FT-Raman Spectroscopy as a Tool to Study the Secondary Structures of Wheat Gliadin Proteins
Abstract
1. Introduction
- Celiac disease—natural gluten intolerance connected with the Human Leukocyte Antigen (HLA) system.
- Gluten enteropathy—consequences of inflammation reaction in a small intestine.
- Food allergy to gluten—IgE-mediated disorders of skin and alimentary tract. Induce symptoms of rhinitis, asthma, and Wheat-Dependent Exercise Induced Anaphylaxis (WDEIA).
- Dermatitis herpetriformis—skin manifestation of gluten intolerance revealed as the skin bladder.
- Gluten ataxia of hands, legs, and vision disorders.
- Non-celiac gluten intolerance (e.g., the contact allergic reaction caused by gluten present in drugs, toothpastes, and cosmetics).
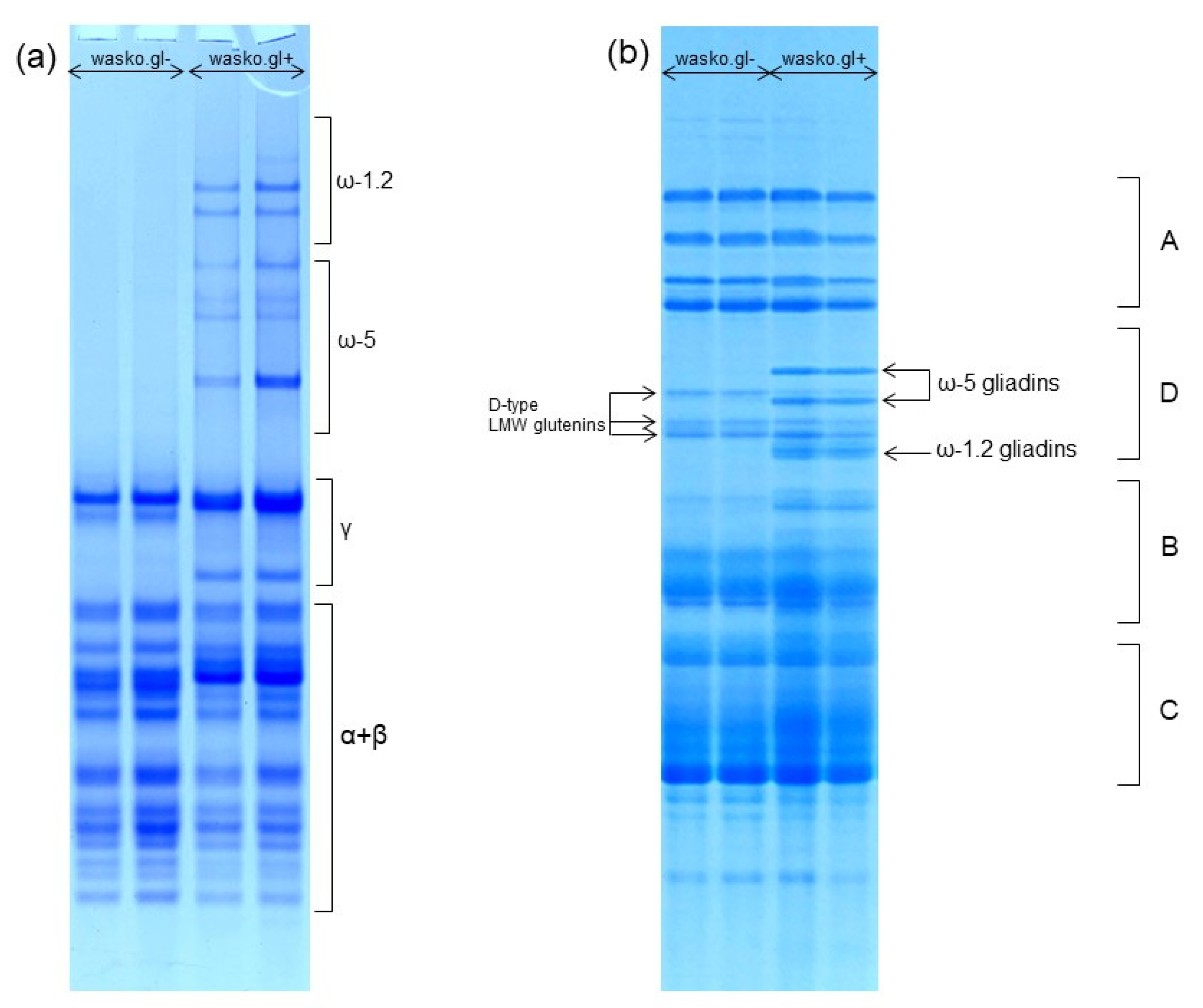
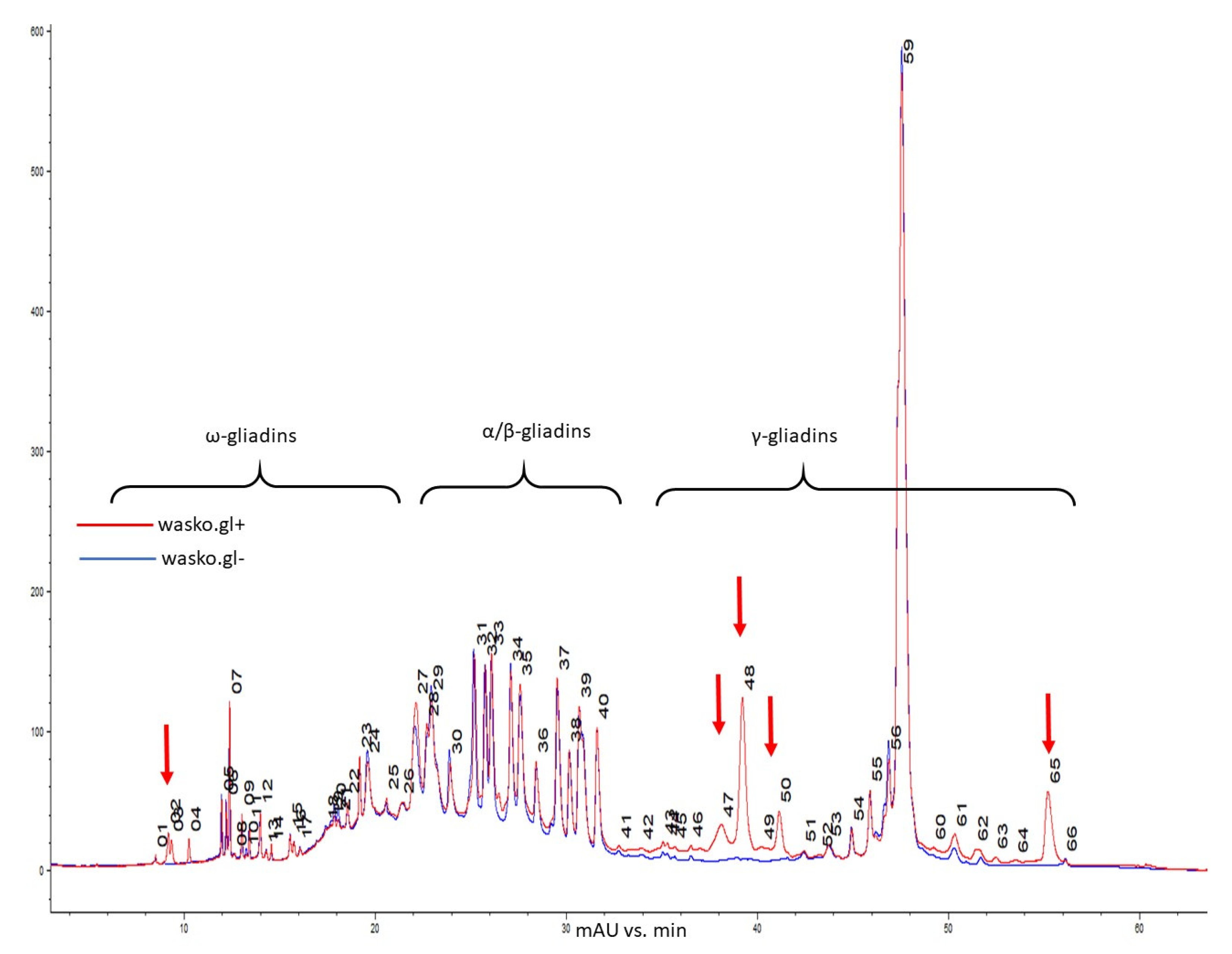
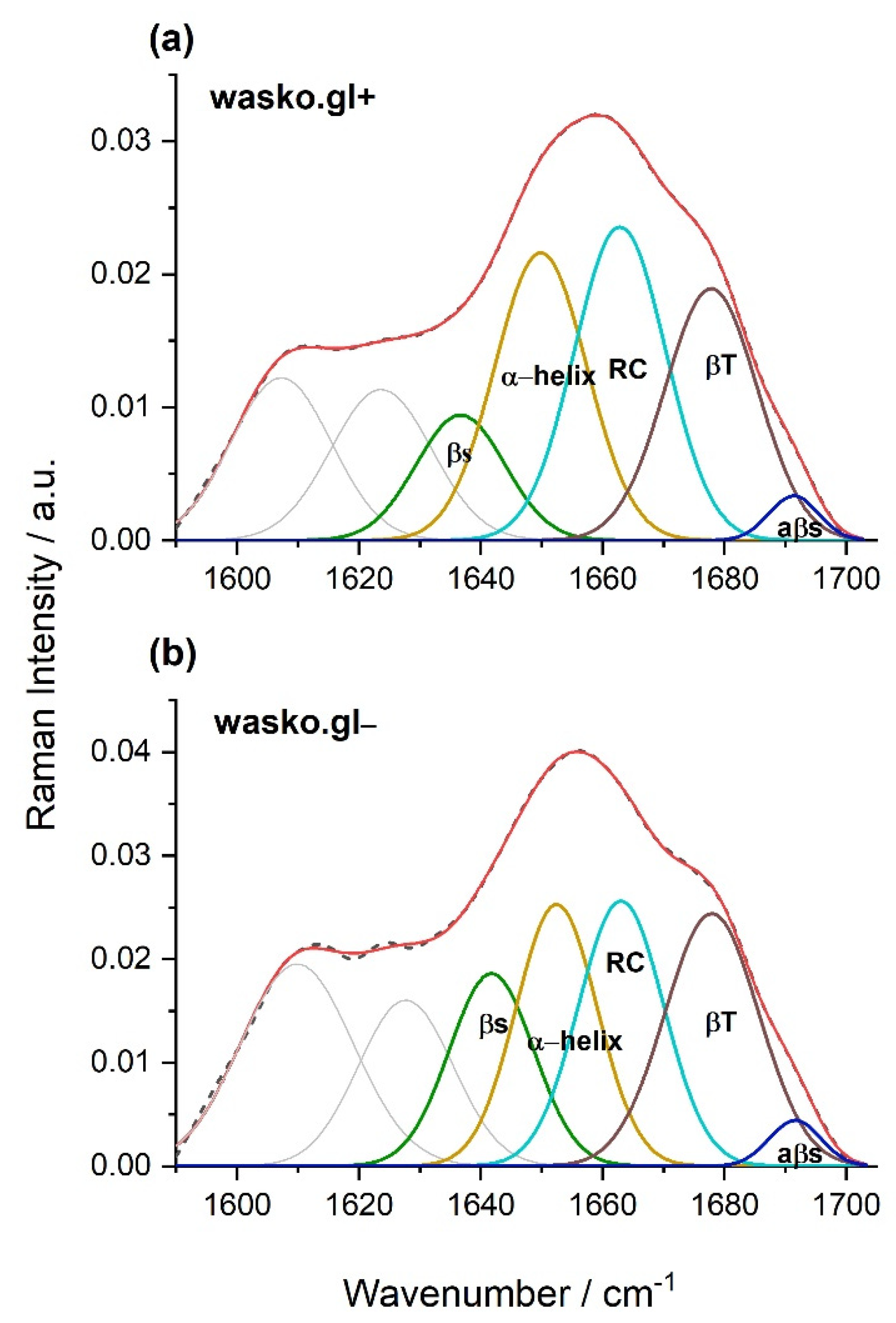
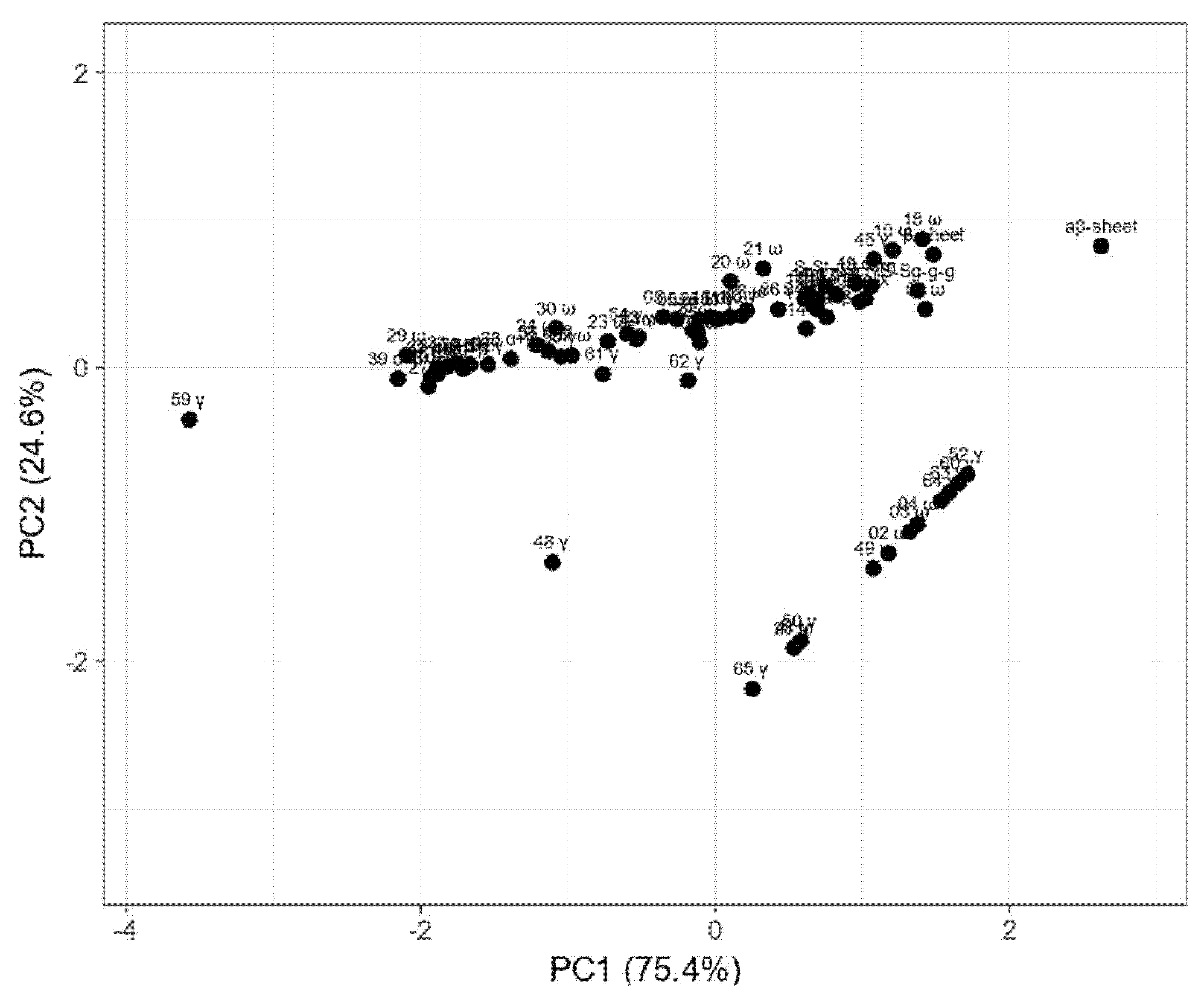
2. Results
3. Discussion
4. Materials and Methods
4.1. Wheat Lines
4.2. Sample Preparation
4.3. Gliadins Extraction
4.4. Acid Polyacrylamide Gel Electrophoresis (A-PAGE) of Wheat Gliadins
4.5. Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) of Wheat Total Proteins
4.6. Reverse-Phase High-Performance Liquid Chromatography (RP-HPLC) of Gliadin Fraction
4.7. Statistical Analysis
4.8. FT-Raman Spectroscopy and Curve Fitting
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Weselucha-Birczynska, A.; Labanowska, M.; Kurdziel, M.; Filek, M. Resonance Raman and EPR spectroscopy studies of untreated spring wheat leaves. Vib. Spectrosc. 2012, 60, 113–117. [Google Scholar] [CrossRef]
- Stawoska, I.; Staszak, A.M.; Ciereszko, I.; Oliwa, J.; Skoczowski, A. Using isothermal calorimetry and FT-Raman spectroscopy for step-by-step monitoring of maize seed germination: Case study. J. Anal. Calorim. 2020, 142, 755–763. [Google Scholar] [CrossRef]
- Saja, D.; Rys, M.; Stawoska, I.; Skoczowski, A. Metabolic response of cornflower (Centaurea cyanus L.) exposed to tribenuron-methyl: One of the active substances of sulfonylurea herbicides. Acta Physiol. Plant. 2016, 38, 13. [Google Scholar] [CrossRef]
- Nawrocka, A.; Krekora, M.; Niewiadomski, Z.; Szymanska-Chargot, M.; Krawecka, A.; Sobota, A.; Mis, A. Effect of moisturizing pre-treatment of dietary fibre preparations on formation of gluten network during model dough mixing—A study with application of FT-IR and FT-Raman spectroscopy. LWT-Food Sci. Technol. 2020, 121, 13. [Google Scholar] [CrossRef]
- Krombholz, R.; Lunter, D. A New Method for In-Situ Skin Penetration Analysis by Confocal Raman Microscopy. Molecules 2020, 25, 4222. [Google Scholar] [CrossRef]
- Saletnik, A.; Saletnik, B.; Puchalski, C. Overview of Popular Techniques of Raman Spectroscopy and Their Potential in the Study of Plant Tissues. Molecules 2021, 26, 1537. [Google Scholar] [CrossRef] [PubMed]
- Labanowska, M.; Weselucha-Birczynska, A.; Kurdziel, M.; Puch, P. Thermal effects on the structure of cereal starches. EPR and Raman spectroscopy studies. Carbohydr. Polym. 2013, 92, 842–848. [Google Scholar] [CrossRef] [PubMed]
- Filek, M.; Labanowska, M.; Kurdziel, M.; Weselucha-Birczynska, A.; Bednarska-Kozakiewicz, E. Structural and biochemical response of chloroplasts in tolerant and sensitive barley genotypes to drought stress. J. Plant. Physiol. 2016, 207, 61–72. [Google Scholar] [CrossRef]
- Moskal, P.; Weselucha-Birczynska, A.; Labanowska, M.; Filek, M. Adaxial and abaxial pattern of Urtica dioica leaves analyzed by 2DCOS ATR-FTIR as a function of their growth time and impact of environmental pollution. Vib. Spectrosc. 2019, 104, 7. [Google Scholar] [CrossRef]
- Kurdziel, M.; Dlubacz, A.; Weselucha-Birczynska, A.; Filek, M.; Labanowska, M. Stable radicals and biochemical compounds in embryos and endosperm of wheat grains differentiating sensitive and tolerant genotypes—EPR and Raman studies. J. Plant. Physiol. 2015, 183, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Labanowska, M.; Kurdziel, M.; Filek, M.; Weselucha-Birczynska, A. The impact of biochemical composition and nature of paramagnetic species in grains on stress tolerance of oat cultivars. J. Plant. Physiol. 2016, 199, 52–66. [Google Scholar] [CrossRef]
- Stawoska, I.; Weselucha-Birczynska, A.; Regonesi, M.E.; Riva, M.; Tortora, P.; Stochel, G. Interaction of selected divalent metal ions with human ataxin-3 Q36. J. Biol. Inorg. Chem. 2009, 14, 1175–1185. [Google Scholar] [CrossRef] [PubMed]
- Peticolas, W.L. Applications of Raman-spectroscopy to biological macromolecules. Biochimie 1975, 57, 417–428. [Google Scholar] [CrossRef]
- Maiti, N.C.; Apetri, M.M.; Zagorski, M.G.; Carey, P.R.; Anderson, V.E. Raman spectroscopic characterization of secondary structure in natively unfolded proteins: Alpha-synuclein. J. Am. Chem. Soc. 2004, 126, 2399–2408. [Google Scholar] [CrossRef] [PubMed]
- Lancelot, E.; Fontaine, J.; Grua-Priol, J.; Assaf, A.; Thouand, G.; Le-Bail, A. Study of structural changes of gluten proteins during bread dough mixing by Raman spectroscopy. Food Chemistry 2021, 358, 7. [Google Scholar] [CrossRef] [PubMed]
- Sadat, A.; Joye, I.J. Peak Fitting Applied to Fourier Transform Infrared and Raman Spectroscopic Analysis of Proteins. Appl. Sci. 2020, 10, 5918. [Google Scholar] [CrossRef]
- Tu, A.T. Raman spectroscopy in biology: Principles and applications; Wiley: New York, NY, USA, 1982. [Google Scholar]
- Lefevre, T.; Rousseau, M.E.; Pezolet, M. Protein secondary structure and orientation in silk as revealed by Raman spectromicroscopy. Biophys. J. 2007, 92, 2885–2895. [Google Scholar] [CrossRef] [PubMed]
- Chi, Z.H.; Chen, X.G.; Holtz, J.S.W.; Asher, S.A. UV resonance Raman-selective amide vibrational enhancement: Quantitative methodology for determining protein secondary structure. Biochemistry 1998, 37, 2854–2864. [Google Scholar] [CrossRef]
- De Gelder, J.; De Gussem, K.; Vandenabeele, P.; Moens, L. Reference database of Raman spectra of biological molecules. J. Raman Spectrosc. 2007, 38, 1133–1147. [Google Scholar] [CrossRef]
- Rygula, A.; Majzner, K.; Marzec, K.M.; Kaczor, A.; Pilarczyk, M.; Baranska, M. Raman spectroscopy of proteins: A review. J. Raman Spectrosc. 2013, 44, 1061–1076. [Google Scholar] [CrossRef]
- Ridgley, D.M.; Claunch, E.C.; Barone, J.R. Characterization of Large Amyloid Fibers and Tapes with Fourier Transform Infrared (FT-IR) and Raman Spectroscopy. Appl. Spectrosc. 2013, 67, 1417–1426. [Google Scholar] [CrossRef]
- Tuma, R. Raman spectroscopy of proteins: From peptides to large assemblies. J. Raman Spectrosc. 2005, 36, 307–319. [Google Scholar] [CrossRef]
- Sapone, A.; Bai, J.C.; Ciacci, C.; Dolinsek, J.; Green, P.H.R.; Hadjivassiliou, M.; Kaukinen, K.; Rostami, K.; Sanders, D.S.; Schumann, M.; et al. Spectrum of gluten-related disorders: Consensus on new nomenclature and classification. BMC Med. 2012, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Wrigley, C.; Békés, F.; Bushuk, W. Gluten: A balance of gliadin and glutenin. In Gliadin and Glutenin. The Unique Balance of Wheat Quality; Wrigley, C., Békés, F., Bushuk, W., Eds.; AACC International: St. Paul, MN, USA, 2006; pp. 3–32. [Google Scholar]
- Shewry, P.R.; Tatham, A.S.; Halford, N.G. The prolamins of the Triticeae. In Seed Proteins; Shewry, P.R., Casey, R., Eds.; Springer: Dordrecht, The Netherlands, 1999; pp. 33–78. [Google Scholar]
- Payne, P.; Holt, L.; Lister, P. Gli-A3 and Gli-B3, two newly designated loci coding for omega-type gliadins and D subunits of glutenin. In Proceedings of the 7th International Wheat Genetics Symposium, Cambridge, UK, 13–18 July 1988; Institute of Plant Science Research: Cambridge, UK; pp. 999–1002. [Google Scholar]
- Payne, P.I.; Corfield, K.G. Subunit composition of wheat glutenin proteins, isolated by gel-filtration in a dissociating medium. Planta 1979, 145, 83–88. [Google Scholar] [CrossRef]
- Wrigley, C.; Bietz, J. Proteins and amino acids. In Wheat: Chemistry and Technology; Pomeranz, Y., Ed.; American Association of Cereal Chemistry: St. Paul, MN, USA, 1988; Volume 1, pp. 159–275. [Google Scholar]
- Muller, S.; Wieser, H. The location of disulphide bonds in monomeric gamma-type gliadins. J. Cereal Sci. 1997, 26, 169–176. [Google Scholar] [CrossRef]
- Klosok, K.; Welc, R.; Fornal, E.; Nawrocka, A. Effects of Physical and Chemical Factors on the Structure of Gluten, Gliadins and Glutenins as Studied with Spectroscopic Methods. Molecules 2021, 26, 508. [Google Scholar] [CrossRef] [PubMed]
- Tatham, A.S. The structures of wheat proteins. In Wheat Structure: Biochemistry and Functionality; Schofield, J.D., Ed.; The Royal Society of Chemistry: Cambridge, UK, 1996; pp. 53–62. [Google Scholar]
- Shewry, P.; Tatham, A. Disulphide bonds in wheat gluten proteins. J. Cereal Sci. 1997, 25, 207–227. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Sokolowska, J.; Darewicz, M. The occurrence of sequences identical with epitopes from the allergen Pen a 1.0102 among food and non-food proteins. Pol. J. Food Nutr. Sci. 2015, 65, 21–29. [Google Scholar] [CrossRef]
- Mameri, H.; Brossard, C.; Gaudin, J.C.; Gohon, Y.; Paty, E.; Beaudouin, E.; Moneret-Vautrin, D.A.; Drouet, M.; Sole, V.; Wien, F.; et al. Structural Basis of IgE Binding to alpha- and gamma-Gliadins: Contribution of Disulfide Bonds and Repetitive and Nonrepetitive Domains. J. Agric. Food Chem. 2015, 63, 6546–6554. [Google Scholar] [CrossRef]
- Matsuo, H.; Morita, E.; Tatham, A.S.; Morimoto, K.; Horikawa, T.; Osuna, H.; Ikezawa, Z.; Kaneko, S.; Kohno, K.; Dekio, S. Identification of the IgE-binding epitope in omega-5 gliadin, a major allergen in wheat-dependent exercise-induced anaphylaxis. J. Biol. Chem. 2004, 279, 12135–12140. [Google Scholar] [CrossRef] [PubMed]
- Battais, F.; Mothes, T.; Moneret-Vautrin, D.A.; Pineau, F.; Kanny, G.; Popineau, Y.; Bodinier, M.; Denery-Papini, S. Identification of IgE-binding epitopes on gliadins for patients with food allergy to wheat. Allergy 2005, 60, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Rustgi, S.; Shewry, P.; Brouns, F.; Deleu, L.; Delcour, J.A. Wheat Seed Proteins: Factor's Influencing Their Content, Composition, and Technological Properties, and Strategies to Reduce Adverse Reactions. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1751–1769. [Google Scholar] [CrossRef] [PubMed]
- Leszczynska, J.; Waga, J.; Lacka, A.; Wolska, K.; Majak, I.; Bartos, A. The effect of enzymatic modification and genetic background on wheat gliadin immunological properties. Food Agric. Immunol. 2013, 24, 217–230. [Google Scholar] [CrossRef]
- Barro, F.; Iehisa, J.C.M.; Gimenez, M.J.; Garcia-Molina, M.D.; Ozuna, C.V.; Comino, I.; Sousa, C.; Gil-Humanes, J. Targeting of prolamins by RNAi in bread wheat: Effectiveness of seven silencing-fragment combinations for obtaining lines devoid of coeliac disease epitopes from highly immunogenic gliadins. Plant. Biotechnol. J. 2016, 14, 986–996. [Google Scholar] [CrossRef] [PubMed]
- Waga, J.; Skoczowski, A. Development and characteristics of ω-gliadin-free wheat genotypes. Euphytica 2014, 195, 105–116. [Google Scholar] [CrossRef]
- Skoczowski, A.; Obtulowicz, K.; Czarnobilska, E.; Dyga, W.; Mazur, M.; Stawoska, I.; Waga, J. Antibody reactivity in patients with IgE-mediated wheat allergy to various subunits and fractions of gluten and non-gluten proteins from omega-gliadin-free wheat genotypes. Ann. Agric. Environ. Med. 2017, 24, 229–236. [Google Scholar] [CrossRef]
- Cho, S.W.; Kang, C.S.; Kang, T.G.; Cho, K.M.; Park, C.S. Influence of different nitrogen application on flour properties, gluten properties by HPLC and end-use quality of Korean wheat. J. Integr. Agric. 2018, 17, 982–993. [Google Scholar] [CrossRef]
- Bietz, J.A. HPLC of cereal endosperm storage proteins. In HPLC of Biological Macromolecules, 2nd Edition, Revised and Expanded; Gooding, K.M., Regnier, F.E., Eds.; Marcel Dekker, Inc.: New York, NY, USA, 2002; pp. 547–587. [Google Scholar]
- Becker, D.; Folck, A.; Wieser, H. Inhibierung der α-Gliadin-Genexpression in hexaploidem Brotweizen. Getreidetechnologie 2006, 60, 153–156. [Google Scholar]
- Kumamarum, T.; Ogawa, M.; Satoh, H.; Okita, T.W. Protein Body Biogenesis in Cereal Endosperms. In Endosperm. Plant Cell Monographs (8); Olsen, O.A., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 8, pp. 141–158. [Google Scholar]
- Fan, D.M.; Ma, W.R.; Wang, L.Y.; Huang, J.L.; Zhao, J.X.; Zhang, H.; Chen, W. Determination of structural changes in microwaved rice starch using Fourier transform infrared and Raman spectroscopy. Starch-Starke 2012, 64, 598–606. [Google Scholar] [CrossRef]
- Kizil, R.; Irudayaraj, J.; Seetharaman, K. Characterization of Irradiated Starches by Using FT-Raman and FTIR Spectroscopy. J. Agric. Food Chem. 2002, 50, 3912–3918. [Google Scholar] [CrossRef]
- Liu, Y.Q.; Xu, Y.; Yan, Y.Z.; Hu, D.D.; Yang, L.Z.; Shen, R.L. Application of Raman spectroscopy in structure analysis and crystallinity calculation of corn starch. Starch-Starke 2015, 67, 612–619. [Google Scholar] [CrossRef]
- Corbett, E.C.; Zichy, V.; Goral, J.; Passingham, C. Fourier transform Raman studies of materials and compounds of biological importance—II. The effect of moisture on the molecular structure of the alpha and beta anomers of d-glucose. Spectrochim. Acta A 1991, 47, 1399–1411. [Google Scholar] [CrossRef]
- Schulz, H.; Baranska, M. Identification and quantification of valuable plant substances by IR and Raman spectroscopy. Vib. Spectrosc. 2007, 43, 13–25. [Google Scholar] [CrossRef]
- Wiercigroch, E.; Szafraniec, E.; Czamara, K.; Pacia, M.Z.; Majzner, K.; Kochan, K.; Kaczor, A.; Baranska, M.; Malek, K. Raman and infrared spectroscopy of carbohydrates: A review. Spectrochim. Acta Pt. A-Mol. Biomol. Spectroc. 2017, 185, 317–335. [Google Scholar] [CrossRef] [PubMed]
- Kacurakova, M.; Mathlouthi, M. FTIR and laser-Raman spectra of oligosaccharides in water: Characterization of the glycosidic bond. Carbohydr. Res. 1996, 284, 145–157. [Google Scholar] [CrossRef]
- Almeida, M.R.; Alves, R.S.; Nascimbem, L.; Stephani, R.; Poppi, R.J.; de Oliveira, L.F.C. Determination of amylose content in starch using Raman spectroscopy and multivariate calibration analysis. Anal. Bioanal. Chem. 2010, 397, 2693–2701. [Google Scholar] [CrossRef] [PubMed]
- Petrou, M.; Edwards, H.G.M.; Janaway, R.C.; Thompson, G.B.; Wilson, A.S. Fourier-Transform Raman spectroscopic study of a Neolithic waterlogged wood assemblage. Anal. Bioanal Chem 2009, 395, 2131–2138. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Durmaz, S.; Ozgenc, O.; Boyaci, I.H.; Yildiz, U.C.; Erisir, E. Examination of the chemical changes in spruce wood degraded by brown-rot fungi using FT-IR and FT-Raman spectroscopy. Vib. Spectrosc. 2016, 85, 202–207. [Google Scholar] [CrossRef]
- Schrader, B.; Klump, H.H.; Schenzel, K.; Schulz, H. Non-destructive NIR FT Raman analysis of plants. J. Mol. Struct. 1999, 509, 201–212. [Google Scholar] [CrossRef]
- Vitek, P.; Novotna, K.; Hodanova, P.; Rapantova, B.; Klem, K. Detection of herbicide effects on pigment composition and PSII photochemistry in Helianthus annuus by Raman spectroscopy and chlorophyll a fluorescence. Spectrochim. Acta Pt. A-Mol. Biomol. Spectroc. 2017, 170, 234–241. [Google Scholar] [CrossRef]
- Heredia-Guerrero, J.A.; Benitez, J.J.; Dominguez, E.; Bayer, I.S.; Cingolani, R.; Athanassiou, A.; Heredia, A. Infrared and Raman spectroscopic features of plant cuticles: A review. Front. Plant. Sci. 2014, 5, 14. [Google Scholar] [CrossRef]
- Baranska, M.; Schulz, H.; Baranski, R.; Nothnagel, T.; Christensen, L.P. In Situ Simultaneous Analysis of Polyacetylenes, Carotenoids and Polysaccharides in Carrot Roots. J. Agric. Food Chem. 2005, 53, 6565–6571. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.Q.; Zhang, L.M. Chemical structural and chain conformational characterization of some bioactive polysaccharides isolated from natural sources. Carbohydr. Polym. 2009, 76, 349–361. [Google Scholar] [CrossRef]
- Mahdad-Benzerdjeb, A.; Taleb-Mokhtari, I.N.; Sekkal-Rahal, M. Normal coordinates analyses of disaccharides constituted by D-glucose, D-galactose and D-fructose units. Spectrochim. Acta Pt. A-Mol. Biomol. Spectroc. 2007, 68, 284–299. [Google Scholar] [CrossRef]
- LiChan, E.C.Y. The applications of Raman spectroscopy in food science. Trends Food Sci. Technol. 1996, 7, 361–370. [Google Scholar] [CrossRef]
- Siamwiza, M.N.; Lord, R.C.; Chen, M.C.; Takamatsu, T.; Harada, I.; Matsuura, H.; Shimanouchi, T. Interpretation of doublet at 850 and 830 cm-1 in Raman spectra of tyrosyl residues in proteins and certain model compounds. Biochemistry 1975, 14, 4870–4876. [Google Scholar] [CrossRef]
- Nawrocka, A.; Szymanska-Chargot, M.; Mis, A.; Ptaszynska, A.A.; Kowalski, R.; Wasko, P.; Gruszecki, W.I. Influence of dietary fibre on gluten proteins structure—a study on model flour with application of FT-Raman spectroscopy. J. Raman Spectroc. 2015, 46, 309–316. [Google Scholar] [CrossRef]
- Ferrer, E.G.; Gomez, A.V.; Anon, M.C.; Puppo, M.C. Structural changes in gluten protein structure after addition of emulsifier. A Raman spectroscopy study. Spectrochim. Acta Pt. A-Mol. Biomol. Spectroc. 2011, 79, 278–281. [Google Scholar] [CrossRef] [PubMed]
- Meng, G.T.; Ma, C.Y.; Phillips, D.L. Raman spectroscopic study of globulin from Phaseolus angularis (red bean). Food Chem. 2003, 81, 411–420. [Google Scholar] [CrossRef]
- Linlaud, N.; Ferrer, E.; Puppo, M.C.; Ferrero, C. Hydrocolloid Interaction with water, protein, and starch in wheat dough. J. Agric. Food Chem. 2011, 59, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Nawrocka, A.; Mis, A.; Szymanska-Chargot, M. Characteristics of relationships between structure of gluten proteins and dough rheology—influence of dietary fibres studied by FT-raman spectroscopy. Food Biophys. 2016, 11, 81–90. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhao, D.; Foster, T.J.; Liu, Y.X.; Wang, Y.; Nirasawa, S.; Tatsumi, E.; Cheng, Y.Q. Konjac glucomannan-induced changes in thiol/disulphide exchange and gluten conformation upon dough mixing. Food Chem. 2014, 143, 163–169. [Google Scholar] [CrossRef]
- Nawrocka, A.; Szymanska-Chargot, M.; Mis, A.; Kowalski, R.; Gruszecki, W.I. Raman studies of gluten proteins aggregation induced by dietary fibres. Food Chem. 2016, 194, 86–94. [Google Scholar] [CrossRef]
- Tatham, A.S.; Shewry, P.R. The conformation of wheat gluten proteins—The secondary structures and thermal stabilities of alpha-gliadins, beta-gliadins, gamma-gliadins and omega-gliadins. J. Cereal Sci. 1985, 3, 103–113. [Google Scholar] [CrossRef]
- Lindsay, M.P.; Skerritt, J.H. The glutenin macropolymer of wheat flour doughs: Structure-function perspectives. Trends Food Sci. Technol. 1999, 10, 247–253. [Google Scholar] [CrossRef]
- Feng, J.; ZHANG, S.; ZHANG, Y.; Jinshui, W. Correlation of gluten molecular conformation with dough viscoelastic properties during storage. Grain Oil Sci. Technol. 2018, 1, 1–7. [Google Scholar]
- Peng, H.; Li, B.; Tian, J. Impact of Punicalagin on the Physicochemical and Structural Properties of Wheat Flour Dough. Foods 2019, 8, 606. [Google Scholar] [CrossRef] [PubMed]
- Bushuk, W.; Zillman, R.R. Wheat cultivar identification by gliadin electrophoregrams. 1. Apparatus, method and nomenclature. Can. J. Plant. Sci. 1978, 58, 505–515. [Google Scholar] [CrossRef]
- Metakovsky, E.; Novoselskaya, A. Gliadin allele identification in common wheat I: Methodological aspects of the analysis of gliadin patterns by one-dimensional polyacrylamide gel electrophoresis. J. Genet. Breed. 1991, 45, 317–324. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- James, L.C.; Roversi, P.; Tawfik, D.S. Antibody multispecificity mediated by conformational diversity. Science 2003, 299, 1362–1367. [Google Scholar] [CrossRef] [PubMed]
- Varjonen, E.; Vainio, E.; Kalimo, K. Antigliadin IgE—indicator of wheat allergy in atopic dermatitis. Allergy 2000, 55, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]


| Peak Number | Wavenumber/cm−1 | Chemical Components | References |
|---|---|---|---|
| 1 | 440 | polysaccharides (skeletal mode of pyranose ring vibrations) | [47,48] |
| 2 | 480 | polysaccharides (marker band to identify presence of starch) | [48,49,50,51,52,53] |
| 3 | 865 | mono-, di- and polysaccharides (stretching or deformation modes referring to the glycosidic bond) | [49,50,51,52] |
| 4 | 940 | polysaccharides (marker band for glycosidic linkage observed in starch) | [48,49,51,52,54] |
| 5 | 1052 | polysaccharides | [48,49,52,54] |
| 6 | 1084 | polysaccharides | [49] |
| 7 | 1127 | polysaccharides | [48,49,52] |
| 8,9 | 1260, 1340 | polysaccharides, lipids, fatty acids | [48,49,52,54] |
| 10 | 1380 | polysaccharides (deformation modes) | [48,49,51,54] |
| 11 | 1457 | polysaccharides, lipids, fatty acids | [48,49,54,55,56] |
| 12 | 1600 | polyphenols, flavonoids (stretching ring vibrations) | [51,57,58,59] |
| 13 | 1660 | amide I | [2,12,51] |
| Seed Lines | SSg-g-g [%] | SSt-g-g [%] | SSt-g-t [%] |
|---|---|---|---|
| wasko.gl+ | 18 (514) | 46 (519, 524) | 36 (531, 545) |
| wasko.gl− | 15 (513) | 39 (519, 525) | 46 (533, 546) |
| wasko.gl− | wasko.gl+ | |
|---|---|---|
| β-sheet (1634–1640 cm−1) | 19 | 12 |
| α-helix (1649–1652 cm−1) | 25 | 29 |
| RC (1663 cm−1) | 26 | 31 |
| β-turn (1677–1678 cm−1) | 27 | 25 |
| aβ-sheet (1690–1692 cm−1) | 3 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stawoska, I.; Wesełucha-Birczyńska, A.; Skoczowski, A.; Dziurka, M.; Waga, J. FT-Raman Spectroscopy as a Tool to Study the Secondary Structures of Wheat Gliadin Proteins. Molecules 2021, 26, 5388. https://doi.org/10.3390/molecules26175388
Stawoska I, Wesełucha-Birczyńska A, Skoczowski A, Dziurka M, Waga J. FT-Raman Spectroscopy as a Tool to Study the Secondary Structures of Wheat Gliadin Proteins. Molecules. 2021; 26(17):5388. https://doi.org/10.3390/molecules26175388
Chicago/Turabian StyleStawoska, Iwona, Aleksandra Wesełucha-Birczyńska, Andrzej Skoczowski, Michał Dziurka, and Jacek Waga. 2021. "FT-Raman Spectroscopy as a Tool to Study the Secondary Structures of Wheat Gliadin Proteins" Molecules 26, no. 17: 5388. https://doi.org/10.3390/molecules26175388
APA StyleStawoska, I., Wesełucha-Birczyńska, A., Skoczowski, A., Dziurka, M., & Waga, J. (2021). FT-Raman Spectroscopy as a Tool to Study the Secondary Structures of Wheat Gliadin Proteins. Molecules, 26(17), 5388. https://doi.org/10.3390/molecules26175388








