. J. Med. Chem. 2018, 61, 10996–11020. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, S.E. Local Nature of Substituent Effects in Stacking Interactions. J. Am. Chem. Soc. 2011, 133, 10262–10274. [Google Scholar] [CrossRef]
- Watt, M.; Hardebeck, L.K.E.; Kirkpatrick, C.C.; Lewis, M. Face-to-Face Arene−Arene Binding Energies: Dominated by Dispersion but Predicted by Electrostatic and Dispersion/Polarizability Substituent Constants. J. Am. Chem. Soc. 2011, 133, 3854–3862. [Google Scholar] [CrossRef]
- Muchowska, K.B.; Adam, C.; Mati, I.K.; Cockroft, S.L. Electrostatic Modulation of Aromatic Rings via Explicit Solvation of Substituents. J. Am. Chem. Soc. 2013, 135, 9976–9979. [Google Scholar] [CrossRef] [Green Version]
- Hwang, J.; Li, P.; Carroll, W.R.; Smith, M.D.; Pellechia, P.J.; Shimizu, K.D. Additivity of Substituent Effects in Aromatic Stacking Interactions. J. Am. Chem. Soc. 2014, 136, 14060–14067. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, S.E.; Houk, K.N. Substituent Effects in the Benzene Dimer are Due to Direct Interactions of the Substituents with the Unsubstituted Benzene. J. Am. Chem. Soc. 2008, 130, 10854–10855. [Google Scholar] [CrossRef] [Green Version]
- Boström, J.; Hogner, A.; Llinàs, A.; Wellner, E.; Plowright, A.T. Oxadiazoles in Medicinal Chemistry. J. Med. Chem. 2012, 55, 1817–1830. [Google Scholar] [CrossRef] [PubMed]
- Biernacki, K.; Daśko, M.; Ciupak, O.; Kubiński, K.; Rachon, J.; Demkowicz, S. Novel 1,2,4-Oxadiazole Derivatives in Drug Discovery. Pharmaceuticals 2020, 13, 111. [Google Scholar] [CrossRef]
- Welch, E.M.; Barton, E.R.; Zhuo, J.; Tomizawa, Y.; Friesen, W.J.; Trifillis, P.; Paushkin, S.; Patel, M.; Trotta, C.R.; Hwang, S.; et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature 2007, 447, 87–91. [Google Scholar] [CrossRef]
- Lanier, G.; Sankholkar, K.; Aronow, W.S. Azilsartan, Aliskiren, and Combination Antihypertensives Utilizing Renin–Angiotensin–Aldosterone System Antagonists. Am. J. Ther. 2014, 21, 419–435. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.J. Opicapone: A Review in Parkinson’s Disease. Drugs 2016, 76, 1293–1300. [Google Scholar] [CrossRef]
- Hale, M.; Wild, J.; Reddy, J.; Yamada, T.; Arjona Ferreira, J.C. Naldemedine versus placebo for opioid-induced constipation (COMPOSE-1 and COMPOSE-2): Two multicentre, phase 3, double-blind, randomised, parallel-group trials. Lancet Gastroenterol. Hepatol. 2017, 2, 555–564. [Google Scholar] [CrossRef]
- Cahn, P.; Sued, O. Raltegravir: A new antiretroviral class for salvage therapy. Lancet 2007, 369, 1235–1236. [Google Scholar] [CrossRef]
- Shetnev, A.; Baykov, S.; Kalinin, S.; Belova, A.; Sharoyko, V.; Rozhkov, A.; Zelenkov, L.; Tarasenko, M.; Sadykov, E.; Korsakov, M.; et al. 1,2,4-Oxadiazole/2-Imidazoline Hybrids: Multi-target-directed Compounds for the Treatment of Infectious Diseases and Cancer. Int. J. Mol. Sci. 2019, 20, 1699. [Google Scholar] [CrossRef] [Green Version]
- Hannoun, M.H.; Hagras, M.; Kotb, A.; El-Attar, A.-A.M.M.; Abulkhair, H.S. Synthesis and antibacterial evaluation of a novel library of 2-(thiazol-5-yl)-1,3,4-oxadiazole derivatives against methicillin-resistant Staphylococcus aureus (MRSA). Bioorg. Chem. 2020, 94, 103364. [Google Scholar] [CrossRef]
- Boudreau, M.A.; Ding, D.; Meisel, J.E.; Janardhanan, J.; Spink, E.; Peng, Z.; Qian, Y.; Yamaguchi, T.; Testero, S.A.; O’Daniel, P.I.; et al. Structure–Activity Relationship for the Oxadiazole Class of Antibacterials. ACS Med. Chem. Lett. 2020, 11, 322–326. [Google Scholar] [CrossRef] [PubMed]
- Tarasenko, M.V.; Presnukhina, S.I.; Baikov, S.V.; Shetnev, A.A. Synthesis and Evaluation of Antibacterial Activity of 1,2,4-Oxadiazole-Containing Biphenylcarboxylic Acids. Russ. J. Gen. Chem. 2020, 90, 1611–1619. [Google Scholar] [CrossRef]
- Tarasenko, M.; Sidneva, V.; Belova, A.; Romanycheva, A.; Sharonova, T.; Baykov, S.; Shetnev, A.; Kofanov, E.; Kuznetsov, M. An efficient synthesis and antimicrobial evaluation of 5-alkenyl- and 5-styryl-1,2,4-oxadiazoles. Arkivoc 2018, 2018, 458–470. [Google Scholar] [CrossRef]
- Hagras, M.; Salama, E.A.; Sayed, A.M.; Abutaleb, N.S.; Kotb, A.; Seleem, M.N.; Mayhoub, A.S. Oxadiazolylthiazoles as novel and selective antifungal agents. Eur. J. Med. Chem. 2020, 189, 112046. [Google Scholar] [CrossRef]
- Bordei Telehoiu, A.T.; Nuță, D.C.; Căproiu, M.T.; Dumitrascu, F.; Zarafu, I.; Ioniță, P.; Bădiceanu, C.D.; Avram, S.; Chifiriuc, M.C.; Bleotu, C.; et al. Design, Synthesis and In Vitro Characterization of Novel Antimicrobial Agents Based on 6-Chloro-9H-carbazol Derivatives and 1,3,4-Oxadiazole Scaffolds. Molecules 2020, 25, 266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krasavin, M.; Shetnev, A.; Sharonova, T.; Baykov, S.; Kalinin, S.; Nocentini, A.; Sharoyko, V.; Poli, G.; Tuccinardi, T.; Presnukhina, S.; et al. Continued exploration of 1,2,4-oxadiazole periphery for carbonic anhydrase-targeting primary arene sulfonamides: Discovery of subnanomolar inhibitors of membrane-bound hCA IX isoform that selectively kill cancer cells in hypoxic environment. Eur. J. Med. Chem. 2019, 164, 92–105. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, X.; Yang, J.; Guo, L.; Wang, X.; Song, B.; Dong, W.; Wang, W. Novel diosgenin derivatives containing 1,3,4-oxadiazole/thiadiazole moieties as potential antitumor agents: Design, synthesis and cytotoxic evaluation. Eur. J. Med. Chem. 2020, 186, 111897. [Google Scholar] [CrossRef]
- Caneschi, W.; Enes, K.B.; Carvalho de Mendonça, C.; de Souza Fernandes, F.; Miguel, F.B.; da Silva Martins, J.; Le Hyaric, M.; Pinho, R.R.; Duarte, L.M.; Leal de Oliveira, M.A.; et al. Synthesis and anticancer evaluation of new lipophilic 1,2,4 and 1,3,4-oxadiazoles. Eur. J. Med. Chem. 2019, 165, 18–30. [Google Scholar] [CrossRef]
- El-Sayed, N.A.; Nour, M.S.; Salem, M.A.; Arafa, R.K. New oxadiazoles with selective-COX-2 and EGFR dual inhibitory activity: Design, synthesis, cytotoxicity evaluation and in silico studies. Eur. J. Med. Chem. 2019, 183, 111693. [Google Scholar] [CrossRef]
- Hamdani, S.S.; Khan, B.A.; Hameed, S.; Batool, F.; Saleem, H.N.; Mughal, E.U.; Saeed, M. Synthesis and evaluation of novel S-benzyl- and S-alkylphthalimide- oxadiazole -benzenesulfonamide hybrids as inhibitors of dengue virus protease. Bioorg. Chem. 2020, 96, 103567. [Google Scholar] [CrossRef]
- Yang, G.; Zheng, H.; Shao, W.; Liu, L.; Wu, Z. Study of the in vivo antiviral activity against TMV treated with novel 1-(t-butyl)-5-amino-4-pyrazole derivatives containing a 1,3,4-oxadiazole sulfide moiety. Pestic. Biochem. Physiol. 2021, 171, 104740. [Google Scholar] [CrossRef] [PubMed]
- Shetnev, A.; Osipyan, A.; Baykov, S.; Sapegin, A.; Chirkova, Z.; Korsakov, M.; Petzer, A.; Engelbrecht, I.; Petzer, J.P. Novel monoamine oxidase inhibitors based on the privileged 2-imidazoline molecular framework. Bioorg. Med. Chem. Lett. 2019, 29, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, B.Z.; Cohen, J.A.; Conway, D.S. Sphingosine 1-Phosphate Receptor Modulators for the Treatment of Multiple Sclerosis. Neurotherapeutics 2017, 14, 859–873. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Choubey, P.K.; Sharma, P.; Seth, A.; Saraf, P.; Shrivastava, S.K. Design, synthesis, and biological evaluation of ferulic acid based 1,3,4-oxadiazole hybrids as multifunctional therapeutics for the treatment of Alzheimer’s disease. Bioorg. Chem. 2020, 95, 103506. [Google Scholar] [CrossRef] [PubMed]
- Bhutani, R.; Pathak, D.P.; Kapoor, G.; Husain, A.; Iqbal, M.A. Novel hybrids of benzothiazole-1,3,4-oxadiazole-4-thiazolidinone: Synthesis, in silico ADME study, molecular docking and in vivo anti-diabetic assessment. Bioorg. Chem. 2019, 83, 6–19. [Google Scholar] [CrossRef] [PubMed]
- Petenzi, M.; Verga, D.; Largy, E.; Hamon, F.; Doria, F.; Teulade-Fichou, M.-P.; Guédin, A.; Mergny, J.-L.; Mella, M.; Freccero, M. Cationic Pentaheteroaryls as Selective G-Quadruplex Ligands by Solvent-Free Microwave-Assisted Synthesis. Chem.-A Eur. J. 2012, 18, 14487–14496. [Google Scholar] [CrossRef] [PubMed]
- Doria, F.; Pirota, V.; Petenzi, M.; Teulade-Fichou, M.-P.; Verga, D.; Freccero, M. Oxadiazole/Pyridine-Based Ligands: A Structural Tuning for Enhancing G-Quadruplex Binding. Molecules 2018, 23, 2162. [Google Scholar] [CrossRef] [Green Version]
- Pibiri, I.; Lentini, L.; Melfi, R.; Gallucci, G.; Pace, A.; Spinello, A.; Barone, G.; Di Leonardo, A. Enhancement of premature stop codon readthrough in the CFTR gene by Ataluren (PTC124) derivatives. Eur. J. Med. Chem. 2015, 101, 236–244. [Google Scholar] [CrossRef]
- Ahmed, M.N.; Nadeem, K.; Andleeb, H.; Sheikhi, M.; Majeed, Z.; Shah, S.W.A.; Tahir, M.N.; Rocha, M.; Gil, D.M. Exploring weak intermolecular interactions in two bis-1,3,4-oxadiazoles derivatives: A combined X-ray diffraction, Hirshfeld surface analysis and theoretical studies. J. Mol. Struct. 2021, 1232, 130030. [Google Scholar] [CrossRef]
- Geyl, K.; Baykov, S.; Tarasenko, M.; Zelenkov, L.E.; Matveevskaya, V.; Boyarskiy, V.P. Convenient entry to N-pyridinylureas with pharmaceutically privileged oxadiazole substituents via the acid-catalyzed C H activation of N-oxides. Tetrahedron Lett. 2019, 60, 151108. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Spackman, M.A.; McKinnon, J.J.; Jayatilaka, D. Electrostatic potentials mapped on Hirshfeld surfaces provide direct insight into intermolecular interactions in crystals. CrystEngComm 2008, 10, 377–388. [Google Scholar] [CrossRef]
- McKinnon, J.J.; Jayatilaka, D.; Spackman, M.A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. Commun. 2007, 3814–3816. [Google Scholar] [CrossRef]
- Bondi, A. van der Waals Volumes and Radii. J. Phys. Chem. 1964, 68, 441–451. [Google Scholar] [CrossRef]
- Contreras-García, J.; Johnson, E.R.; Keinan, S.; Chaudret, R.; Piquemal, J.-P.; Beratan, D.N.; Yang, W. NCIPLOT: A Program for Plotting Noncovalent Interaction Regions. J. Chem. Theory Comput. 2011, 7, 625–632. [Google Scholar] [CrossRef]
- Johnson, E.R.; Keinan, S.; Mori-Sánchez, P.; Contreras-García, J.; Cohen, A.J.; Yang, W. Revealing Noncovalent Interactions. J. Am. Chem. Soc. 2010, 132, 6498–6506. [Google Scholar] [CrossRef] [Green Version]
- Narth, C.; Maroun, Z.; Boto, R.A.; Chaudret, R.; Bonnet, M.-L.; Piquemal, J.-P.; Contreras-García, J. A Complete NCI Perspective: From New Bonds to Reactivity. In Applications of Topological Methods in Molecular Chemistry. Challenges and Advances in Computational Chemistry and Physics; Springer: Cham, Switzerland, 2016; Volume 22, pp. 491–527. [Google Scholar]
- Krawczyk, M.S.; Sroka, A.; Majerz, I. The Crystal Structure and Intermolecular Interactions in Fenamic Acids–Acridine Complexes. Molecules 2021, 26, 2956. [Google Scholar] [CrossRef]
- Belmont-Sánchez, J.; García-Rubiño, M.; Frontera, A.; González-Pérez, J.; Castiñeiras, A.; Niclós-Gutiérrez, J. H-Bonds, π-Stacking and (Water)O-H/π Interactions in (µ4-EDTA)Bis(Imidazole) Dicopper(II) Dihydrate. Crystals 2021, 11, 48. [Google Scholar] [CrossRef]
- Bader, R.F.W. A quantum theory of molecular structure and its applications. Chem. Rev. 1991, 91, 893–928. [Google Scholar] [CrossRef]
- Espinosa, E.; Alkorta, I.; Elguero, J.; Molins, E. From weak to strong interactions: A comprehensive analysis of the topological and energetic properties of the electron density distribution involving X–H⋯F–Y systems. J. Chem. Phys. 2002, 117, 5529–5542. [Google Scholar] [CrossRef]
- Macchioni, A.; Ciancaleoni, G.; Zuccaccia, C.; Zuccaccia, D. Determining accurate molecular sizes in solution through NMR diffusion spectroscopy. Chem. Soc. Rev. 2008, 37, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Bellachioma, G.; Ciancaleoni, G.; Zuccaccia, C.; Zuccaccia, D.; Macchioni, A. NMR investigation of non-covalent aggregation of coordination compounds ranging from dimers and ion pairs up to nano-aggregates. Coord. Chem. Rev. 2008, 252, 2224–2238. [Google Scholar] [CrossRef]
- Sivchik, V.V.; Grachova, E.V.; Melnikov, A.S.; Smirnov, S.N.; Ivanov, A.Y.; Hirva, P.; Tunik, S.P.; Koshevoy, I.O. Solid-State and Solution Metallophilic Aggregation of a Cationic [Pt(NCN)L] + Cyclometalated Complex. Inorg. Chem. 2016, 55, 3351–3363. [Google Scholar] [CrossRef]
- Cohen, Y.; Avram, L.; Frish, L. Diffusion NMR Spectroscopy in Supramolecular and Combinatorial Chemistry: An Old Parameter?New Insights. Angew. Chem. Int. Ed. 2005, 44, 520–554. [Google Scholar] [CrossRef]
- Mikherdov, A.S.; Novikov, A.S.; Kinzhalov, M.A.; Boyarskiy, V.P.; Starova, G.L.; Ivanov, A.Y.; Kukushkin, V.Y. Halides Held by Bifurcated Chalcogen–Hydrogen Bonds. Effect of μ (S,N–H) Cl Contacts on Dimerization of Cl(carbene)Pd II Species. Inorg. Chem. 2018, 57, 3420–3433. [Google Scholar] [CrossRef]
- Temesgen, Z. Raltegravir: First in class HIV integrase inhibitor. Ther. Clin. Risk Manag. 2008, 4, 493–500. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://www.annualreports.com/ (accessed on 7 September 2021).
- Tomkinson, H.K.; Kemp, J.V.; Wollseifen, T.; Morris, T.; Oliver, S.D. An open-label, randomized, single-center, two-period, phase I, crossover study of the effect of zibotentan (ZD4054) on the pharmacokinetics of midazolam in healthy male volunteers. Clin. Ther. 2010, 32, 1372–1386. [Google Scholar] [CrossRef]
- Haque, S.; Dashwood, M.R.; Heetun, M.; Shiwen, X.; Farooqui, N.; Ramesh, B.; Welch, H.; Savage, F.J.; Ogunbiyi, O.; Abraham, D.J.; et al. Efficacy of the Specific Endothelin A Receptor Antagonist Zibotentan (ZD4054) in Colorectal Cancer: A Preclinical Study. Mol. Cancer Ther. 2013, 12, 1556–1567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Burke, V.; Totrov, M.; Williams, C.; Cardozo, T.; Gorny, M.K.; Zolla-Pazner, S.; Kong, X.-P. Conserved structural elements in the V3 crown of HIV-1 gp120. Nat. Struct. Mol. Biol. 2010, 17, 955–961. [Google Scholar] [CrossRef]
- Brylinski, M. Aromatic interactions at the ligand-protein interface: Implications for the development of docking scoring functions. Chem. Biol. Drug Des. 2018, 91, 380–390. [Google Scholar] [CrossRef]
- Cheng, T.; Li, Q.; Zhou, Z.; Wang, Y.; Bryant, S.H. Structure-Based Virtual Screening for Drug Discovery: A Problem-Centric Review. AAPS J. 2012, 14, 133–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheldrick, G.M. SHELXT–Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- CrysAlis Pro. Data Collection and Processing Software for Agilent X-Ray Diffractometers; Aglient Technologies: Yarnton, UK, 2013. [Google Scholar]
- Spackman, P.R.; Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer: A program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Crystallogr. 2021, 54, 1006–1011. [Google Scholar] [CrossRef]
- Zhao, Y.; Truhlar, D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other function. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar] [CrossRef] [Green Version]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision C.01; Gaussian Inc.: Wallingford, CT, USA, 2010. [Google Scholar]
- Chai, J.-D.; Head-Gordon, M. Long-range corrected hybrid density functionals with damped atom–atom dispersion corrections. Phys. Chem. Chem. Phys. 2008, 10, 6615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Mikhailov, I.E.; Popov, L.D.; Tkachev, V.V.; Aldoshin, S.M.; Dushenko, G.A.; Revinskii, Y.V.; Minkin, V.I. Synthesis and crystal structure of novel fluorescent 1,3,4-oxadiazole-containing carboxylate ligands. J. Mol. Struct. 2018, 1157, 374–380. [Google Scholar] [CrossRef]
- Chen, F.; Tian, T.; Bai, B.; Wang, J.; Wang, H.; Li, M. Crystal structures, intermolecular interactions and fluorescence properties of a series of symmetrical bi-1,3,4-oxadiazole derivatives. J. Mater. Chem. C 2016, 4, 4451–4458. [Google Scholar] [CrossRef]
- Wang, H.; Chen, F.; Jia, X.; Liu, H.; Ran, X.; Ravva, M.K.; Bai, F.-Q.; Qu, S.; Li, M.; Zhang, H.-X.; et al. Controllable molecular aggregation and fluorescence properties of 1,3,4-oxadiazole derivatives. J. Mater. Chem. C 2015, 3, 11681–11688. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Wang, Y.; Zhang, W.; Tian, T.; Bai, B.; Wang, H.; Bai, F.-Q.; Li, M. Role of Intermolecular Interactions in Molecular Packing of Alkoxy-Substituted Bis-1,3,4-oxadiazole Derivatives. Cryst. Growth Des. 2019, 19, 6100–6113. [Google Scholar] [CrossRef]
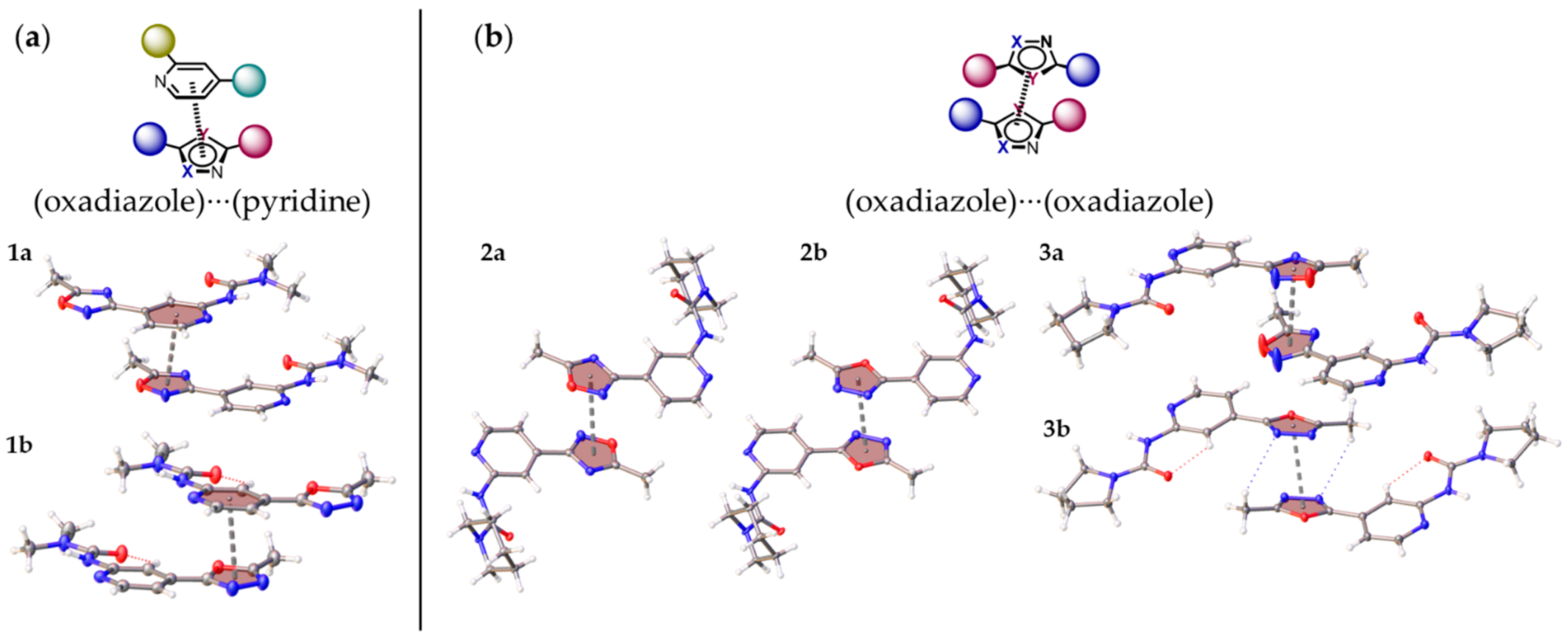
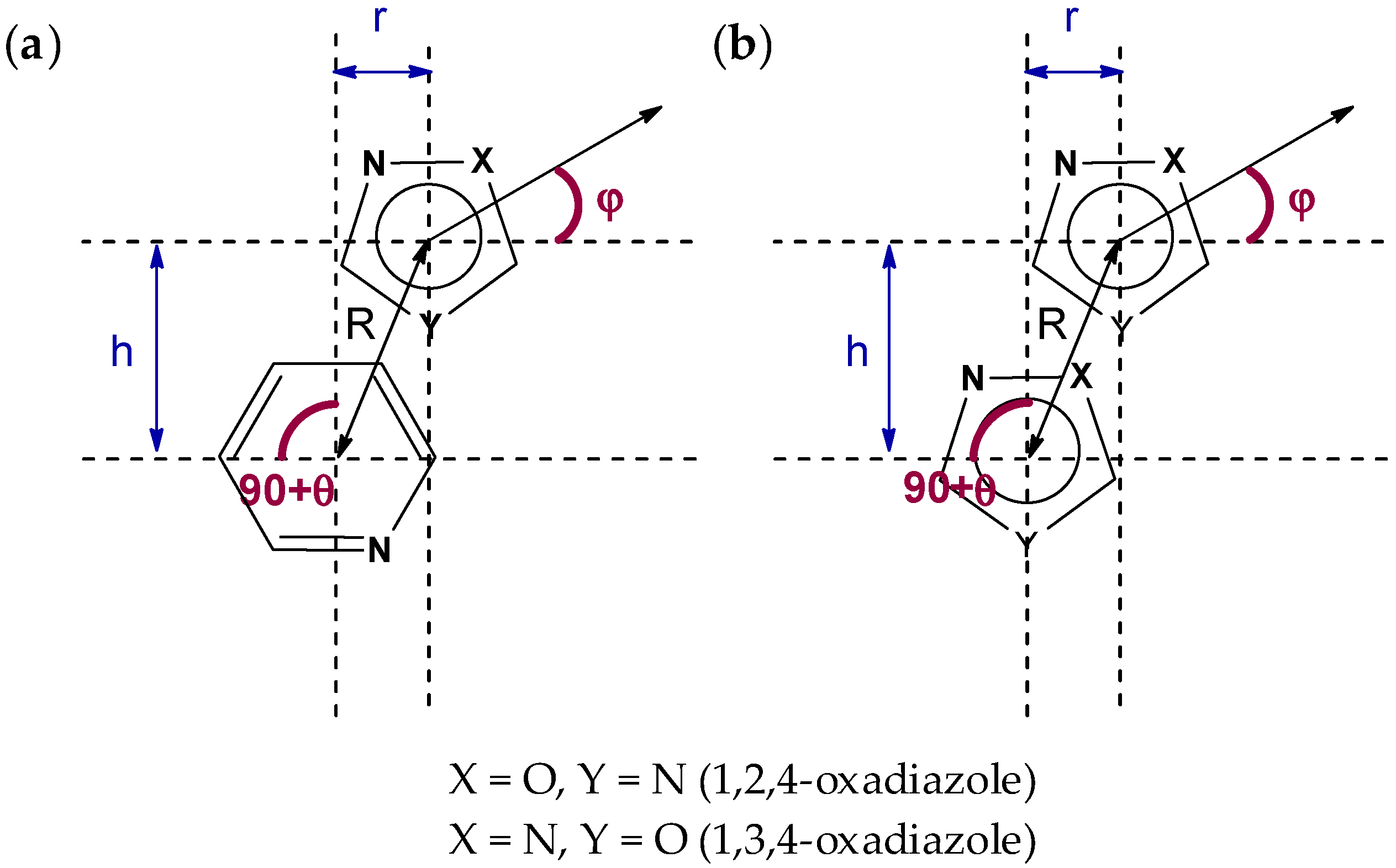
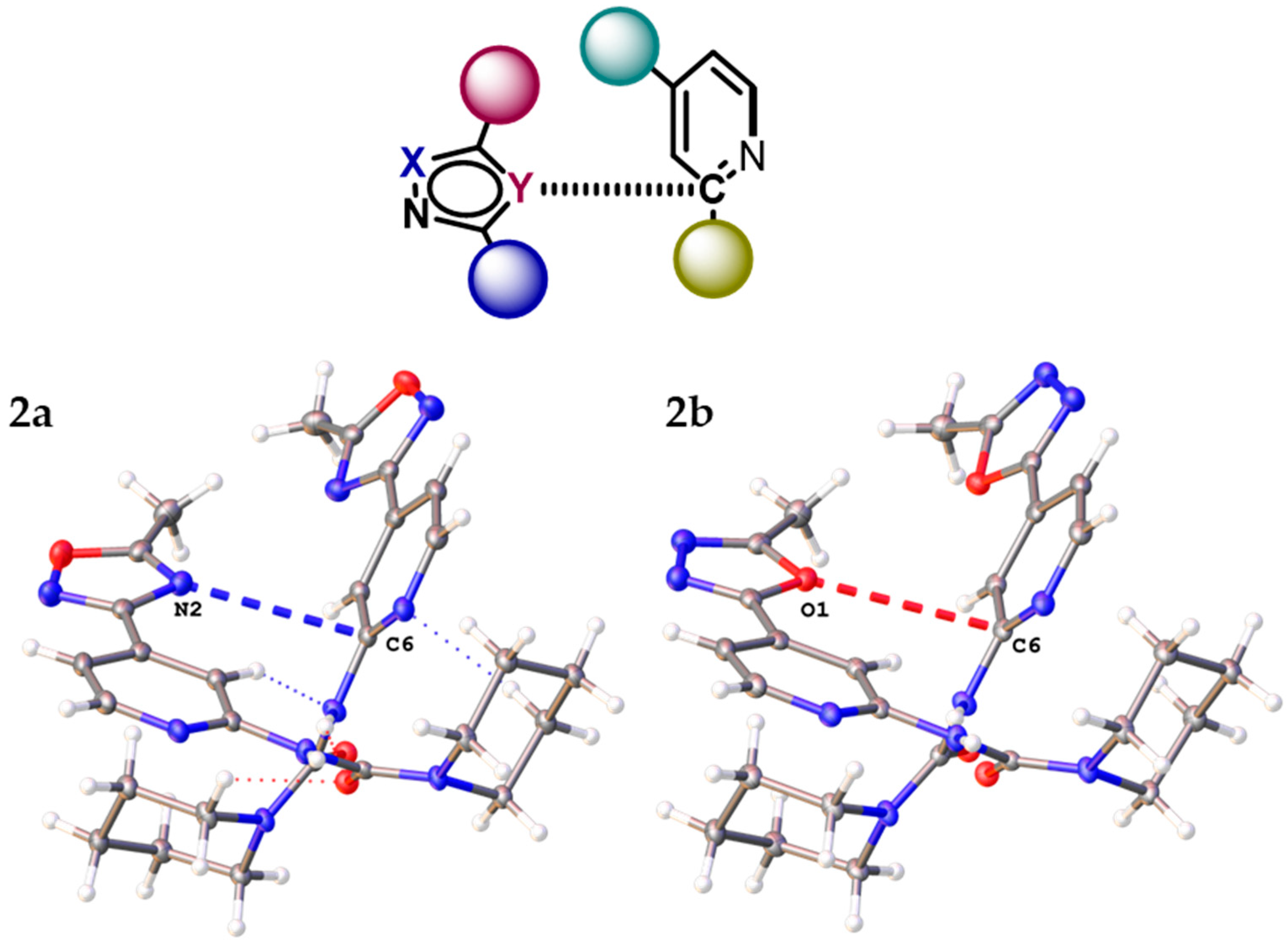

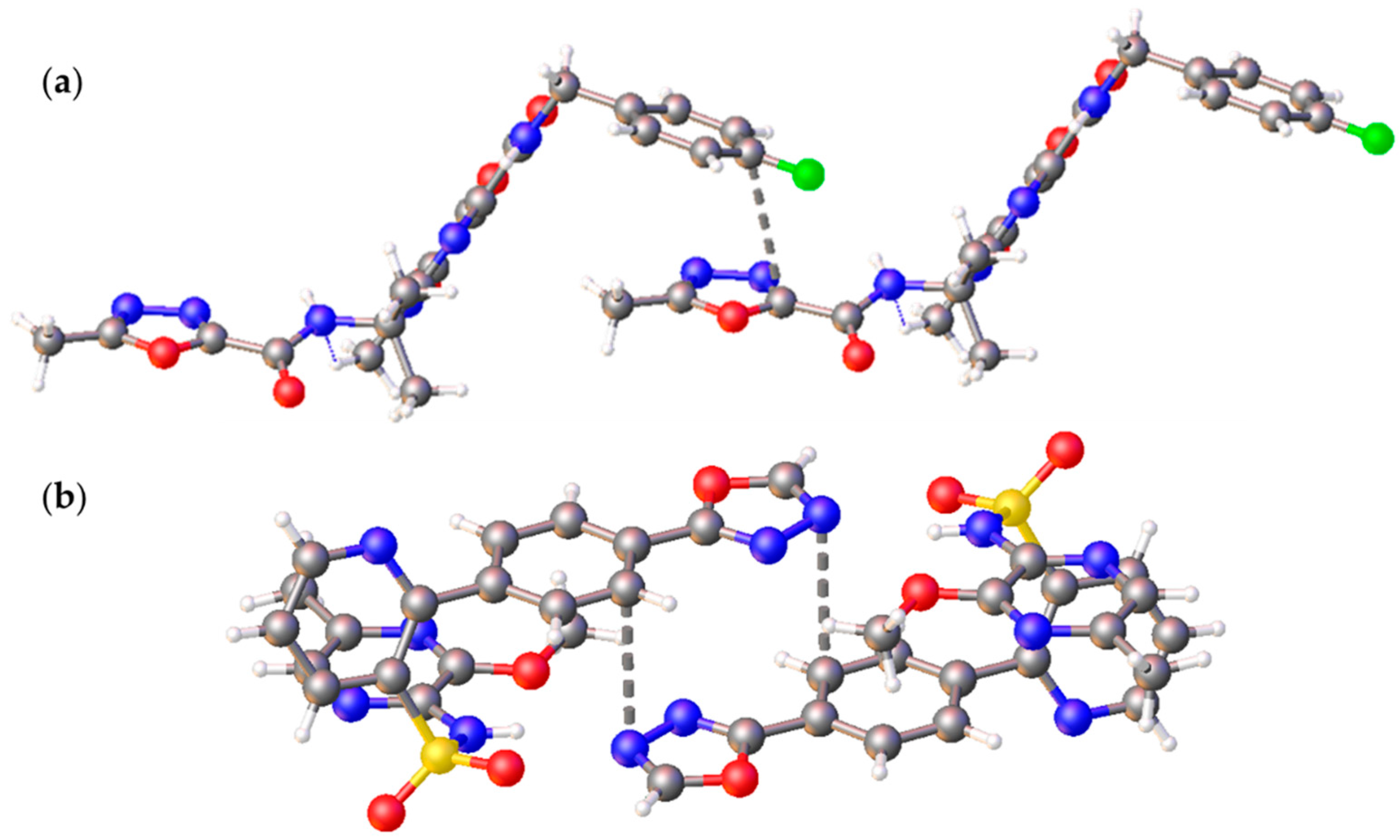
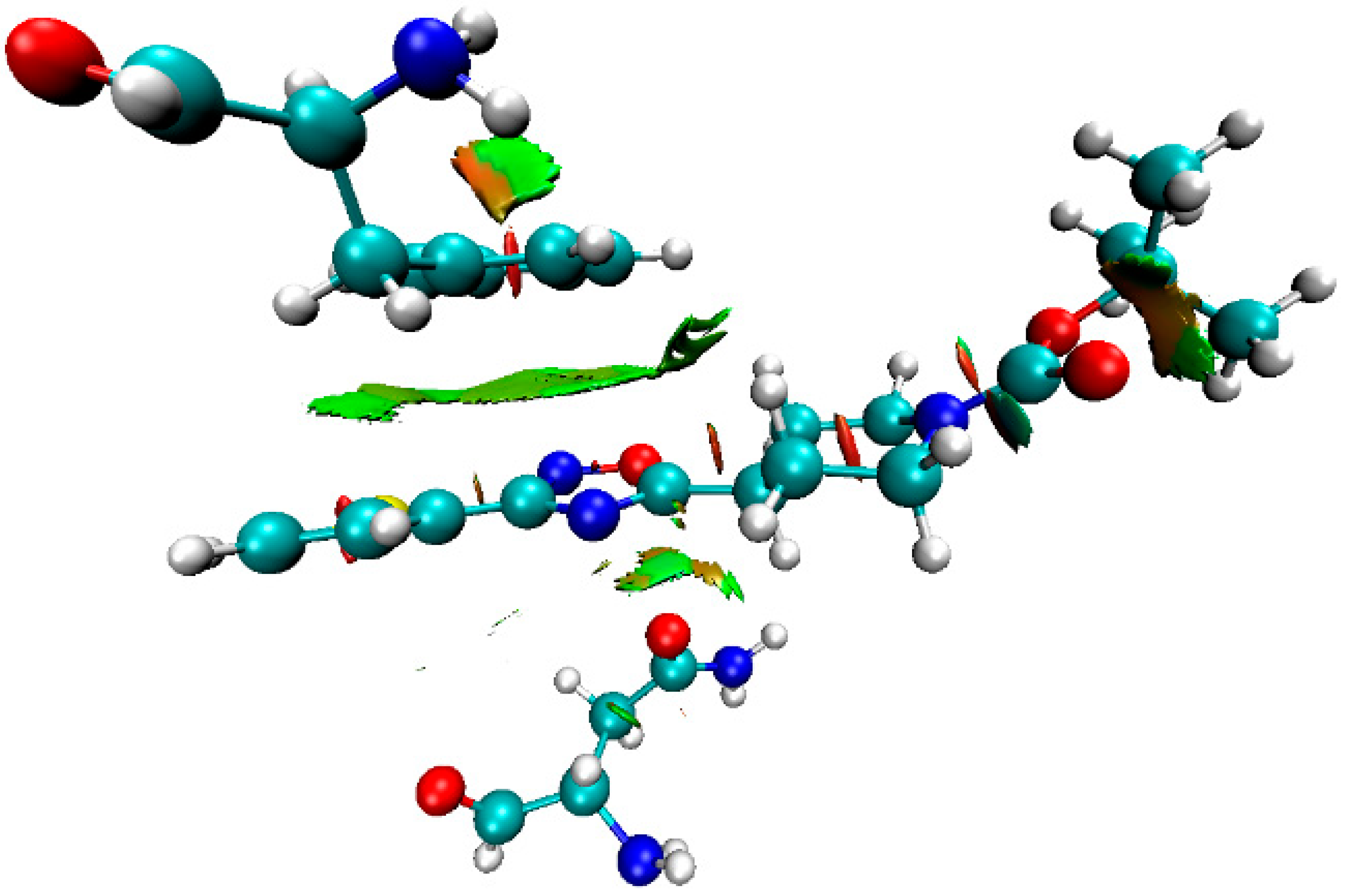
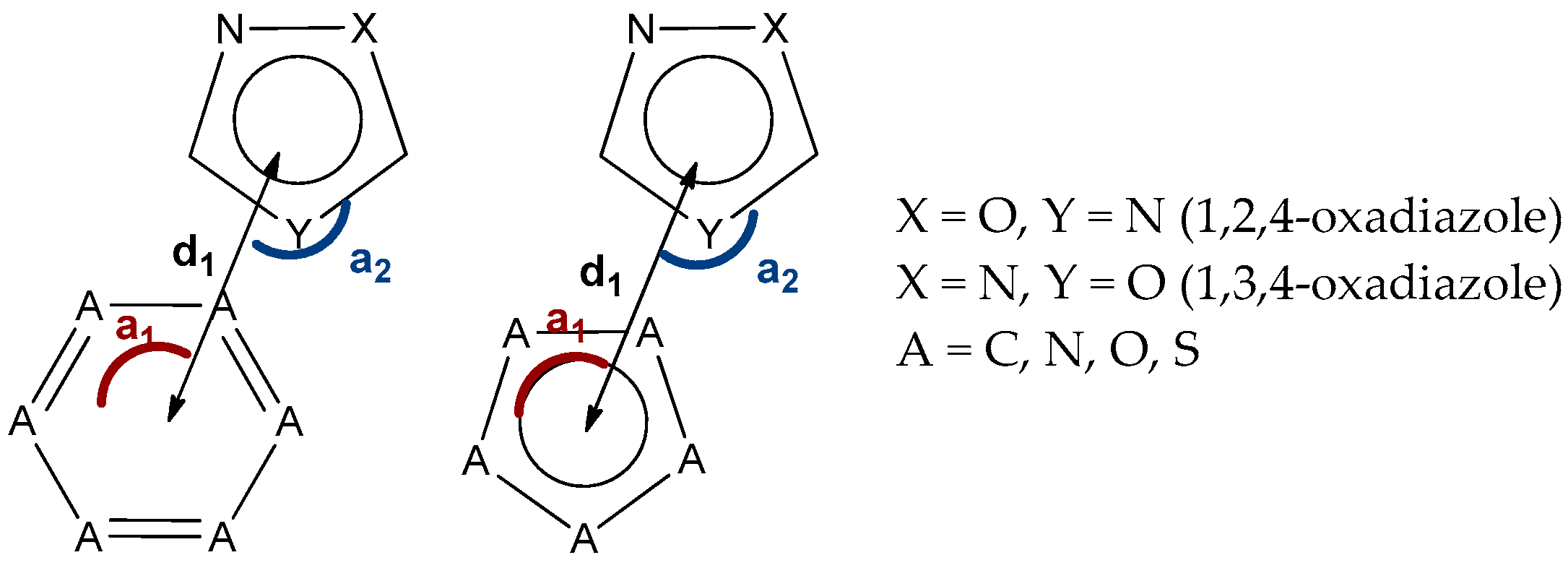
| Structure | Type | R | h1, Å | h2, Å | r1, Å | r2, Å | φ, ° | θ, ° |
|---|---|---|---|---|---|---|---|---|
| 1a | a | 3.6230(8) | 3.5157(10) | 3.4774(10) | 0.875(3) | 1.017(2) | 10.7(2) | 3.69(5) |
| 1b | a | 3.5203(9) | 3.4735(11) | 3.4031(11) | 0.572(3) | 0.900(2) | 8.5(3) | 5.75(6) |
| 2a | b | 3.6838(12) | 3.3492(16) | 3.3492(16) | 1.534(3) | 1.534(3) | 0.00(14) | 0.00(14) |
| 2b | b | 3.4848(11) | 3.2480(14) | 3.2480(14) | 1.262(2) | 1.262(2) | 0.00(19) | 0.00(18) |
| 3a | b | 3.5016(11) | 3.2481(15) | 3.189(2) | 3.4064(11) | 0.812(3) | 68.1(6) | 12.56(8) |
| 3b | b | 3.3978(9) | 3.2675(10) | 3.2675(10) | 0.9321(19) | 0.9321(19) | 0.00(19) | 0.00(13) |
| π-System | 1,2,4-Oxadiazoles | 1,3,4-Oxadiazoles | ||||||
|---|---|---|---|---|---|---|---|---|
| Total | C | N | O | Total | C | N | O | |
| six-membered ring | 74 | 74 (C3 68; C5 53) | 22 (N1 16; N4 10) | 28 | 233 | 229 | 45 | 74 |
| five-membered rings | 76 (58 oxa-oxa) | 76 (C3 66; C5 63) | 58 (N1 46; N4 26) | 38 | 97 (76 oxa-oxa) | 95 | 58 | 14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baykov, S.V.; Mikherdov, A.S.; Novikov, A.S.; Geyl, K.K.; Tarasenko, M.V.; Gureev, M.A.; Boyarskiy, V.P. π–π Noncovalent Interaction Involving 1,2,4- and 1,3,4-Oxadiazole Systems: The Combined Experimental, Theoretical, and Database Study. Molecules 2021, 26, 5672. https://doi.org/10.3390/molecules26185672
Baykov SV, Mikherdov AS, Novikov AS, Geyl KK, Tarasenko MV, Gureev MA, Boyarskiy VP. π–π Noncovalent Interaction Involving 1,2,4- and 1,3,4-Oxadiazole Systems: The Combined Experimental, Theoretical, and Database Study. Molecules. 2021; 26(18):5672. https://doi.org/10.3390/molecules26185672
Chicago/Turabian StyleBaykov, Sergey V., Alexander S. Mikherdov, Alexander S. Novikov, Kirill K. Geyl, Marina V. Tarasenko, Maxim A. Gureev, and Vadim P. Boyarskiy. 2021. "π–π Noncovalent Interaction Involving 1,2,4- and 1,3,4-Oxadiazole Systems: The Combined Experimental, Theoretical, and Database Study" Molecules 26, no. 18: 5672. https://doi.org/10.3390/molecules26185672
APA StyleBaykov, S. V., Mikherdov, A. S., Novikov, A. S., Geyl, K. K., Tarasenko, M. V., Gureev, M. A., & Boyarskiy, V. P. (2021). π–π Noncovalent Interaction Involving 1,2,4- and 1,3,4-Oxadiazole Systems: The Combined Experimental, Theoretical, and Database Study. Molecules, 26(18), 5672. https://doi.org/10.3390/molecules26185672









