Exploring the Role of L10 Loop in New Delhi Metallo-β-lactamase (NDM-1): Kinetic and Dynamic Studies
Abstract
1. Introduction
2. Results
2.1. Determination of kcat and Km
2.1.1. Imipenem
2.1.2. Meropenem
2.1.3. Benzylpenicillin
2.1.4. Carbenicillin
2.1.5. Cefazolin
2.1.6. Cefoxitin
2.1.7. Cefotaxime and Ceftazidime
2.1.8. Cefepime
2.2. Effect of pH on Km and kcat
2.3. Thermofluor Stability of NDM-1 and Its Mutants
2.4. Fluorescence Spectra
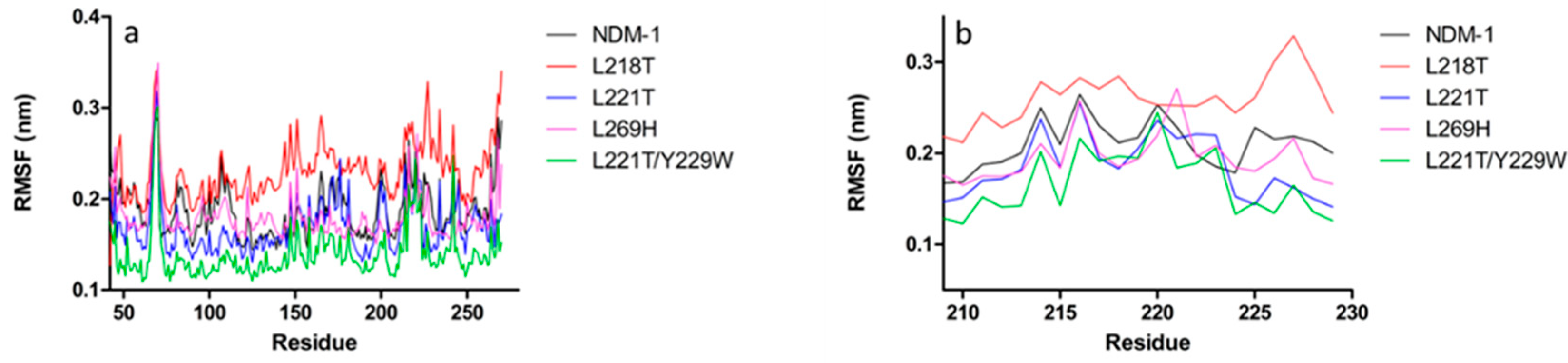
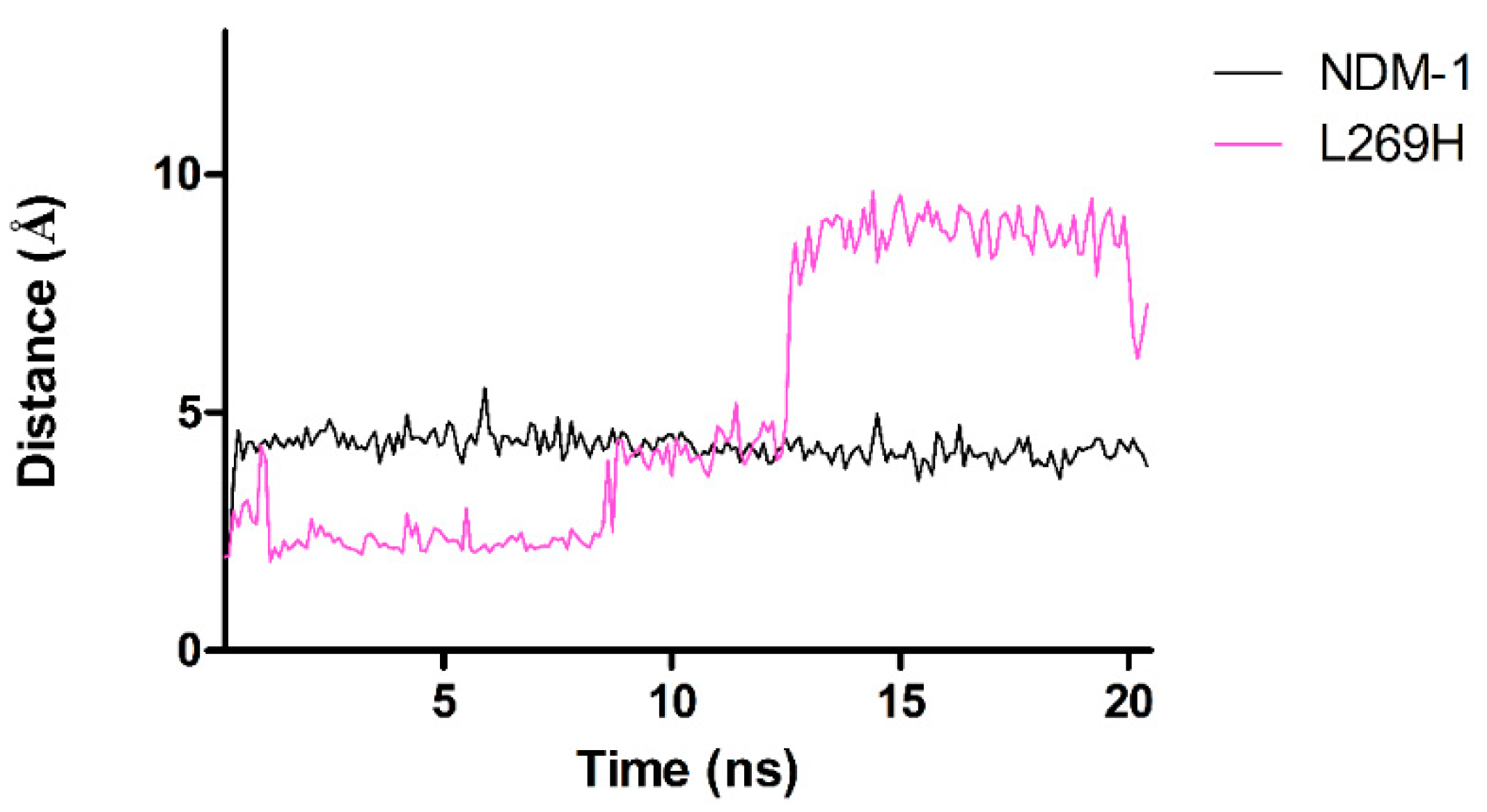
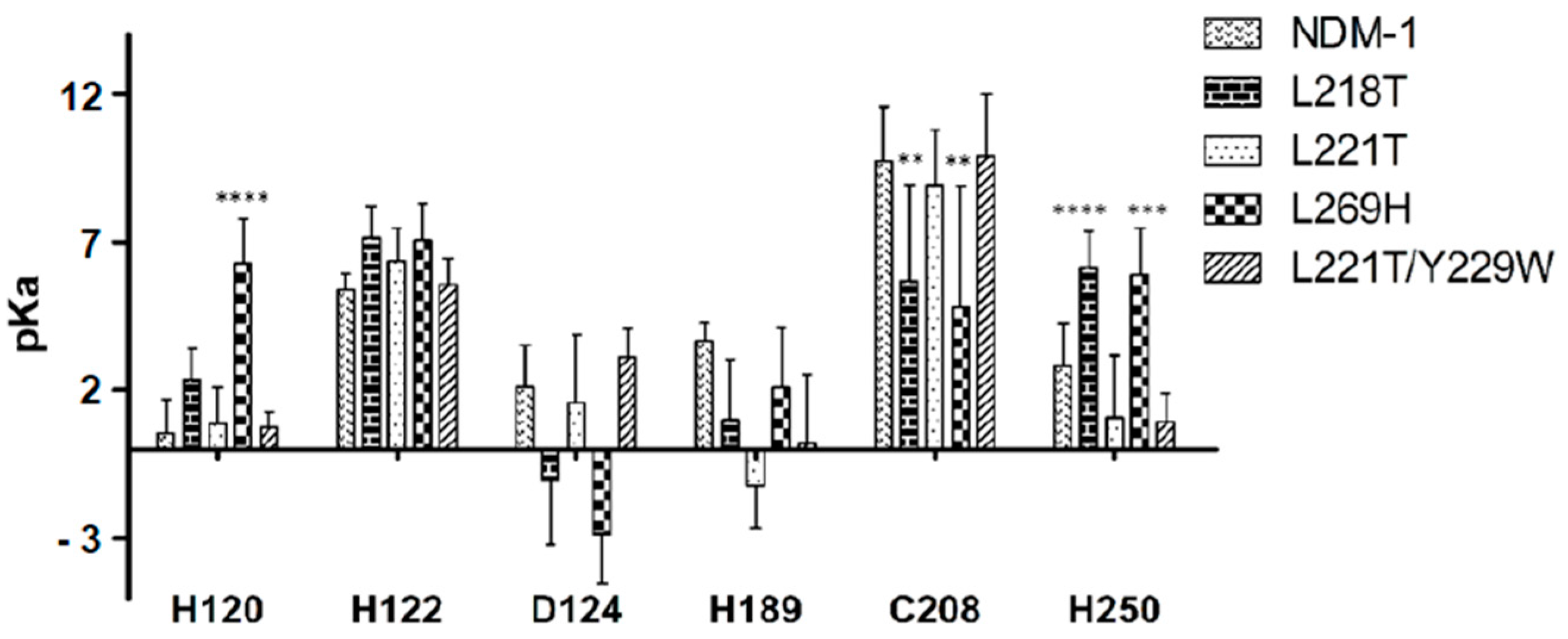
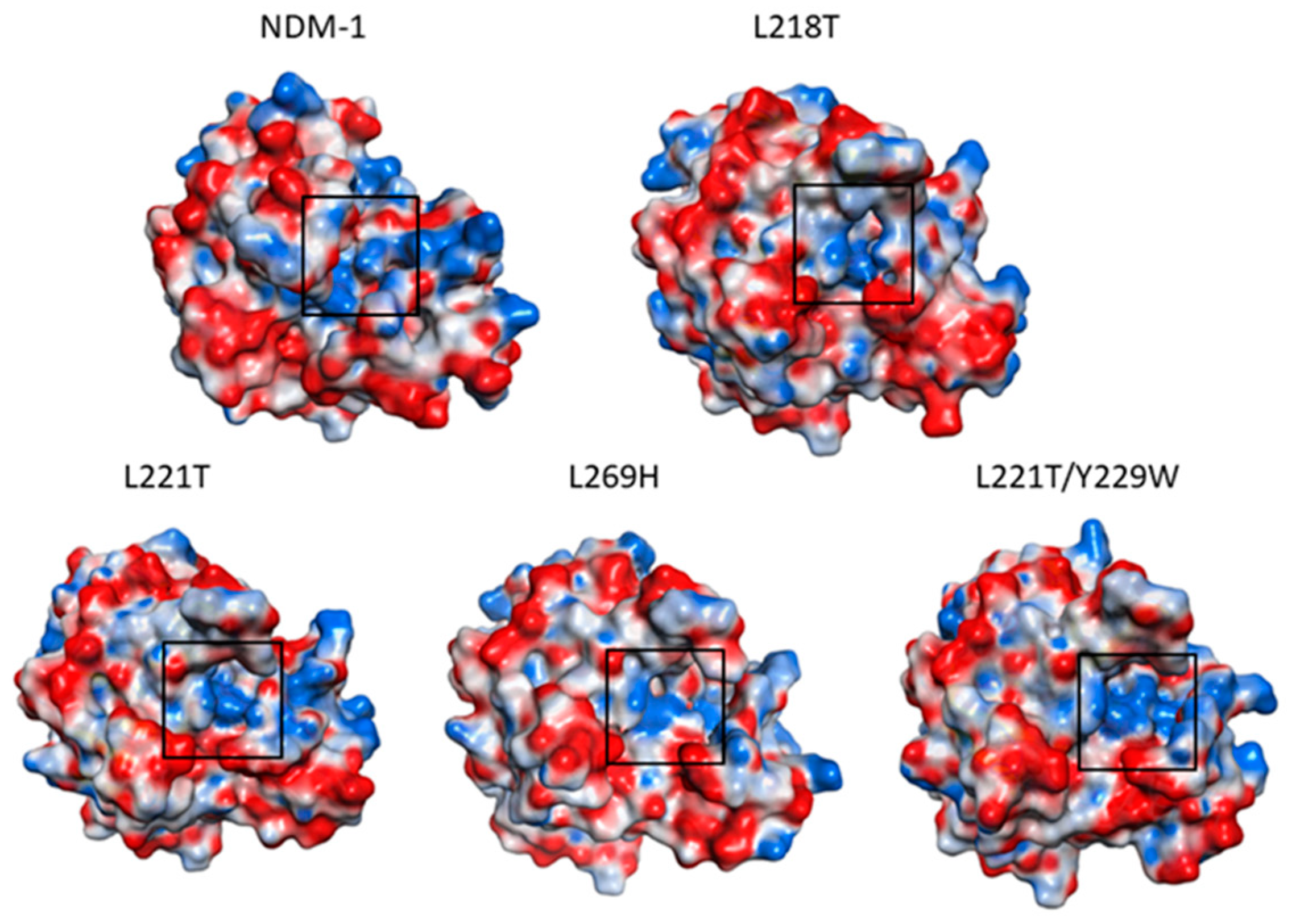
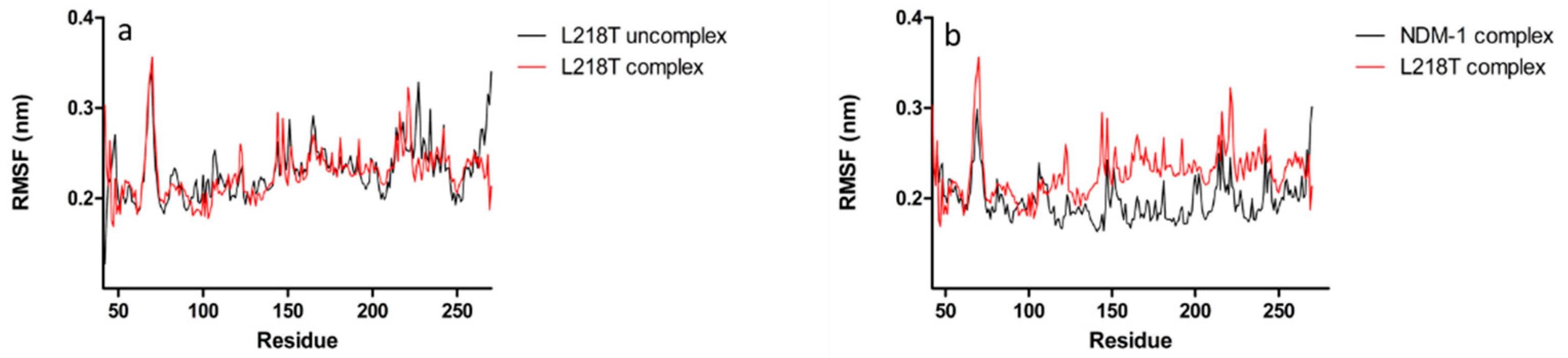
2.5. Molecular Dynamics Simulations
2.6. Molecular Dynamics Simulations for NDM-1 and L218T in Complex with Meropenem
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Site-Directed Mutagenesis
5.2. Cloning of blaL218T, blaL221T, blaL269H and blaL221T/Y229W
5.3. Production and Purification of L218T, L221T, L269H and L221T-Y229W Enzymes
5.4. Determination of Kinetic Parameters
5.5. Fluorescence Assays
5.6. Thermofluor Assay
5.7. Molecular Dynamics Simulations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bush, K.; Bradford, P.A. Epidemiology of β-lactamase-producing pathogens. Clin. Microbiol. Rev. 2020, 33, e00047-19. [Google Scholar] [CrossRef] [PubMed]
- Papp-Wallace, K.M.; Endimiani, A.; Taracila, M.A.; Bonomo, R.A. Carbapenems: Past, present, and future. Antimicrob. Agents Chemother. 2011, 55, 4943–4960. [Google Scholar] [CrossRef] [PubMed]
- Ambler, R.P. The structure of β-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1980, 289, 321–331. [Google Scholar]
- Hall, B.G.; Barlow, M. Revised Ambler classification of β-lactamases. J. Antimicrob. Chemother. 2005, 55, 1050–1051. [Google Scholar] [CrossRef] [PubMed]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef]
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; MaNally, A.; Zong, Z. NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 2019, 32, e00115-18. [Google Scholar] [CrossRef]
- Iredell, J.; Brown, J.; Tagg, K. Antibiotic resistance in Enterobacteriaceae: Mechanisms and clinical implications. BMJ 2016, 352, h6420. [Google Scholar] [CrossRef]
- Chatterjee, S.; Mondal, A.; Mitra, S.; Basu, S. Acinetobacter baumannii transfers the blaNDM-1 gene via outer membrane vesicles. J. Antimicrob. Chemother. 2017, 72, 2201–2207. [Google Scholar] [CrossRef] [PubMed]
- Krahn, T.; Wibberg, D.; Maus, I.; Winkler, A.; Bontron, S.; Sczyrba, A.; Nordmann, P.; Pulher, A.; Poirel, L.; Schluter, A. Intraspecies transfer of the chromosomal Acinetobacter baumannii blaNDM-1 carbapenemase gene. Antimicrob. Agents Chemother. 2016, 60, 3032–3040. [Google Scholar] [CrossRef]
- Ramirez, M.S.; Bonomo, R.A.; Tolmasky, M.E. Carbapenemases: Transforming Acinetobacter baumannii into a yet more dangerous menace. Biomolecules 2020, 10, 720. [Google Scholar] [CrossRef]
- Tanner, W.D.; Atkinson, R.M.; Goel, R.K.; Toleman, M.A.; Benson, L.S.; Porucznik, C.A.; Van Derslice, J.A. Horizontal transfer of the blaNDM-1 gene to Pseudomonas aeruginosa and Acinetobacter baumannii in biofilms. FEMS Microbiol. Lett. 2017, 1, 364. [Google Scholar] [CrossRef]
- Kumarasamy, K.K.; Toleman, M.A.; Walsh, T.R.; Bagaria, J.; Butt, F.; Balakrishnan, R. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010, 10, 597–602. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L.; Walsh, T.R.; Livermore, D.M. The emerging NDM carbapenemases. Trends Microbiol. 2011, 19, 588–595. [Google Scholar] [CrossRef]
- Garau, G.; Garcia-Saez, I.; Bebrone, C.; Anne, C.; Mercuri, P.; Galleni, M.; Frere, J.M.; Dideberg, O. Update of the standard numbering scheme for class β-lactamases. Antimicrob. Agents Chemother. 2004, 48, 2347–2349. [Google Scholar] [CrossRef]
- González, J.M.; Meini, M.R.; Tomatis, P.E.; Medrano Martín, F.J.; Cricco, J.A.; Vila, A.J. Metallo-β-lactamases withstand low Zn(II) conditions by tuning metal-ligand interactions. Nat. Chem. Biol. 2012, 8, 698–700. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Cunningham, M.A.; Mire, J.; Tesar, C.; Sacchettini, J.; Joachimiak, A. NDM-1, the ultimate promiscuous enzyme: Substrate recognition and catalytic mechanism. FASEB J. 2013, 27, 1917–1927. [Google Scholar] [CrossRef] [PubMed]
- Lisa, M.N.; Palacios, A.R.; Aitha, M.; Gonzalez, M.M.; Moreno, D.M.; Crowder, M.W.; Bonomo, R.A.; Spencer, J.; Tierney, D.L.; Llarrull, L.I.; et al. A general reaction mechanism for carbapenem hydrolysis by mononuclear and binuclear metallo-β-lactamases. Nat. Commun. 2017, 8, 538. [Google Scholar] [CrossRef]
- Zhang, H.; Hao, Q. Crystal structure of NDM-1 reveals a common β-lactam hydrolysis mechanism. FASEB J. 2011, 25, 2574–2582. [Google Scholar] [CrossRef]
- Gou, Y.; Wang, J.; Niu, G.; Shui, W.; Sun, Y.; Zhou, H.; Zhang, Y.; Yang, C.; Lou, Z.; Rao, Z. A structural view of the antibiotic degradation enzyme NDM-1 from a superbug. Protein Cell 2011, 2, 384–394. [Google Scholar]
- Khan, S.; Ali, A.; Khan, A.U. Structural and functional insight of New Delhi metallo-β-lactamase-1 variants. Future Med. Chem. 2018, 10, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, H.; Shi, Y.; Hu, F.; Lao, X.; Gao, X.; Zheng, H.; Yao, W. Probing the effect of the non-active-site mutation Y229W in New Delhi metallo-β-lactamase-1 by site-directed mutagenesis, kinetic studies, and molecular dynamics simulations. PLoS ONE 2013, 8, e82080. [Google Scholar] [CrossRef] [PubMed]
- Moali, C.; Anne, C.; Lamotte-Brasseur, J.; Groslambert, S.; Devreese, B.; Van Beeumen, J.; Galleni, M.; Frère, J.M. Analysis of the importance of the metallo-β-lactamase active site loop in substrate binding and catalysis. Chem. Biol. 2003, 10, 319–329. [Google Scholar] [CrossRef]
- Furuyama, T.; Nonomura, H.; Ishii, Y.; Hanson, N.D.; Shimizu-Ibuka, A. Structural and mutagenic analysis of metallo-β-lactamase IMP-18. Antimicrob. Agents Chemother. 2016, 60, 5521–5526. [Google Scholar] [CrossRef][Green Version]
- Palacios, A.R.; Mojica, M.F.; Giannini, E.; Taracila, M.A.; Bethel, C.R.; Alzari, P.M.; Otero, L.H.; Klinke, S.; Llarrull, L.I.; Bonomo, R.A.; et al. The reaction mechanism of metallo-β-lactamases is tuned by the conformation of an active-site mobile loop. Antimicrob. Agents Chemother. 2019, 63, e01754-18. [Google Scholar] [CrossRef]
- Piccirilli, A.; Brisdelli, F.; Aschi, M.; Celenza, G.; Amicosante, G.; Perilli, M. Kinetic profile and molecular dynamic studies show that Y229W substitution in an NDM-1/L209F variant restores the hydrolytic activity of the enzyme toward penicillins, cephalosporins, and carbapenems. Antimicrob. Agents Chemother. 2019, 63, e02270-18. [Google Scholar] [CrossRef] [PubMed]
- Green, V.L.; Verma, A.; Owens, R.J.; Phillips, S.E.V.; Carr, S.B. Structure of New Delhi metallo-β-lactamase 1 (NDM-1). Acta Cryst. 2011, 67, 1160–1164. [Google Scholar] [CrossRef]
- Marcoccia, F.; Leiros, H.K.S.; Aschi, M.; Amicosante, G.; Perilli, M. Exploring the role of L209 residue in active site of NDM-1 a metallo-β-lactamase. PLoS ONE 2018, 13, e0189686. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, G.; Zhu, Y.; Zeng, L.; Ahmad, A.; Wang, C.; Pang, B.; Fang, H.; Zhao, L.; Hao, Q. Active-site conformational fluctuations promote the enzymatic activity of NDM-1. Antimicrob. Agents Chemother. 2018, 62, e01579-18. [Google Scholar] [CrossRef]
- Gonzalez, J.M.; Medrano Martin, F.J.; Costello, A.L.; Tierney, D.L.; Vila, A.J. The Zn2 position in metallo-β-lactamases is critical for activity: A study on chimeric metal sites on a conserved protein scaffold. J. Mol. Biol. 2007, 373, 1141–1156. [Google Scholar] [CrossRef]
- Tomatis, P.E.; Fabiane, S.M.; Simona, F.; Carloni, P.; Sutton, B.J.; Vila, A.J. Adaptive protein evolution grants organismal fitness by improving catalysis and flexibility. Proc. Natl. Acad. Sci. USA 2008, 105, 20605–20610. [Google Scholar] [CrossRef]
- Marcoccia, F.; Bottoni, C.; Sabatini, A.; Colapietro, M.; Mercuri, P.S.; Galleni, M.; Kerff, F.; Matagne, A.; Celenza, G.; Amicosante, G.; et al. Kinetic Study of Laboratory Mutants of NDM-1 Metallo-β-Lactamase and the Importance of an Isoleucine at Position 35. Antimicrob. Agents Chemother. 2016, 60, 2366–2372. [Google Scholar] [CrossRef] [PubMed]
- Piccirilli, A.; Mercuri, P.S.; Galleni, M.; Aschi, M.; Matagne, A.; Amicosante, G.; Perilli, M. P174E Substitution in GES-1 and GES-5 β-lactamases improves catalytic efficiency toward carbapenems. Antimicrob. Agents Chemother. 2018, 62, e01851-17. [Google Scholar] [CrossRef] [PubMed]
- Segel, I.H. Biochemical Calculations, 2nd ed.; Dawes, E.A., Ed.; John Wiley & Sons: New York, NY, USA, 1976; Volume 4, pp. 236–241. [Google Scholar]
- Frère, J.M.; Verlaine, O. First-order kinetics in the study of enzymes: Application for the reporter substrate method and to the estimation of kcat/Km. Curr. Biotechnol. 2020, 9, 171–176. [Google Scholar] [CrossRef]
- Molecular Operating Environment (MOE), 2019.01; Chemical Computing Group ULC, 1010 Sherbooke St. West, Suite #910, Montreal, QC H3A 2R7, Canada. 2021.
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Feng, H.; Liu, X.; Wang, S.; Fleming, J.; Wang, D.C.; Liu, W. The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis. Nat. Commun. 2017, 8, 2242. [Google Scholar] [CrossRef]
| Substrates | Variant | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| Imipenem | NDM-1 * | 35 ± 1 | 64 ± 3 | 1.8 × 106 |
| L218T | 13 ± 1 | 2 ± 0.1 | 1.5 × 105 | |
| L221T | 59 ± 4 | 8 ± 1 | 1.3 × 105 | |
| L269H | 149 ± 9 | 40 ± 3 | 2.7 × 105 | |
| L221T/Y229W | 129 ± 6 | 29 ± 1 | 2.2 × 105 | |
| Y229W * | 81 ± 3 | 38 ± 1 | 4.7 × 105 | |
| Meropenem | NDM-1 * | 80 ± 2 | 75 ± 2 | 9.4 × 105 |
| L218T | 22 ± 2 | 1 ± 0.1 | 4.5 × 104 | |
| L221T | 20 ± 1 | 9 ± 1 | 4.5 × 105 | |
| L269H | 117 ± 7 | 109 ± 8 | 9.3 × 105 | |
| L221T/Y229W | 174 ± 6 | 135 ± 3 | 7.7 × 105 | |
| Y229W * | 259 ± 18 | 820 ± 5 | 3.2 × 106 | |
| Benzylpenicillin | NDM-1 * | 250 ± 10 | 105 ± 5 | 4.2 × 105 |
| L218T | 832 ± 20 | 294 ± 2 | 3.5 × 105 | |
| L221T | 937 ± 25 | 78 ± 5 | 8.3 × 104 | |
| L269H | 541 ± 12 | 549 ± 5 | 1.0 × 106 | |
| L221T/Y229W | >2000 | >1000 | 0.29 | |
| Y229W * | 841 ± 35 | 552 ± 3 | 6.6 × 105 | |
| Carbenicillin | NDM-1 * | 285 ± 5 | 108 ± 4 | 3.8 × 105 |
| L218T | 265 ± 10 | 59 ± 3 | 2.2 × 105 | |
| L221T | 398 ± 15 | 48 ± 3 | 1.2 × 105 | |
| L269H | 339 ± 9 | 347 ± 5 | 1.0 × 106 | |
| L221T/Y229W | >2000 | >1000 | 0.29 | |
| Y229W * | 1176 ± 25 | 866 ± 6 | 7.3 × 105 | |
| Cefazolin | NDM-1 * | 20 ± 1 | 42 ± 1 | 2.1 × 106 |
| L218T | 74 ± 5 | 9 ± 1 | 1.2 × 105 | |
| L221T | 25 ± 3 | 8 ± 1 | 3.2 × 105 | |
| L269H | 41 ± 2 | 83 ± 2 | 2.0 × 106 | |
| L221T/Y229W | 9 ± 1 | 14 ± 1 | 1.5 × 106 | |
| Y229W* | 48 ± 2 | 191 ± 4 | 4.0 × 106 | |
| Cefoxitin | NDM-1 * | 26 ± 1 | 23 ± 1 | 8.8 × 105 |
| L218T | 31 ± 3 | 3 ± 0.2 | 9.6 × 104 | |
| L221T | 31 ± 3 | 1.5 ± 0.1 | 4.8 × 104 | |
| L269H | 100 ± 5 | 11 ± 1 | 1.1 × 105 | |
| L221T/Y229W | >5000 | >11 | 3.1 × 103 | |
| Y229W * | 2100 ± 25 | 4 ± 0.2 | 2.0 × 103 | |
| Cefotaxime | NDM-1 * | 14 ± 1 | 20 ± 1 | 1.4 × 106 |
| L218T | 24 ± 2 | 30 ± 2 | 1.2 × 106 | |
| L221T | 10 ± 1 | 2 ± 0.1 | 2.0 × 10 | |
| L269H | 37 ± 2 | 37 ± 1 | 1.0 × 106 | |
| L221T/Y229W | 10 ± 1 | 14 ± 1 | 1.4 × 106 | |
| Y229W * | 52 ± 4 | 107 ± 3 | 2.1 × 106 | |
| Ceftazidime | NDM-1 * | 50 ± 4 | 18 ± 0.5 | 3.7 × 105 |
| L218T | 19 ± 1 | 45 ± 3 | 2.4 × 106 | |
| L221T | 32 ± 1 | 19 ± 1 | 5.9 × 105 | |
| L269H | 81 ± 4 | 10 ± 1 | 1.2 × 105 | |
| L221T/Y229W | 222 ± 10 | 7 ± 1 | 3.0 × 104 | |
| Y229W * | 250 ± 15 | 10 ± 1 | 4.0 × 104 | |
| Cefepime | NDM-1 * | 35 ± 5 | 13 ± 1 | 3.7 × 105 |
| L218T | 39 ± 2 | 3 ± 0.1 | 7.7 × 104 | |
| L221T | 40 ± 3 | 2 ± 0.2 | 5.0 × 104 | |
| L269H | 64 ± 3 | 11 ± 1 | 1.7 × 105 | |
| L221T/Y229W | 152 ± 8 | 3 ± 0.5 | 2.0 × 104 | |
| Y229W * | 117 ± 8 | 2.5 ± 0.1 | 2.0 × 104 |
| Meropenem | HEPES 20 mM 20 μM ZnCl2 pH 7.00 | BIS TRIS 20 mM 20 μM ZnCl2 pH 6.5 | MES 20 mM 20 μM ZnCl2 pH 6.0 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) | Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) | Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) | |
| NDM-1 | 80 ± 2 | 75 | 0.94 | 46 ± 2 | 18 | 0.39 | 98 ± 3 | 40 | 0.41 |
| L218T | 22 ± 2 | 1 | 0.04 | 166 ± 7 | 11 | 0.06 | 189 ± 8 | 15 | 0.08 |
| L221T | 20 ± 1 | 9 | 0.45 | 30 ± 2 | 5 | 0.16 | 28 ± 2 | 4 | 0.14 |
| L269H | 117 ± 7 | 109 | 0.93 | 42 ± 3 | 96 | 2.28 | 182 ± 7 | 167 | 0.92 |
| L221T/Y229W | 174 ± 6 | 135 | 0.77 | 298 ± 18 | 67 | 0.22 | 101 ± 4 | 36 | 0.36 |
| Y229W | 259 ± 18 | 820 | 3.17 | 72 ± 4 | 67 | 0.93 | 104 ± 5 | 60 | 0.57 |
| Enzymes | TmB (°C) | ΔTmB (°C) | TmD (°C) | ΔTmD (°C) |
|---|---|---|---|---|
| NDM-1 | 50.66 | − | 53.73 | − |
| L218T | 50.38 | −0.28 | 52.41 | 1.32 |
| L221T | 49.06 | −1.60 | 46.55 | −7.18 |
| L269H | 42.87 | −7.79 | 44.51 | −9.22 |
| Y229W | 46.62 | −4.04 | 47.86 | −5.87 |
| L221T/Y229W | 45.02 | −5.64 | 46.36 | −7.37 |
| NDM-1 | L218T | L221T | L269H | L221T/Y229W | |
|---|---|---|---|---|---|
| Zn1-Zn2 | 3.26 ± 0.05 | 3.28 ± 0.05 | 3.27 ± 0.05 | 3.18 ± 0.10 | 3.27 ± 0.06 |
| Zn1-OH- | 1.70 ± 0.03 | 1.71 ± 0.03 | 1.70 ± 0.03 | 1.68 ± 0.03 | 1.71 ± 0.03 |
| Zn2-OH- | 1.66 ± 0.02 | 1.68 ± 0.03 | 1.68 ± 0.03 | 1.69 ± 0.03 | 1.66 ± 0.02 |
| Zn1-H120 | 4.30 ± 0.49 | 4.13 ± 0.28 | 4.29 ± 0.29 | 1.84 ± 0.04 | 4.18 ± 0.26 |
| Zn1-H122 | 4.98 ± 0.84 | 4.40 ± 0.46 | 4.59 ± 0.47 | 4.37 ± 0.95 | 4.64 ± 0.52 |
| Zn1-H189 | 1.96 ± 0.07 | 1.93 ± 0.06 | 1.96 ± 0.07 | 4.27 ± 0.66 | 1.94 ± 0.07 |
| Zn2-D124 | 3.27 ± 0.51 | 1.89 ± 0.10 | 2.18 ± 0.63 | 2.90 ± 0.99 | 1.83 ± 0.30 |
| Zn2-C208 | 1.95 ± 0.03 | 2.00 ± 0.05 | 1.99 ± 0.05 | 1.98 ± 0.05 | 1.95 ± 0.04 |
| Zn2-H250 | 4.26 ± 0.35 | 4.28 ± 0.31 | 4.32 ± 0.32 | 5.19 ± 2.92 | 4.40 ± 0.30 |
| NDM-1/Meropenem Complex | L218T/Meropenem Complex | |
|---|---|---|
| Zn1-Zn2 | 4.54 ± 0.15 | 5.87 ± 0.42 |
| Zn1-H120 | 3.63 ± 0.56 | 4.04 ± 0.78 |
| Zn1-H122 | 1.95 ± 0.06 | 4.34 ± 0.98 |
| Zn1-H189 | 2.05 ± 0.10 | 1.92 ± 0.07 |
| Zn2-D124 | 1.73 ± 0.04 | 1.84 ± 0.33 |
| Zn2-C208 | 1.93 ± 0.03 | 1.96 ± 0.04 |
| Zn2-H250 | 1.97 ± 0.07 | 2.36 ± 0.31 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piccirilli, A.; Criscuolo, E.; Brisdelli, F.; Mercuri, P.S.; Cherubini, S.; De Sciscio, M.L.; Maccarrone, M.; Galleni, M.; Amicosante, G.; Perilli, M. Exploring the Role of L10 Loop in New Delhi Metallo-β-lactamase (NDM-1): Kinetic and Dynamic Studies. Molecules 2021, 26, 5489. https://doi.org/10.3390/molecules26185489
Piccirilli A, Criscuolo E, Brisdelli F, Mercuri PS, Cherubini S, De Sciscio ML, Maccarrone M, Galleni M, Amicosante G, Perilli M. Exploring the Role of L10 Loop in New Delhi Metallo-β-lactamase (NDM-1): Kinetic and Dynamic Studies. Molecules. 2021; 26(18):5489. https://doi.org/10.3390/molecules26185489
Chicago/Turabian StylePiccirilli, Alessandra, Emanuele Criscuolo, Fabrizia Brisdelli, Paola Sandra Mercuri, Sabrina Cherubini, Maria Laura De Sciscio, Mauro Maccarrone, Moreno Galleni, Gianfranco Amicosante, and Mariagrazia Perilli. 2021. "Exploring the Role of L10 Loop in New Delhi Metallo-β-lactamase (NDM-1): Kinetic and Dynamic Studies" Molecules 26, no. 18: 5489. https://doi.org/10.3390/molecules26185489
APA StylePiccirilli, A., Criscuolo, E., Brisdelli, F., Mercuri, P. S., Cherubini, S., De Sciscio, M. L., Maccarrone, M., Galleni, M., Amicosante, G., & Perilli, M. (2021). Exploring the Role of L10 Loop in New Delhi Metallo-β-lactamase (NDM-1): Kinetic and Dynamic Studies. Molecules, 26(18), 5489. https://doi.org/10.3390/molecules26185489









