Recent Advances about the Applications of Click Reaction in Chemical Proteomics
Abstract
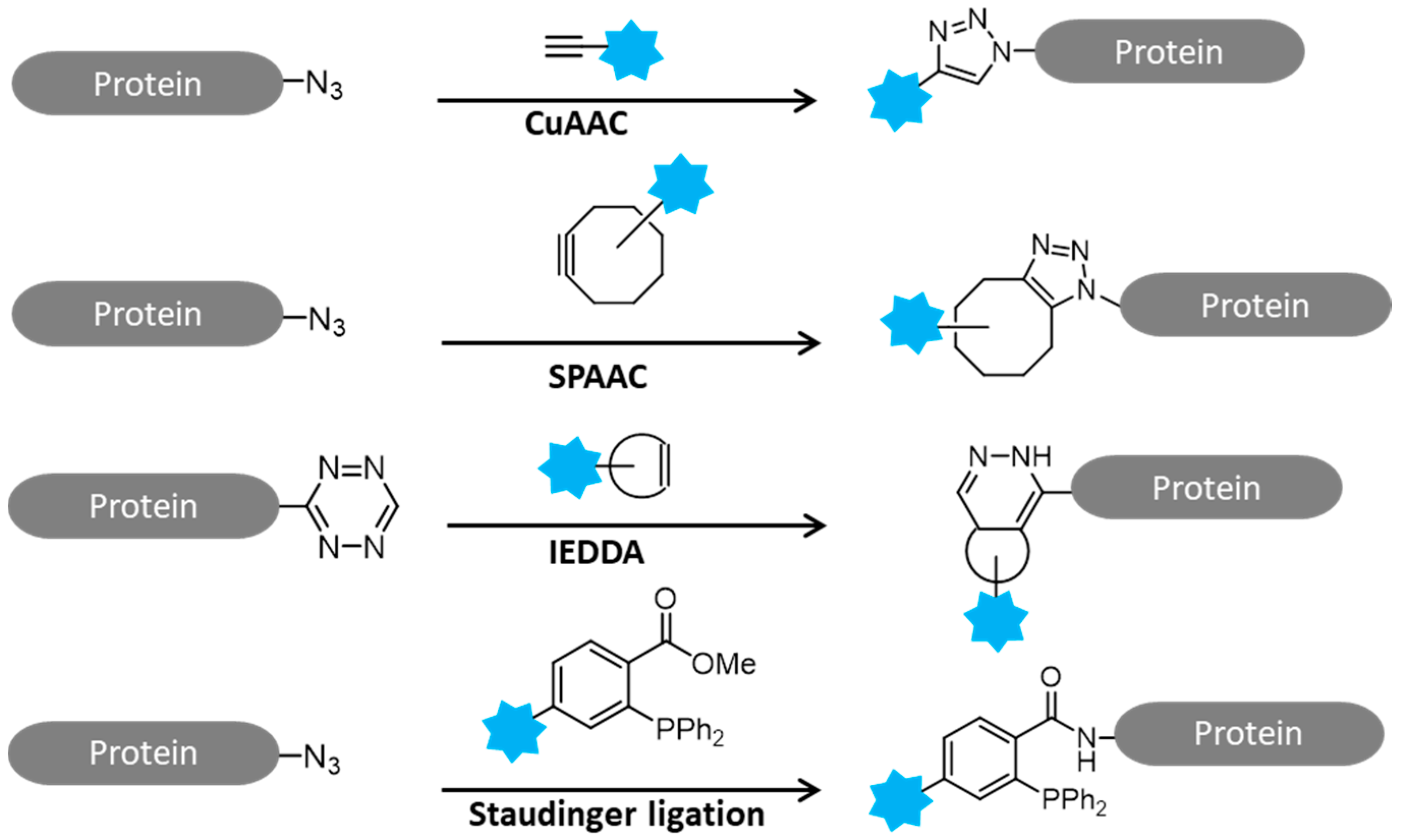
:1. Introduction
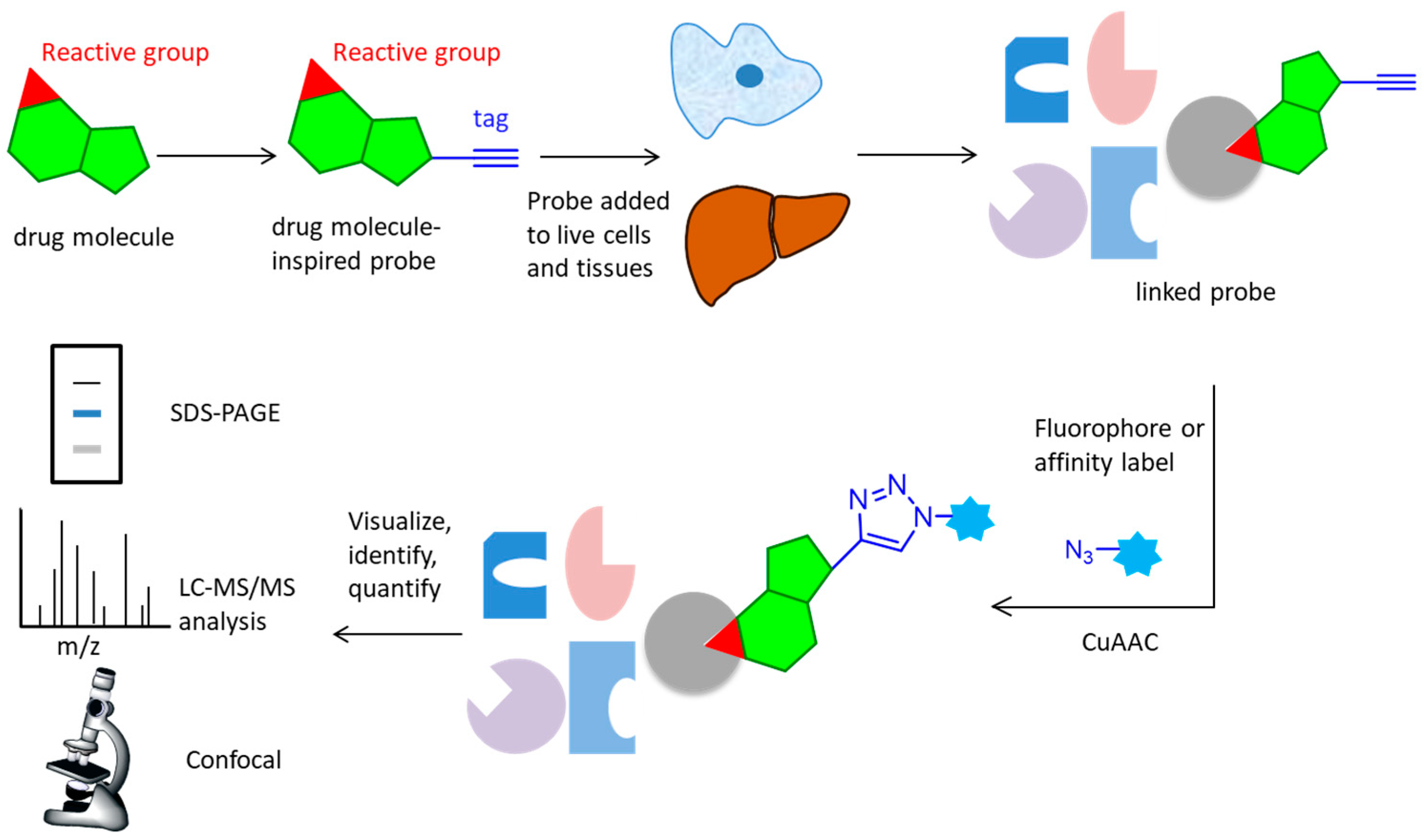
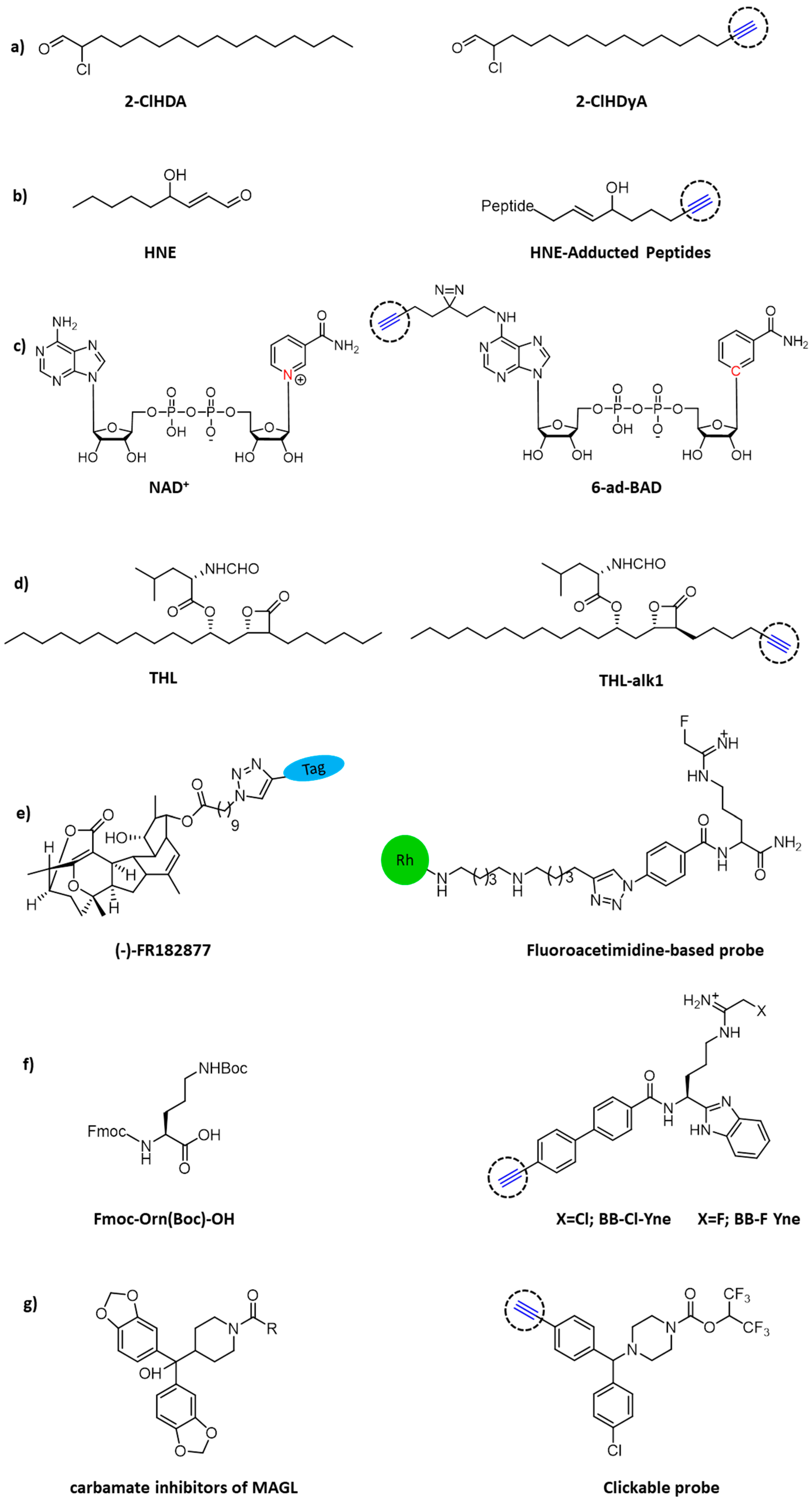
2. Activity-Based Protein Profiling
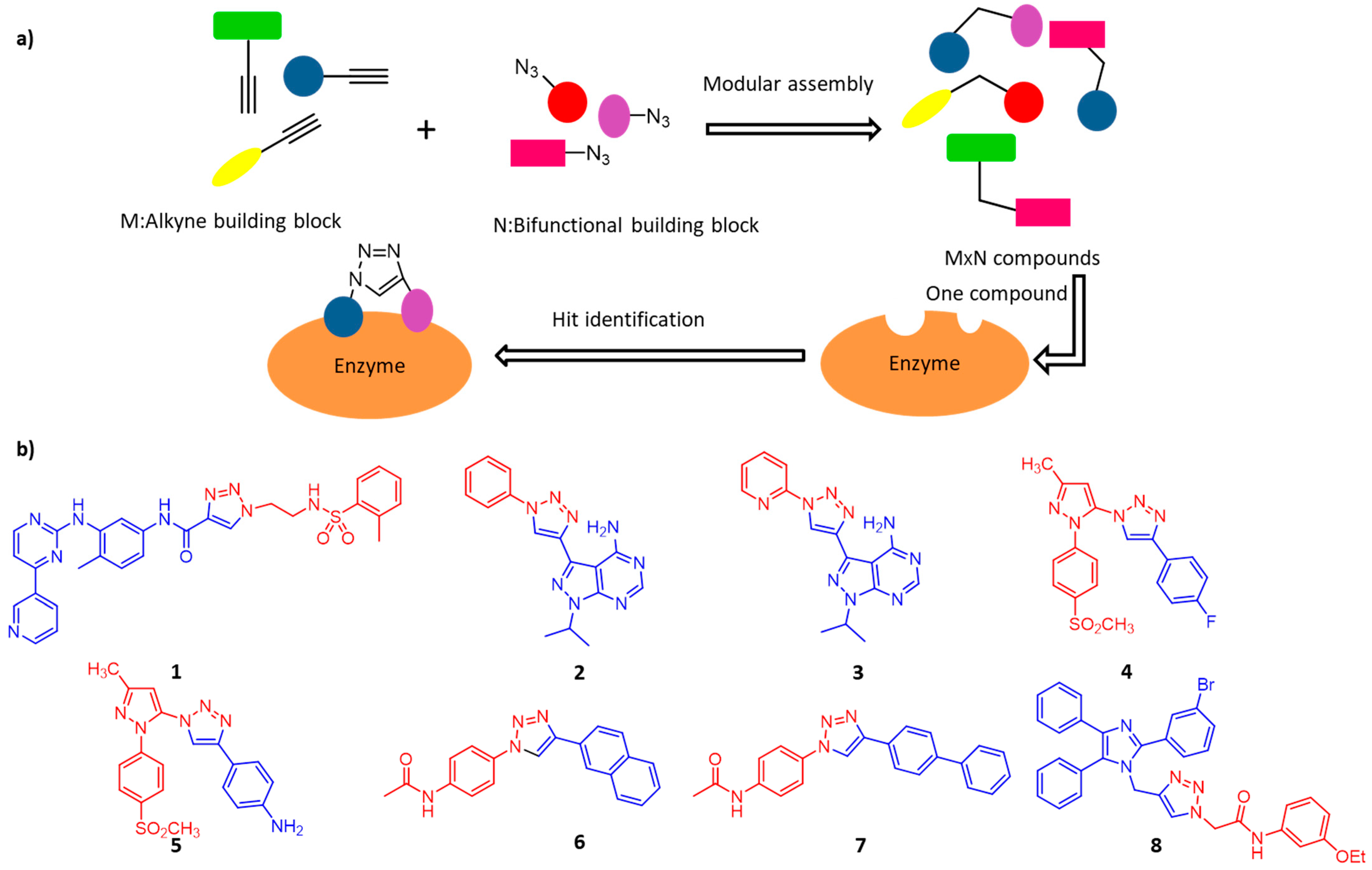
3. Enzyme-Inhibitors Screening
3.1. Protein Kinases
3.2. Cyclooxygenase-2
3.3. O-GlcNAC Transferase
3.4. α-Glucosidases
4. Protein Labeling and Modifications
4.1. Labeling of New Proteins
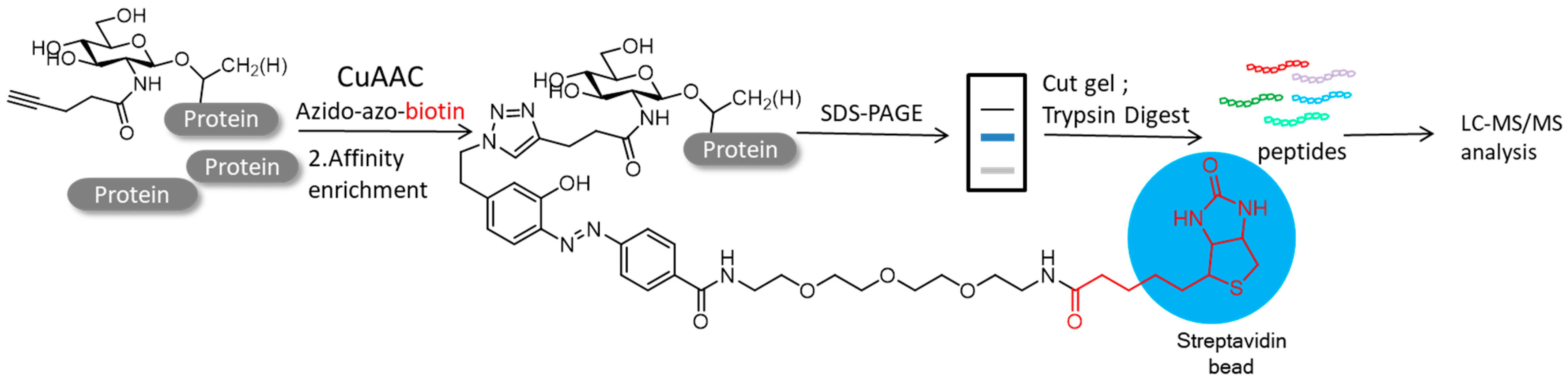
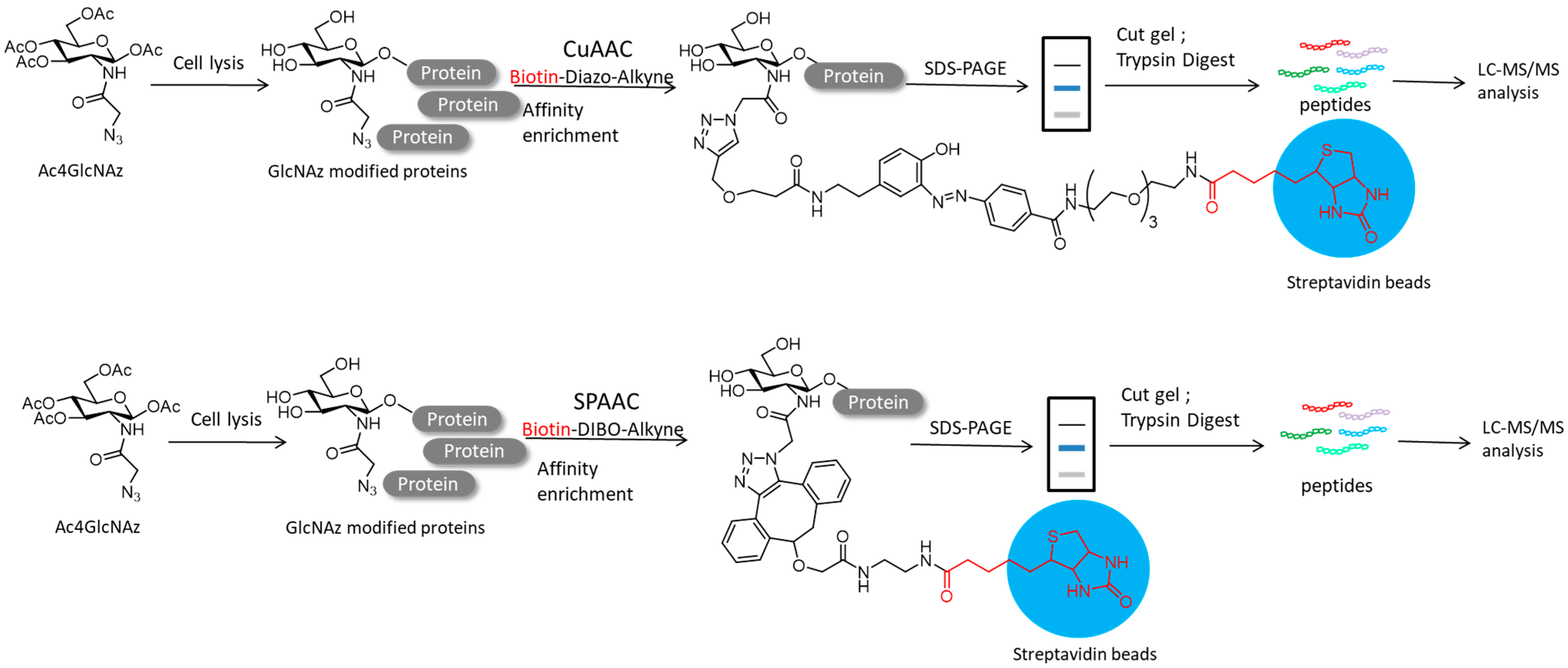
4.2. Protein Post-Translational Modifications
4.3. Protein Engineering
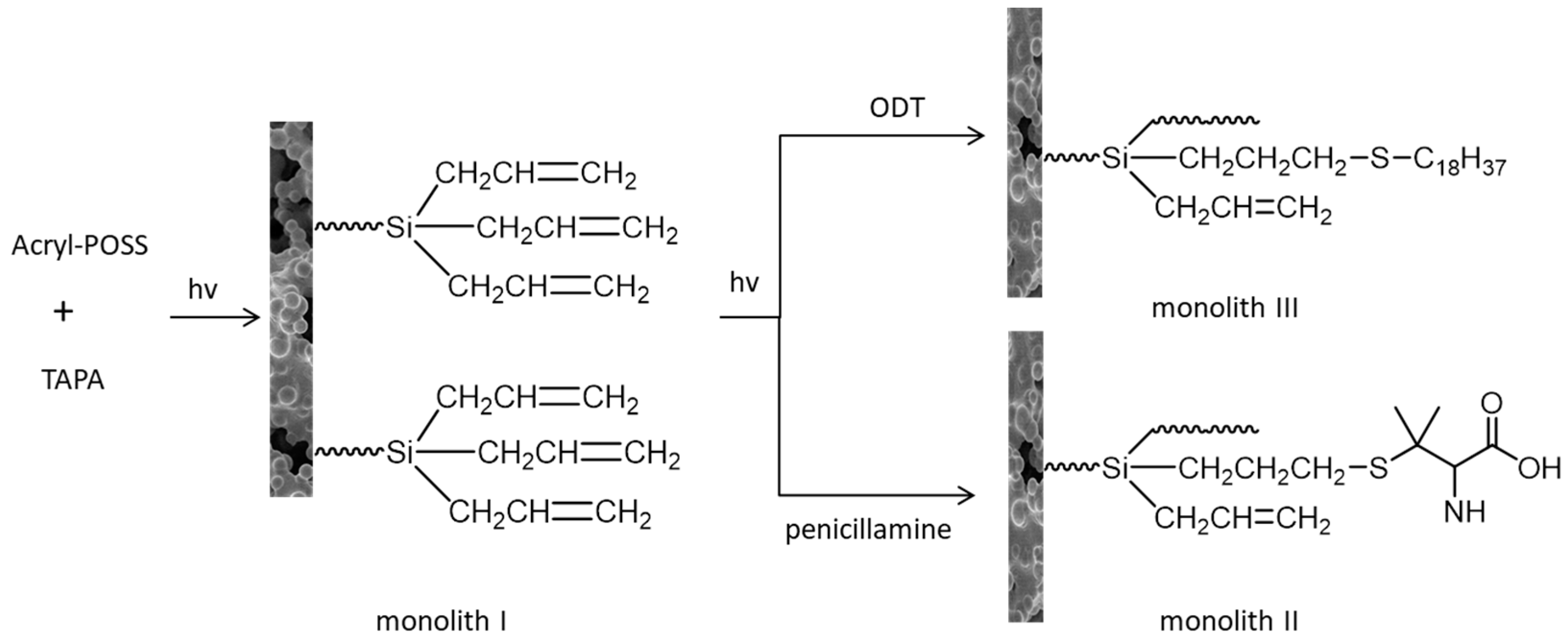
5. Hybrid Monolithic Column in Proteomic Analysis
6. Considerations and Looking Forward
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Parker, C.G.; Pratt, M.R. Click Chemistry in Proteomic Investigations. Cell 2020, 180, 605–632. [Google Scholar] [CrossRef]
- Sam, H. Disease proteomics. Nature 2003, 422, 226–232. [Google Scholar]
- Hartmuth, C.; Kolb, M.G.; Finn, K.; Sharpless, B. Click-Chemie: Diverse chemische Funktionalität mit einer Handvoll guter Reaktionen. Angew. Chem. 2001, 113, 2056–2075. [Google Scholar]
- Meyer, J.P.; Adumeau, P.; Lewis, J.S.; Zeglis, B.M. Click Chemistry and Radiochemistry: The First 10 Years. Bioconj. Chem. 2016, 27, 2791–2807. [Google Scholar] [CrossRef] [Green Version]
- Kalesh, K.A.; Shi, H.; Ge, J.; Yao, S.Q. The use of click chemistry in the emerging field of catalomics. Org. Biomol. Chem. 2010, 8, 1749–1762. [Google Scholar] [CrossRef]
- Evans, M.J.; Cravatt, B.F. Mechanism-Based Profiling of Enzyme Families. Chem. Rev. 2006, 106, 3279–3301. [Google Scholar] [CrossRef]
- Uttamchandani, M.; Li, J.; Sun, H.; Yao, S.Q. Activity-based protein profiling: New developments and directions in functional proteomics. ChemBioChem 2008, 9, 667–675. [Google Scholar] [CrossRef]
- Willems, L.I.; van der Linden, W.A.; Li, N.; Li, K.-Y.; Liu, N.; Hoogendoorn, S.; van der Marel, G.A.; Florea, B.I.; Overkleeft, H.S. Bioorthogonal Chemistry: Applications in Activity-Based Protein Profiling. Acc. Chem. Res. 2011, 44, 718–729. [Google Scholar] [CrossRef]
- Nemmara, V.V.; Subramanian, V.; Muth, A.; Mondal, S.; Salinger, A.J.; Maurais, A.J.; Tilvawala, R.; Weerapana, E.; Thompson, P.R. The Development of Benzimidazole-Based Clickable Probes for the Efficient Labeling of Cellular Protein Arginine Deiminases (PADs). ACS Chem. Biol. 2018, 13, 712–722. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Statsyuk, A.V. Development of activity-based probes for ubiquitin and ubiquitin-like protein signaling pathways. J. Am. Chem Soc. 2013, 135, 16948–16962. [Google Scholar] [CrossRef]
- Wright, A.T.; Cravatt, B.F. Chemical proteomic probes for profiling cytochrome p450 activities and drug interactions in vivo. Chem. Biol. 2007, 14, 1043–1051. [Google Scholar] [CrossRef] [Green Version]
- Tsai, C.S.; Yen, H.Y.; Lin, M.I.; Tsai, T.I.; Wang, S.Y.; Huang, W.I.; Hsu, T.L.; Cheng, Y.S.; Fang, J.M.; Wong, C.H. Cell-permeable probe for identification and imaging of sialidases. Proc. Natl. Acad. Sci. USA 2013, 110, 2466–2471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bantscheff, M.; Eberhard, D.; Abraham, Y.; Bastuck, S.; Boesche, M.; Hobson, S.; Mathieson, T.; Perrin, J.; Raida, M.; Rau, C.; et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 2007, 25, 1035–1044. [Google Scholar] [CrossRef]
- Patricelli, M.P.; Szardenings, A.K.; Liyanage, M.; Nomanbhoy, T.K.; Wu, M.; Weissig, H.; Aban, A.; Chun, D.; Tanner, S.; Kozarich, J.W. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry 2007, 46, 350–358. [Google Scholar] [CrossRef]
- Lapinsky, D.J.; Johnson, D.S. Recent developments and applications of clickable photoprobes in medicinal chemistry and chemical biology. Future Med. Chem. 2015, 7, 2143–2171. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hao, P.; Li, L.; Tan, C.Y.; Cheng, X.; Chen, G.Y.; Sze, S.K.; Shen, H.M.; Yao, S.Q. Design and synthesis of minimalist terminal alkyne-containing diazirine photo-crosslinkers and their incorporation into kinase inhibitors for cell- and tissue-based proteome profiling. Angew. Chem. Int. Ed. Engl. 2013, 52, 8551–8556. [Google Scholar] [CrossRef]
- Li, Z.; Wang, D.; Li, L.; Pan, S.; Na, Z.; Tan, C.Y.; Yao, S.Q. “Minimalist” cyclopropene-containing photo-cross-linkers suitable for live-cell imaging and affinity-based protein labeling. J. Am. Chem. Soc. 2014, 136, 9990–9998. [Google Scholar] [CrossRef]
- Li, J.; Wang, J.; Wen, L.; Zhu, H.; Li, S.; Huang, K.; Jiang, K.; Li, X.; Ma, C.; Qu, J.; et al. An OGA-Resistant Probe Allows Specific Visualization and Accurate Identification of O-GlcNAc-Modified Proteins in Cells. ACS Chem. Biol. 2016, 11, 3002–3006. [Google Scholar] [CrossRef]
- Ballard, T.E.; Murrey, H.E.; Geoghegan, K.F.; am Ende, C.W.; Johnson, D.S. Investigating γ-secretase protein interactions in live cells using active site-directed clickable dual-photoaffinity probes. MedChemComm 2014, 5, 321–327. [Google Scholar] [CrossRef]
- Zuhl, A.M.; Nolan, C.E.; Brodney, M.A.; Niessen, S.; Atchison, K.; Houle, C.; Karanian, D.A.; Ambroise, C.; Brulet, J.W.; Beck, E.M.; et al. Chemoproteomic profiling reveals that cathepsin D off-target activity drives ocular toxicity of beta-secretase inhibitors. Nat. Commun. 2016, 7, 13042. [Google Scholar] [CrossRef] [PubMed]
- Eirich, J.; Orth, R.; Sieber, S.A. Unraveling the protein targets of vancomycin in living S. aureus and E. faecalis cells. J. Am. Chem. Soc. 2011, 133, 12144–12153. [Google Scholar] [CrossRef]
- Gao, J.; Mfuh, A.; Amako, Y.; Woo, C.M. Small Molecule Interactome Mapping by Photoaffinity Labeling Reveals Binding Site Hotspots for the NSAIDs. J. Am. Chem. Soc. 2018, 140, 4259–4268. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, D.C.; Sorum, A.W.; Meier, J.L. Chemoproteomic profiling of lysine acetyltransferases highlights an expanded landscape of catalytic acetylation. J. Am. Chem. Soc. 2014, 136, 8669–8676. [Google Scholar] [CrossRef] [Green Version]
- Wright, M.H.; Sieber, S.A. Chemical proteomics approaches for identifying the cellular targets of natural products. Nat. Prod. Rep. 2016, 33, 681–708. [Google Scholar] [CrossRef] [Green Version]
- Niphakis, M.J.; Lum, K.M.; Cognetta, A.B., 3rd; Correia, B.E.; Ichu, T.A.; Olucha, J.; Brown, S.J.; Kundu, S.; Piscitelli, F.; Rosen, H.; et al. A Global Map of Lipid-Binding Proteins and Their Ligandability in Cells. Cell 2015, 161, 1668–1680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hulce, J.J.; Cognetta, A.B.; Niphakis, M.J.; Tully, S.E.; Cravatt, B.F. Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat. Methods 2013, 10, 259–264. [Google Scholar] [CrossRef] [Green Version]
- Kidd, D.; Liu, Y.; Cravatt, B.F. Profiling serine hydrolase activities in complex proteomes. Biochemistry 2001, 40, 4005–4015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Patricelli, M.P.; Cravatt, B.F. Activity-based protein profiling: The serine hydrolases. Proc. Natl. Acad. Sci. USA 1999, 96, 14694–14699. [Google Scholar] [CrossRef] [Green Version]
- Kato, D.; Boatright, K.M.; Berger, A.B.; Nazif, T.; Blum, G.; Ryan, C.; Chehade, K.A.; Salvesen, G.S.; Bogyo, M. Activity-based probes that target diverse cysteine protease families. Nat. Chem. Biol. 2005, 1, 33–38. [Google Scholar] [CrossRef]
- Hewings, D.S.; Heideker, J.; Ma, T.P.; AhYoung, A.P.; El Oualid, F.; Amore, A.; Costakes, G.T.; Kirchhofer, D.; Brasher, B.; Pillow, T.; et al. Reactive-site-centric chemoproteomics identifies a distinct class of deubiquitinase enzymes. Nat. Commun. 2018, 9, 1162. [Google Scholar] [CrossRef]
- Ward, J.A.; McLellan, L.; Stockley, M.; Gibson, K.R.; Whitlock, G.A.; Knights, C.; Harrigan, J.A.; Jacq, X.; Tate, E.W. Quantitative Chemical Proteomic Profiling of Ubiquitin Specific Proteases in Intact Cancer Cells. ACS Chem. Biol. 2016, 11, 3268–3272. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Ouyang, X.; Wan, X.; Gajiwala, K.S.; Kath, J.C.; Jones, L.H.; Burlingame, A.L.; Taunton, J. Broad-Spectrum Kinase Profiling in Live Cells with Lysine-Targeted Sulfonyl Fluoride Probes. J. Am. Chem. Soc. 2017, 139, 680–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebraud, H.; Wright, D.J.; East, C.E.; Holding, F.P.; O’Reilly, M.; Heightman, T.D. In-gel activity-based protein profiling of a clickable covalent ERK1/2 inhibitor. Mol. Biosyst. 2016, 12, 2867–2874. [Google Scholar] [CrossRef]
- Tan, J.; Yang, X.; Zhuang, L.; Jiang, X.; Chen, W.; Lee, P.L.; Karuturi, R.K.; Tan, P.B.; Liu, E.T.; Yu, Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007, 21, 1050–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chase, A.; Cross, N.C. Aberrations of EZH2 in cancer. Clin. Cancer Res. 2011, 17, 2613–2618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, J.A.; Lange, C.A. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat. Res. 2008, 647, 21–29. [Google Scholar] [CrossRef]
- Yang, P.Y.; Liu, K.; Ngai, M.H.; Lear, M.J.; Wenk, M.R.; Yao, S.Q. Activity-based proteome profiling of potential cellular targets of Orlistat-an FDA-approved drug with anti-tumor activities. J. Am. Chem. Soc. 2010, 132, 656–666. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Cheng, X.; Sze, S.K.; Yao, S.Q. Proteome profiling reveals potential cellular targets of staurosporine using a clickable cell-permeable probe. Chem. Commun. 2011, 47, 11306–11308. [Google Scholar] [CrossRef]
- Shi, H.; Zhang, C.J.; Chen, G.Y.; Yao, S.Q. Cell-based proteome profiling of potential dasatinib targets by use of affinity-based probes. J. Am. Chem. Soc. 2012, 134, 3001–3014. [Google Scholar] [CrossRef]
- Yang, P.Y.; Wang, M.; He, C.Y.; Yao, S.Q. Proteomic profiling and potential cellular target identification of K11777, a clinical cysteine protease inhibitor, in Trypanosoma brucei. Chem. Commun. 2012, 48, 835–837. [Google Scholar] [CrossRef]
- Tam, E.K.; Li, Z.; Goh, Y.L.; Cheng, X.; Wong, S.Y.; Santhanakrishnan, S.; Chai, C.L.; Yao, S.Q. Cell-based proteome profiling using an affinity-based probe (AfBP) derived from 3-deazaneplanocin A (DzNep). Chem. Asian J. 2013, 8, 1818–1828. [Google Scholar] [CrossRef]
- Prasch, J.; Bernhart, E.; Reicher, H.; Kollroser, M.; Rechberger, G.N.; Koyani, C.N.; Trummer, C.; Rech, L.; Rainer, P.P.; Hammer, A.; et al. Myeloperoxidase-Derived 2-Chlorohexadecanal Is Generated in Mouse Heart during Endotoxemia and Induces Modification of Distinct Cardiomyocyte Protein Subsets In Vitro. Int. J. Mol. Sci. 2020, 21, 9235. [Google Scholar] [CrossRef]
- Ji, C.; Kozak, K.R.; Marnett, L.J. IkappaB kinase, a molecular target for inhibition by 4-hydroxy-2-nonenal. J. Biol. Chem. 2001, 276, 18223–18228. [Google Scholar] [CrossRef] [Green Version]
- Neely, M.; Sidell, K.R.; Graham, D.G.; Montine, T.J. The Lipid Peroxidation Product 4-Hydroxynonenal Inhibits Neurite Outgrowth, Disrupts Neuronal Microtubules, and Modifies Cellular Tubulin. J. Neurochem. 2010, 72, 2323–2333. [Google Scholar] [CrossRef] [Green Version]
- Dinkova-Kostova, A.T.; Holtzclaw, W.D.; Cole, R.N.; Itoh, K.; Wakabayashi, N.; Katoh, Y.; Yamamoto, M.; Talalay, P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. USA 2002, 99, 11908–11913. [Google Scholar] [CrossRef] [Green Version]
- Levonen, A.L.; Landar, A.; Ramachandran, A.; Ceaser, E.K.; Darley-Usmar, V.M. Cellular mechanisms of redox cell signalling: Role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem. J. 2004, 378, 373–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vila, A.; Tallman, K.A.; Jacobs, A.T.; Liebler, D.C.; Porter, N.A.; Marnett, L.J. Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chem. Res. Toxicol. 2008, 21, 432–444. [Google Scholar] [CrossRef]
- Cohen, M.S. Interplay between compartmentalized NAD+ synthesis and consumption: A focus on the PARP family. Genes Dev. 2020, 34, 254–262. [Google Scholar] [CrossRef]
- Sileikyte, J.; Sundalam, S.; David, L.L.; Cohen, M.S. Chemical Proteomics Approach for Profiling the NAD Interactome. J. Am. Chem. Soc. 2021, 143, 6787–6791. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, M.S.; Wenk, M.R. Activity-Based Lipid Esterase Profiling of M. bovis BCG at Different Metabolic States Using Tetrahydrolipstatin (THL) as Bait. Methods Mol. Biol. 2017, 1491, 75–85. [Google Scholar] [PubMed]
- Adam, G.C.; Vanderwal, C.D.; Sorensen, E.J.; Cravatt, B.F. (-)-FR182877 is a potent and selective inhibitor of carboxylesterase-1. Angew. Chem. Int. Ed. Engl. 2003, 42, 5480–5484. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Knuckley, B.; Bhatia, M.; Pellechia, P.J.; Thompson, P.R. Activity-based protein profiling reagents for protein arginine deiminase 4 (PAD4): Synthesis and in vitro evaluation of a fluorescently labeled probe. J. Am. Chem. Soc. 2006, 128, 14468–14469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, J.E.; Causey, C.P.; Knuckley, B.; Slack-Noyes, J.L.; Thompson, P.R. Protein arginine deiminase 4 (PAD4): Current understanding and future therapeutic potential. Curr. Opin. Drug Discov. Dev. 2009, 12, 616–627. [Google Scholar]
- Vossenaar, E.R.; Zendman, A.J.; van Venrooij, W.J.; Pruijn, G.J. PAD, a growing family of citrullinating enzymes: Genes, features and involvement in disease. Bioessays 2003, 25, 1106–1118. [Google Scholar] [CrossRef]
- Stone, E.M.; Schaller, T.H.; Bianchi, H.; Person, M.D.; Fast, W. Inactivation of Two Diverse Enzymes in the Amidinotransferase Superfamily by 2-Chloroacetamidine: Dimethylargininase and Peptidylarginine Deiminase. Biochemistry 2005, 44, 13744–13752. [Google Scholar] [CrossRef] [PubMed]
- Bicker, K.L.; Thompson, P.R. The protein arginine deiminases: Structure, function, inhibition, and disease. Biopolymers 2013, 99, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Ghari, F.; Quirke, A.M.; Munro, S.; Kawalkowska, J.; Picaud, S.; Mcgouran, J.; Subramanian, V.; Muth, A.; Williams, R.; Kessler, B. Citrullination-acetylation interplay guides E2F-1 activity during the inflammatory response. Sci. Adv. 2016, 2, 1501257. [Google Scholar] [CrossRef] [Green Version]
- Knight, J.S.; Subramanian, V.; O’Dell, A.A.; Yalavarthi, S.; Zhao, W.; Smith, C.K.; Hodgin, J.B.; Thompson, P.R.; Kaplan, M.J. Peptidylarginine deiminase inhibition disrupts NET formation and protects against kidney, skin and vascular disease in lupus-prone MRL/lpr mice. Ann. Rheum. Dis. 2015, 74, 2199–2206. [Google Scholar] [CrossRef] [Green Version]
- Kawalkowska, J.; Quirke, A.M.; Ghari, F.; Davis, S.; Subramanian, V.; Thompson, P.R.; Williams, R.O.; Fischer, R.; La Thangue, N.B.; Venables, P.J. Abrogation of collagen-induced arthritis by a peptidyl arginine deiminase inhibitor is associated with modulation of T cell-mediated immune responses. Sci Rep. 2016, 6, 26430. [Google Scholar] [CrossRef] [Green Version]
- Speers, A.E.; Adam, G.C.; Cravatt, B. Activity-Based Protein Profiling in Vivo Using a Copper(I)-Catalyzed Azide-Alkyne (3 + 2) Cycloaddition. J. Am. Chem. Soc. 2003, 125, 4686–4687. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.W.; Cognetta, A.B.; Niphakis, M.J.; Cravatt, B.F. Proteome-wide reactivity profiling identifies diverse carbamate chemotypes tuned for serine hydrolase inhibition. ACS Chem. Biol. 2013, 8, 1590–1599. [Google Scholar] [CrossRef] [Green Version]
- Erlanson, D.A.; Braisted, A.C.; Raphael, D.R.; Randal, M.; Stroud, R.M.; Gordon, E.M.; Wells, J.A. Site-directed ligand discovery. Proc. Natl. Acad. Sci. USA 2000, 97, 9367–9372. [Google Scholar] [CrossRef] [Green Version]
- Mamidyala, S.K.; Finn, M.G. In situ click chemistry: Probing the binding landscapes of biological molecules. Chem. Soc. Rev. 2010, 39, 1252–1261. [Google Scholar] [CrossRef]
- Capdeville, R.; Buchdunger, E.; Zimmermann, J.; Matter, A. Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat. Rev. Drug Discov. 2002, 1, 493–502. [Google Scholar] [CrossRef]
- Brik, A.; Wu, C.Y.; Wong, C.H. Microtiter plate based chemistry and in situ screening: A useful approach for rapid inhibitor discovery. Org. Biomol. Chem. 2006, 4, 1446–1457. [Google Scholar] [CrossRef]
- Cohen, P. Protein kinases—The major drug targets of the twenty-first century? Nat. Rev. Drug Discov. 2002, 1, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Kalesh, K.A.; Liu, K.; Yao, S.Q. Rapid synthesis of Abelson tyrosine kinase inhibitors using click chemistry. Org. Biomol. Chem. 2009, 7, 5129–5136. [Google Scholar] [CrossRef]
- Klein, M.; Diner, P.; Dorin-Semblat, D.; Doerig, C.; Grotli, M. Synthesis of 3-(1,2,3-triazol-1-yl)- and 3-(1,2,3-triazol-4-yl)-substituted pyrazolo[3,4-d]pyrimidin-4-amines via click chemistry: Potential inhibitors of the Plasmodium falciparum PfPK7 protein kinase. Org. Biomol. Chem. 2009, 7, 3421–3429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, W.L.; Urade, Y.; Jakobsson, P.J. Enzymes of the cyclooxygenase pathways of prostanoid biosynthesis. Chem. Rev. 2011, 111, 5821–5865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhardwaj, A.; Kaur, J.; Wuest, M.; Wuest, F. In situ click chemistry generation of cyclooxygenase-2 inhibitors. Nat. Commun. 2017, 8, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres, C.R.; Hart, G.W. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 1984, 259, 3308–3317. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, J.; Zhang, L. Discovery of cell-permeable O-GlcNAc transferase inhibitors via tethering in situ click chemistry. J. Med. Chem. 2017, 60, 263–272. [Google Scholar] [CrossRef]
- Hirsh, A.J.; Yao, S.Y.M.; Young, J.D.; Cheeseman, C.I. Inhibition of glucose absorption in the rat jejunum: A novel action of alpha-D-glucosidase inhibitors. Gastroenterology 1997, 113, 205–211. [Google Scholar] [CrossRef]
- Yamagishi, S.-I.; Matsui, T.; Ueda, S.; Fukami, K.; Okuda, S. Clinical Utility of Acarbose, an α-Glucosidase Inhibitor in Cardiometabolic Disorders. Curr. Drug Metab. 2009, 10, 159–163. [Google Scholar] [CrossRef]
- Hara, T.; Nakamura, J.; Koh, N.; Sakakibara, F.; Hotta, N. An importance of carbohydrate ingestion for the expression of the effect of alpha-glucosidase inhibitor in NIDDM. Diabetes Care 1996, 19, 642–647. [Google Scholar] [CrossRef]
- Wang, G.; Peng, Z.; Wang, J.; Li, J.; Li, X. Synthesis and biological evaluation of novel 2,4,5-triarylimidazole–1,2,3-triazole derivatives via click chemistry as α-glucosidase inhibitors. Bioorg. Med. Chem. Lett. 2016, 26, 5719–5723. [Google Scholar] [CrossRef]
- Obermaier, C.; Griebel, A.; Westermeier, R. Principles of protein labeling techniques. Methods Mol. Biol. 2015, 1295, 153–165. [Google Scholar]
- Singh, R.K.; Lee, J.-K.; Chandrabose, S.; Selvaraj, C.; Singh, R.; Li, J.; Kim, S.-Y. Protein Engineering Approaches in the Post-Genomic Era. Curr. Protein Pept. Sci. 2018, 19, 5–15. [Google Scholar]
- Yang, M.; Li, J.; Chen, P.R. Transition metal-mediated bioorthogonal protein chemistry in living cells. Chem. Soc. Rev. 2014, 43, 6511–6526. [Google Scholar] [CrossRef] [PubMed]
- Beatty, K.E.; Xie, F.; Wang, Q.; Tirrell, D.A. Selective Dye-Labeling of Newly Synthesized Proteins in Bacterial Cells. J. Am. Chem. Soc. 2005, 127, 14150–14151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aslebagh, R.; Wormwood, K.L.; Channaveerappa, D.; Wetie, A.; Darie, C.C. Identification of Posttranslational Modifications (PTMs) of Proteins by Mass Spectrometry. Adv. Exp. Med. Biol 2019, 1140, 199–224. [Google Scholar]
- Sletten, E.M.; Bertozzi, C.R. Bioorthogonal chemistry: Fishing for selectivity in a sea of functionality. Angew. Chem. Int. Ed. Engl. 2009, 48, 6974–6998. [Google Scholar] [CrossRef] [Green Version]
- Agard, N.J.; Prescher, J.A.; Bertozzi, C.R. A Strain-Promoted [3+2] Azide−Alkyne Cycloaddition for Covalent Modification of Biomolecules in Living Systems. J. Am. Chem. Soc. 2004, 126, 15046–15047. [Google Scholar] [CrossRef]
- Zaro, B.W.; Hang, H.C.; Pratt, M.R. Incorporation of unnatural sugars for the identification of glycoproteins. Methods Mol. Biol 2013, 951, 57–67. [Google Scholar] [PubMed] [Green Version]
- Li, S.; Zhu, H.; Wang, J.; Wang, X.; Li, X.; Ma, C.; Wen, L.; Yu, B.; Wang, Y.; Li, J.; et al. Comparative analysis of Cu (I)-catalyzed alkyne-azide cycloaddition (CuAAC) and strain-promoted alkyne-azide cycloaddition (SPAAC) in O-GlcNAc proteomics. Electrophoresis 2016, 37, 1431–1436. [Google Scholar] [CrossRef] [Green Version]
- Deng, L.; Meng, T.; Chen, L.; Wei, W.; Wang, P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal. Transduct. Target. Ther. 2020, 5, 11. [Google Scholar] [CrossRef] [Green Version]
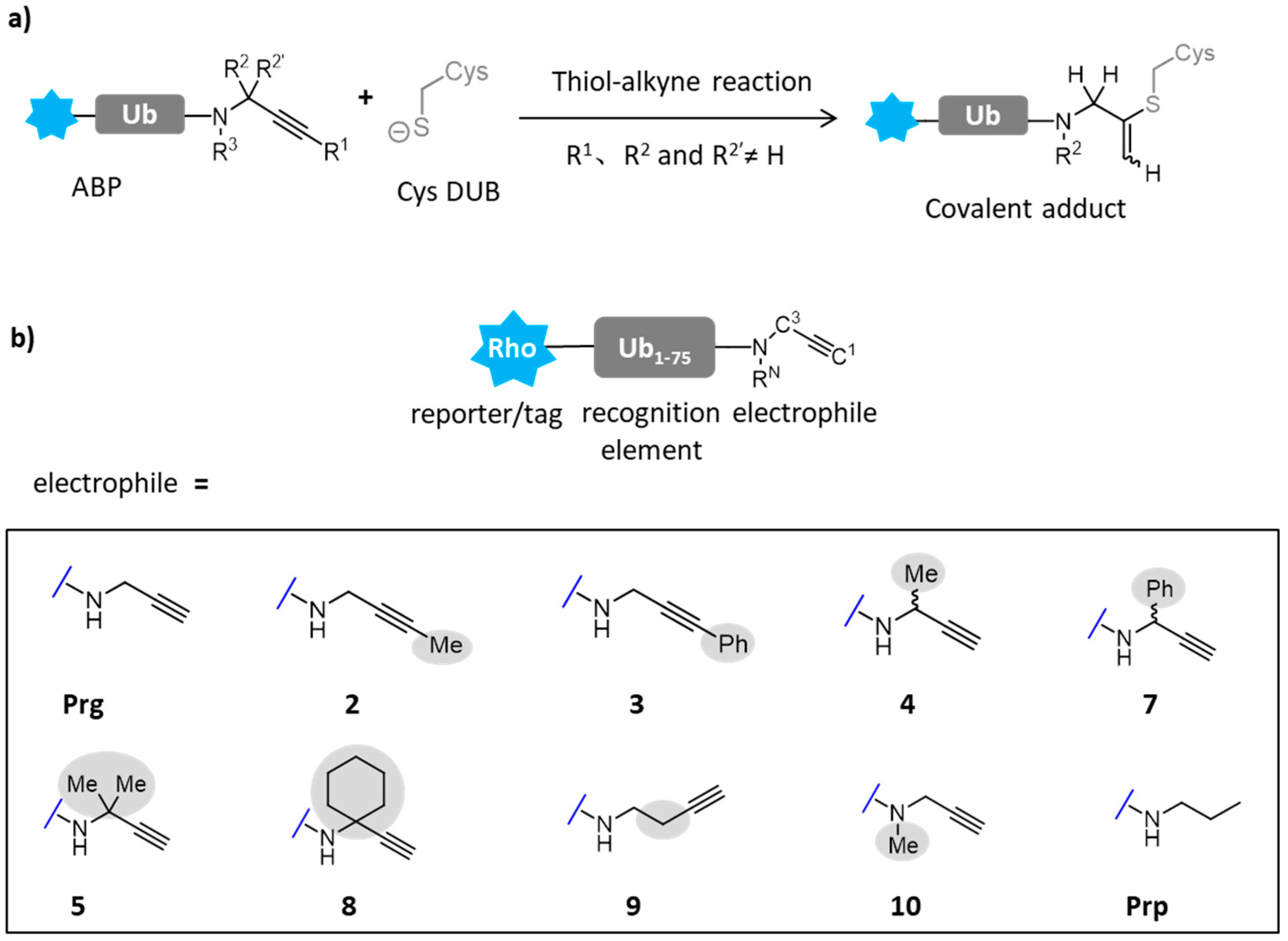
- Mons, E.; Kim, R.Q.; van Doodewaerd, B.R.; van Veelen, P.A.; Mulder, M.P.C.; Ovaa, H. Exploring the Versatility of the Covalent Thiol-Alkyne Reaction with Substituted Propargyl Warheads: A Deciding Role for the Cysteine Protease. J. Am. Chem. Soc. 2021, 143, 6423–6433. [Google Scholar] [CrossRef] [PubMed]
- Kralj, J.M.; Hochbaum, D.R.; Douglass, A.D.; Cohen, A.E. Electrical Spiking in Escherichia coli Probed with a Fluorescent Voltage-Indicating Protein. Science 2011, 333, 345–348. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Lin, C.; Xu, Y.; Luo, H.; Peng, L.; Zeng, X.; Zheng, H.; Chen, P.R.; Zou, P. A far-red hybrid voltage indicator enabled by bioorthogonal engineering of rhodopsin on live neurons. Nat. Chem. 2021, 13, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Hjertén, S.; Liao, J.L.; Zhang, R. High-performance liquid chromatography on continuous polymer beds. J. Chromatogr. A 1989, 473, 273–275. [Google Scholar] [CrossRef]
- Svec, F.; Frechet, J.M.J. Continuous rods of macroporous polymer as high-performance liquid chromatography separation media. Anal. Chem. 1992, 64, 820–822. [Google Scholar] [CrossRef]
- Lv, X.; Tan, W.; Chen, Y.; Chen, Y.; Ma, M.; Chen, B.; Yao, S. Facile “one-pot” synthesis of poly(methacrylic acid)-based hybrid monolith via thiol-ene click reaction for hydrophilic interaction chromatography. J. Chromatogr. A 2016, 1454, 49–57. [Google Scholar] [CrossRef]
- Svec, F.; Lv, Y. Advances and recent trends in the field of monolithic columns for chromatography. Anal. Chem. 2015, 87, 250–273. [Google Scholar] [CrossRef]
- Teisseyre, T.Z.; Urban, J.; Halpern-Manners, N.W.; Chambers, S.D.; Bajaj, V.S.; Svec, F.; Pines, A. Remotely detected NMR for the characterization of flow and fast chromatographic separations using organic polymer monoliths. Anal. Chem. 2011, 83, 6004–6010. [Google Scholar] [CrossRef] [PubMed]
- Eeltink, S.; Wouters, S.; Dores-Sousa, J.L.; Svec, F. Advances in organic polymer-based monolithic column technology for high-resolution liquid chromatography-mass spectrometry profiling of antibodies, intact proteins, oligonucleotides, and peptides. J. Chromatogr. A 2017, 1498, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Oberacher, H.; Premstaller, A.; Huber, C.G. Characterization of some physical and chromatographic properties of monolithic poly(styrene-co-divinylbenzene) columns. J. Chromatogr. A 2004, 1030, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Liang, Z.; Qiao, X.; De Ng, Q.; Tao, D.; Zhang, L.; Zhang, Y. Organic-inorganic hybrid silica monolith based immobilized trypsin reactor with high enzymatic activity. Anal. Chem. 2008, 80, 2949–2956. [Google Scholar] [CrossRef]
- Colon, H.; Zhang, X.; Murphy, J.K.; Rivera, J.G.; Colon, L.A. Allyl-functionalized hybrid silica monoliths. Chem. Commun. 2005, 22, 2826–2828. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, J.; Liu, Z.; Wang, H.; Ou, J.; Ye, M.; Zou, H. Functionalization of hybrid monolithic columns via thiol-ene click reaction for proteomics analysis. J. Chromatogr. A 2017, 1498, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Hsu, C.H.; Lin, Z.; Shan, W.; Wang, J.; Jiang, J.; Huang, M.; Lotz, B.; Yu, X.; Zhang, W.B.; et al. Two dimensional nanocrystals of molecular Janus particles. J. Am. Chem. Soc. 2014, 136, 10691–10699. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, J.; Jiang, S.; Wang, Z.S. POSS with eight imidazolium iodide arms for efficient solid-state dye-sensitized solar cells. Chem. Commun. 2014, 50, 1685–1687. [Google Scholar] [CrossRef]
- Lin, H.; Ou, J.; Zhang, Z.; Dong, J.; Zou, H. Ring-opening polymerization reaction of polyhedral oligomeric silsesquioxanes (POSSs) for preparation of well-controlled 3D skeletal hybrid monoliths. Chem. Commun. 2013, 49, 231–233. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Zhang, L.; Qiao, X.; Wang, Y.; Yan, H.; Bai, L. Facile one-pot preparation of an imidazolium embedded C8 hybrid monolith using polyhedral oligomeric silsesquioxane for capillary liquid chromatography. RSC Adv. 2015, 5, 91436–91440. [Google Scholar] [CrossRef]
- Ma, C.; Ma, S.; Chen, Y.; Wang, Y.; Ou, J.; Zhang, J.; Ye, M. Fast fabrication and modification of polyoctahedral silsesquioxane-containing monolithic columns via two-step photo-initiated reactions and their application in proteome analysis of tryptic digests. Talanta 2020, 209, 120526–120558. [Google Scholar] [CrossRef]
- Wu, M.; Wu, R.; Li, R.; Qin, H.; Dong, J.; Zhang, Z.; Zou, H. Polyhedral oligomeric silsesquioxane as a cross-linker for preparation of inorganic-organic hybrid monolithic columns. Anal. Chem. 2010, 82, 5447–5454. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Dai, Q.; Li, X.; Zhu, X.; Ma, T.; Qiao, X.; Shen, S.; Liu, X. Dipentaerythritol penta-/hexa-acrylate based-highly cross-linked hybrid monolithic column: Preparation and its applications for ultrahigh efficiency separation of proteins. Anal. Chim. Acta 2017, 963, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Hong, T.; Yang, X.; Xu, Y.; Ji, Y. Recent advances in the preparation and application of monolithic capillary columns in separation science. Anal. Chim. Acta 2016, 931, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Li, W.; Chen, R.; Han, Y.; Liu, X.; Wang, T.; Guo, H.; Qiao, X. Amino acid and ionic liquid modified polyhedral oligomeric silsesquioxane-based hybrid monolithic column for high-efficiency capillary liquid chromatography. J. Chromatogr. A 2018, 1572, 82–89. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, T.; Xu, X.; Huang, R. Recent Advances about the Applications of Click Reaction in Chemical Proteomics. Molecules 2021, 26, 5368. https://doi.org/10.3390/molecules26175368
Yao T, Xu X, Huang R. Recent Advances about the Applications of Click Reaction in Chemical Proteomics. Molecules. 2021; 26(17):5368. https://doi.org/10.3390/molecules26175368
Chicago/Turabian StyleYao, Tingting, Xiaowei Xu, and Rong Huang. 2021. "Recent Advances about the Applications of Click Reaction in Chemical Proteomics" Molecules 26, no. 17: 5368. https://doi.org/10.3390/molecules26175368
APA StyleYao, T., Xu, X., & Huang, R. (2021). Recent Advances about the Applications of Click Reaction in Chemical Proteomics. Molecules, 26(17), 5368. https://doi.org/10.3390/molecules26175368






