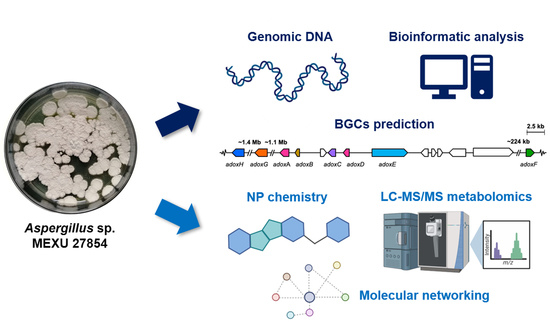
Genome Mining and Molecular Networking-Based Metabolomics of the Marine Facultative Aspergillus sp. MEXU 27854
Abstract
:1. Introduction
2. Results and Discussion
2.1. General Genomic Features of Aspergillus sp. MEXU 27854
2.2. BGCs in Aspergillus sp. MEXU 27854
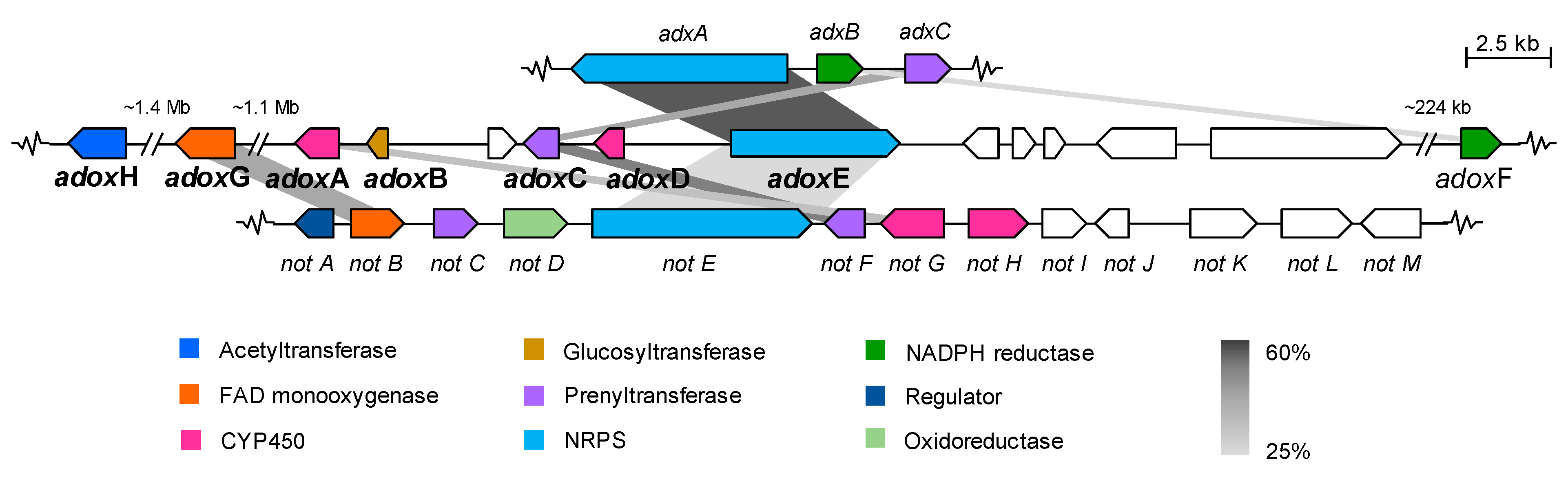
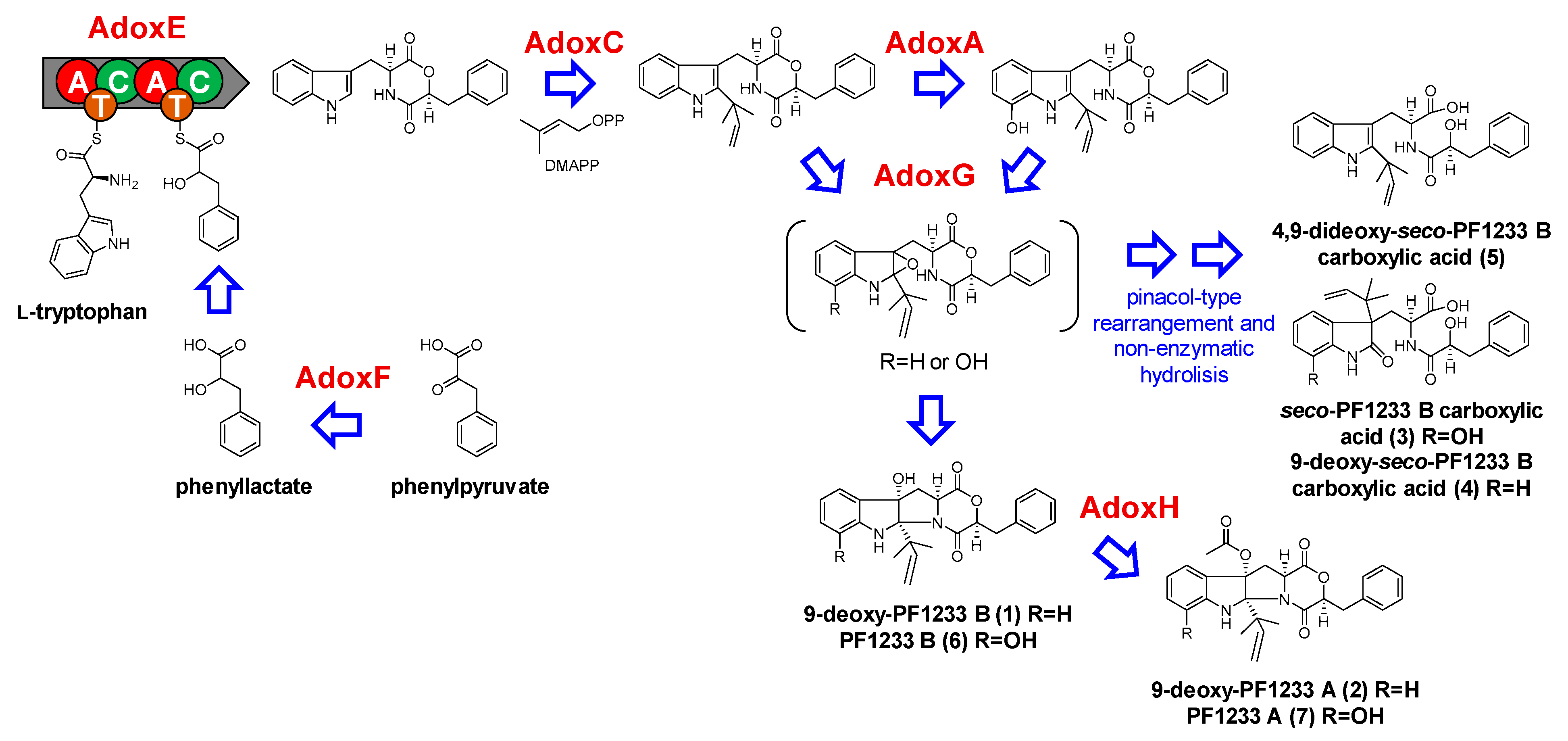
2.2.1. Dioxomorpholines
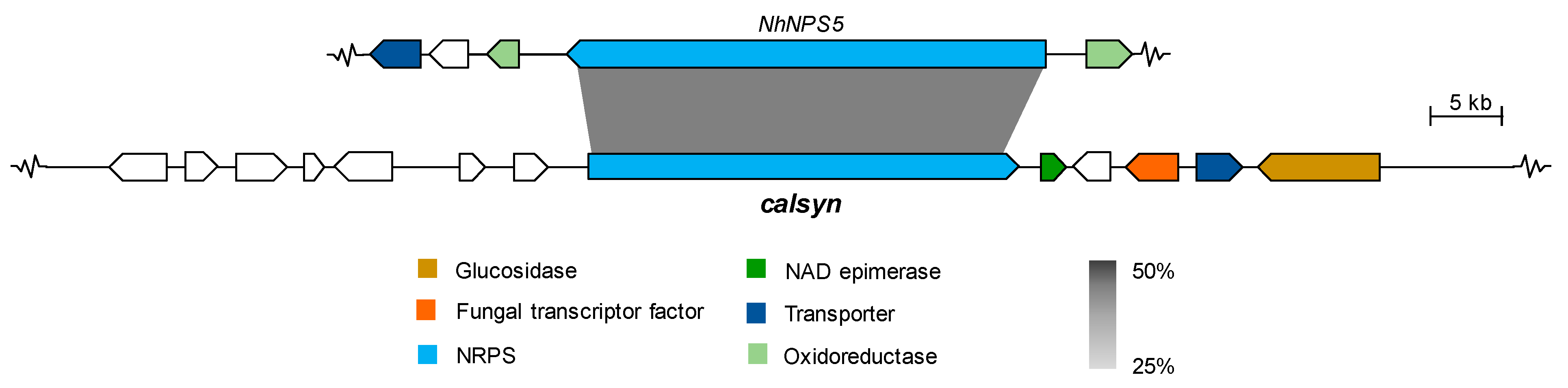
2.2.2. Cyclic Peptides
2.2.3. Butyrolactones
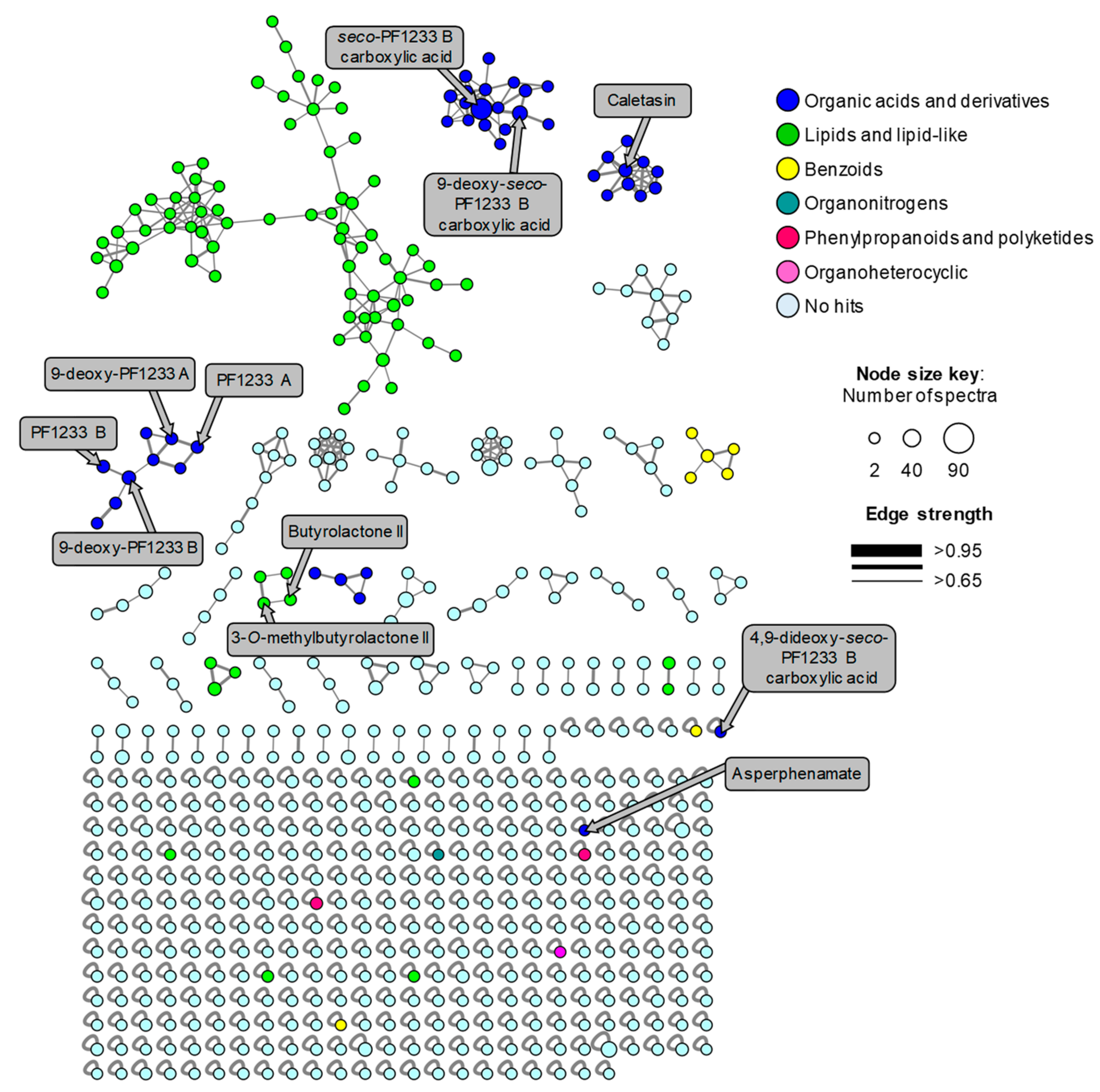
2.3. Mass-Spectrometry-Based Metabolomics Analysis
3. Materials and Methods
3.1. Strain and DNA Isolation
3.2. Sequencing and Assembly
3.3. Genome Annotation and BGC Prediction
3.4. Extract Preparation and Chemical Study
3.5. LC-MS/MS Analysis
3.6. Metabolomic Analysis
3.7. Data Availability
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Hagestad, O.C.; Andersen, J.H.; Altermark, B.; Hansen, E.; Rämä, T. Cultivable marine fungi from the arctic archipelago of Svalbard and their antibacterial activity. Mycology 2020, 11, 230–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Li, Z.; Gao, J.; He, H.; Dai, H.; Xia, X.; Liu, C.; Zhang, L.; Song, F. New diketopiperazines from a marine-derived fungus strain Aspergillus versicolor MF180151. Mar. Drugs 2019, 17, 262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagano, Y.; Miura, T.; Tsubouchi, T.; Lima, A.O.; Kawato, M.; Fujiwara, Y.; Fujikura, K. Cryptic fungal diversity revealed in deep-sea sediments associated with whale-fall chemosynthetic ecosystems. Mycology 2020, 11, 263–278. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, H.; Zhu, W. New natural products from the marine-derived Aspergillus fungi—A review. Acta Microbiol. Sin. 2016, 56, 331–362. [Google Scholar]
- Horton, T.; Kroh, A.; Ahyong, S.; Bailly, N.; Boyko, C.B.; Brandão, S.N.; Gofas, S.; Hooper, J.N.A.; Hernandez, F.; Holovachov, O.; et al. World Register of Marine Species (WoRMS). Available online: https://www.marinespecies.org (accessed on 26 August 2021).
- Galagan, J.E.; Calvo, S.E.; Cuomo, C.; Ma, L.J.; Wortman, J.R.; Batzoglou, S.; Lee, S.I.; Baştürkmen, M.; Spevak, C.C.; Clutterbuck, J.; et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 2005, 438, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Machida, M.; Asai, K.; Sano, M.; Tanaka, T.; Kumagai, T.; Terai, G.; Kusumoto, K.; Arima, T.; Akita, O.; Kashiwagi, Y.; et al. Genome sequencing and analysis of Aspergillus oryzae. Nature 2005, 438, 1157–1161. [Google Scholar] [CrossRef] [Green Version]
- Nierman, W.C.; Pain, A.; Anderson, M.J.; Wortman, J.R.; Kim, H.S.; Arroyo, J.; Berriman, M.; Abe, K.; Archer, D.B.; Bermejo, C.; et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 2005, 438, 1151–1156. [Google Scholar] [CrossRef]
- Coordinators, N.R. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2016, 44, D7–D19. [Google Scholar] [CrossRef] [Green Version]
- Aparicio-Cuevas, M.A.; Rivero-Cruz, I.; Sánchez-Castellanos, M.; Menéndez, D.; Raja, H.A.; Joseph-Nathan, P.; González, M.D.C.; Figueroa, M. Dioxomorpholines and derivatives from a marine-facultative Aspergillus species. J. Nat. Prod. 2017, 80, 2311–2318. [Google Scholar] [CrossRef] [Green Version]
- Aparicio-Cuevas, M.A.; González, M.D.C.; Raja, H.; Rivero-Cruz, I.; Kurina, S.J.; Burdette, J.E.; Oberlies, N.H.; Figueroa, M. Metabolites from the marine-facultative Aspergillus sp. MEXU 27854 and Gymnoascus hyalinosporus MEXU 29901 from Caleta Bay, Mexico. Tetrahedron Lett. 2019, 60, 1649–1652. [Google Scholar] [CrossRef]
- Robey, M.T.; Ye, R.; Bok, J.W.; Clevenger, K.D.; Islam, M.N.; Chen, C.; Gupta, R.; Swyers, M.; Wu, E.; Gao, P.; et al. Identification of the first diketomorpholine biosynthetic pathway using FAC-MS technology. ACS Chem. Biol. 2018, 13, 1142–1147. [Google Scholar] [CrossRef]
- Khalil, Z.G.; Huang, X.C.; Raju, R.; Piggott, A.M.; Capon, R.J. Shornephine A: Structure, chemical stability, and P-glycoprotein inhibitory properties of a rare diketomorpholine from an australian marine-derived Aspergillus sp. J. Org. Chem. 2014, 79, 8700–8705. [Google Scholar] [CrossRef] [Green Version]
- Samson, R.A.; Peterson, S.W.; Frisvad, J.C.; Varga, J. New species in Aspergillus section Terrei. Stud. Mycol. 2011, 69, 39–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Govic, Y.; Papon, N.; Le Gal, S.; Bouchara, J.P.; Vandeputte, P. Non-ribosomal peptide synthetase gene clusters in the human pathogenic fungus Scedosporium apiospermum. Front. Microbiol. 2019, 10, 2062. [Google Scholar] [CrossRef]
- Martínez-Núñez, M.A.; López, V.E.L. Nonribosomal peptides synthetases and their applications in industry. Sustain. Chem. Process. 2016, 4, 13. [Google Scholar] [CrossRef]
- Von Bargen, K.W.; Niehaus, E.M.; Krug, I.; Bergander, K.; Würthwein, E.U.; Tudzynski, B.; Humpf, H.U. Isolation and structure elucidation of fujikurins A-D: Products of the PKS19 gene cluster in Fusarium fujikuroi. J. Nat Prod. 2015, 78, 1809–1815. [Google Scholar] [CrossRef] [PubMed]
- Hajjaj, H.; Klaebe, A.; Loret, M.O.; Goma, G.; Blanc, P.J.; Francois, J. Biosynthetic pathway of citrinin in the filamentous fungus Monascus ruber as revealed by 13C nuclear magnetic resonance. Appl. Environ. Microbiol. 1999, 65, 311–314. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Gilchrist, C.L.M.; Lacey, H.J.; Crombie, A.; Vuong, D.; Pitt, J.I.; Lacey, E.; Chooi, Y.H.; Piggott, A.M. Discovery and heterologous biosynthesis of the burnettramic acids: Rare PKS-NRPS-derived bolaamphiphilic pyrrolizidinediones from an australian fungus, Aspergillus burnettii. Org. Lett. 2019, 21, 1287–1291. [Google Scholar] [CrossRef]
- Kjærbølling, I.; Vesth, T.C.; Frisvad, J.C.; Nybo, J.L.; Theobald, S.; Kuo, A.; Bowyer, P.; Matsuda, Y.; Mondo, S.; Lyhne, E.K.; et al. Linking secondary metabolites to gene clusters through genome sequencing of six diverse Aspergillus species. Proc. Natl. Acad. Sci. USA. 2018, 115, e753–e761. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Srinivasan, K.; Tran, H.; Yu, F.; Finefield, J.M.; Sunderhaus, J.D.; McAfoos, T.J.; Tsukamoto, S.; Williams, R.M.; Sherman, D.H. Comparative analysis of the biosynthetic systems for fungal bicyclo [2.2.2] diazaoctane indole alkaloids: The (+)/(−)-notoamide, paraherquamide and malbrancheamide pathways. Med. Chem. Commun. 2012, 3, 987–996. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, B.; Banerjee, A.; Tiwari, H.; Jana, S.; Bose, S.; Chakrabarti, S. Structural and functional characterization of the vindoline biosynthesis pathway enzymes of Catharanthus roseus. J. Mol. Model. 2018, 24, 53. [Google Scholar] [CrossRef]
- Ma, X.; Koepke, J.; Panjikar, S.; Fritzsch, G.; Stöckigt, J. Crystal structure of vinorine synthase, the first representative of the BAHD superfamily. J. Biol. Chem. 2005, 280, 13576–13583. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Du, L.; Zhang, X.; Newmister, S.A.; McCauley, M.; Alegre-Requena, J.V.; Zhang, W.; Mu, S.; Minami, A.; Fraley, A.E.; et al. Fungal-derived brevianamide assembly by a stereoselective semipinacolase. Nat. Catal. 2020, 3, 497–506. [Google Scholar] [CrossRef]
- Wang, X.; Lin, M.; Xu, D.; Lai, D.; Zhou, L. Structural diversity and biological activities of fungal cyclic peptides, excluding cyclodipeptides. Molecules 2017, 22, 2069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fremlin, L.J.; Piggott, A.M.; Lacey, E.; Capon, R.J. Cottoquinazoline A and cotteslosins A and B, metabolites from an australian marine-derived strain of Aspergillus versicolor. J. Nat Prod. 2009, 72, 666–670. [Google Scholar] [CrossRef]
- Romans-Fuertes, P.; Sondergaard, T.E.; Sandmann, M.I.H.; Wollenberg, R.D.; Nielsen, K.F.; Hansen, F.T.; Giese, H.; Brodersen, D.E.; Sørensen, J.L. Identification of the non-ribosomal peptide synthetase responsible for biosynthesis of the potential anti-cancer drug sansalvamide in Fusarium solani. Curr. Genet. 2016, 62, 799–807. [Google Scholar] [CrossRef]
- Chen, M.; Wang, K.-L.; Liu, M.; She, Z.-G.; Wang, C.-Y. Bioactive steroid derivatives and butyrolactone derivatives from a Gorgonian-derived Aspergillus sp. fungus. Chem. Biodivers. 2015, 12, 1398–1406. [Google Scholar] [CrossRef]
- Guo, C.J.; Knox, B.P.; Sanchez, J.F.; Chiang, Y.M.; Bruno, K.S.; Wang, C.C. Application of an efficient gene targeting system linking secondary metabolites to their biosynthetic genes in Aspergillus terreus. Org. Lett. 2013, 15, 3562–3565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, C.J.; Sun, W.W.; Bruno, K.S.; Oakley, B.R.; Keller, N.P.; Wang, C.C.C. Spatial regulation of a common precursor from two distinct genes generates metabolite diversity. Chem. Sci. 2015, 6, 5913–5921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Dijk, J.W.A.; Wang, C.C.C. Expanding the chemical space of nonribosomal peptide synthetase-like enzymes by domain and tailoring enzyme recombination. Org. Lett. 2018, 20, 5082–5085. [Google Scholar] [CrossRef]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F.; et al. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Mugunthan, M. Evaluation of three DNA extraction methods from fungal cultures. Med. J. Armed Forces India 2018, 74, 333–336. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [Green Version]
- Stanke, M.; Diekhans, M.; Baertsch, R.; Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 2008, 24, 637–644. [Google Scholar] [CrossRef] [Green Version]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [Green Version]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Ernst, M.; Kang, K.B.; Caraballo-Rodríguez, A.M.; Nothias, L.F.; Wandy, J.; Chen, C.; Wang, M.; Rogers, S.; Medema, M.H.; Dorrestein, P.C.; et al. MolNetEnhancer: Enhanced molecular networks by integrating metabolome mining and annotation tools. Metabolites 2019, 9, 144. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Marquès, M.; Mari, M.; Audí-Miró, C.; Sierra, J.; Soler, A.; Nadal, M.; Domingo, J.L. Climate change impact on the PAH photodegradation in soils: Characterization and metabolites identification. Environ. Int. 2016, 89–90, 155–165. [Google Scholar] [CrossRef] [PubMed]
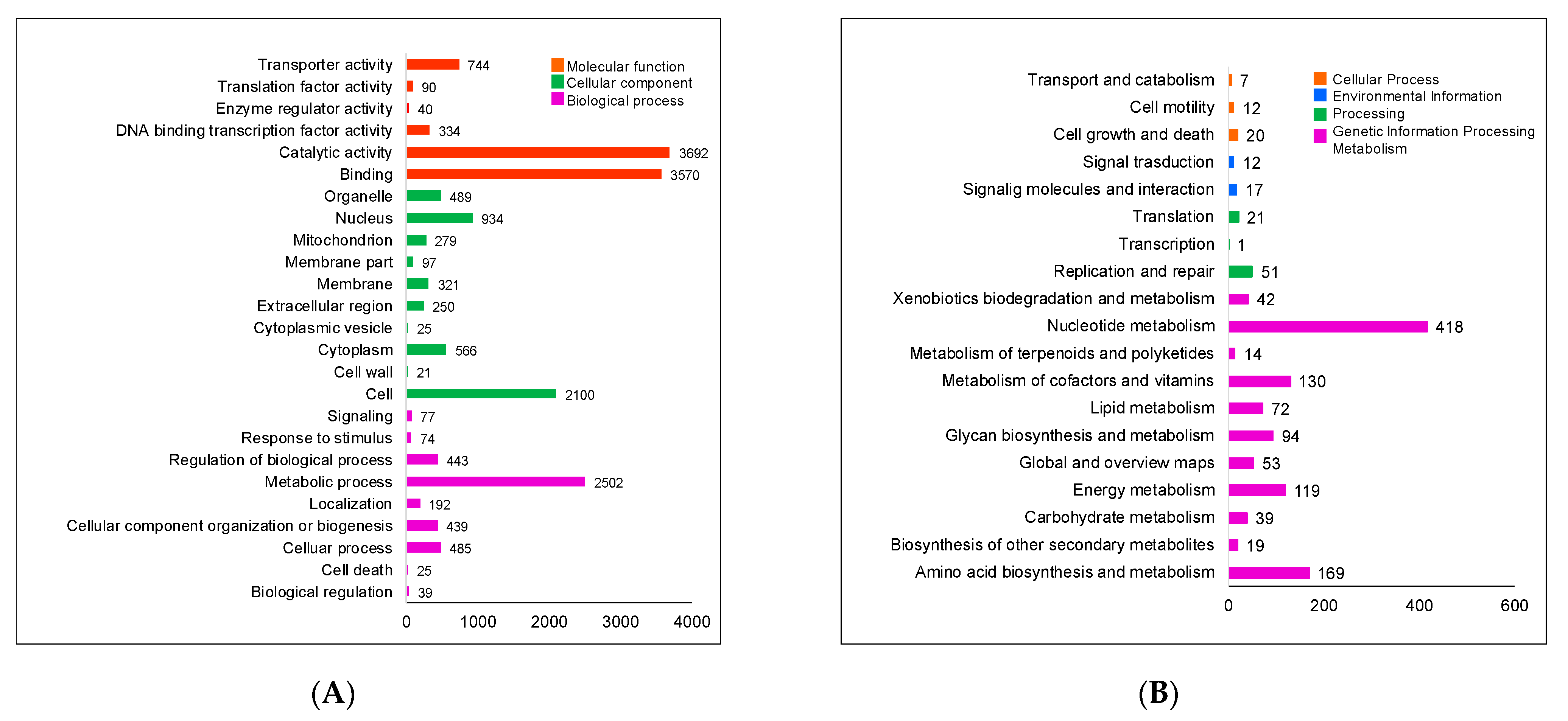
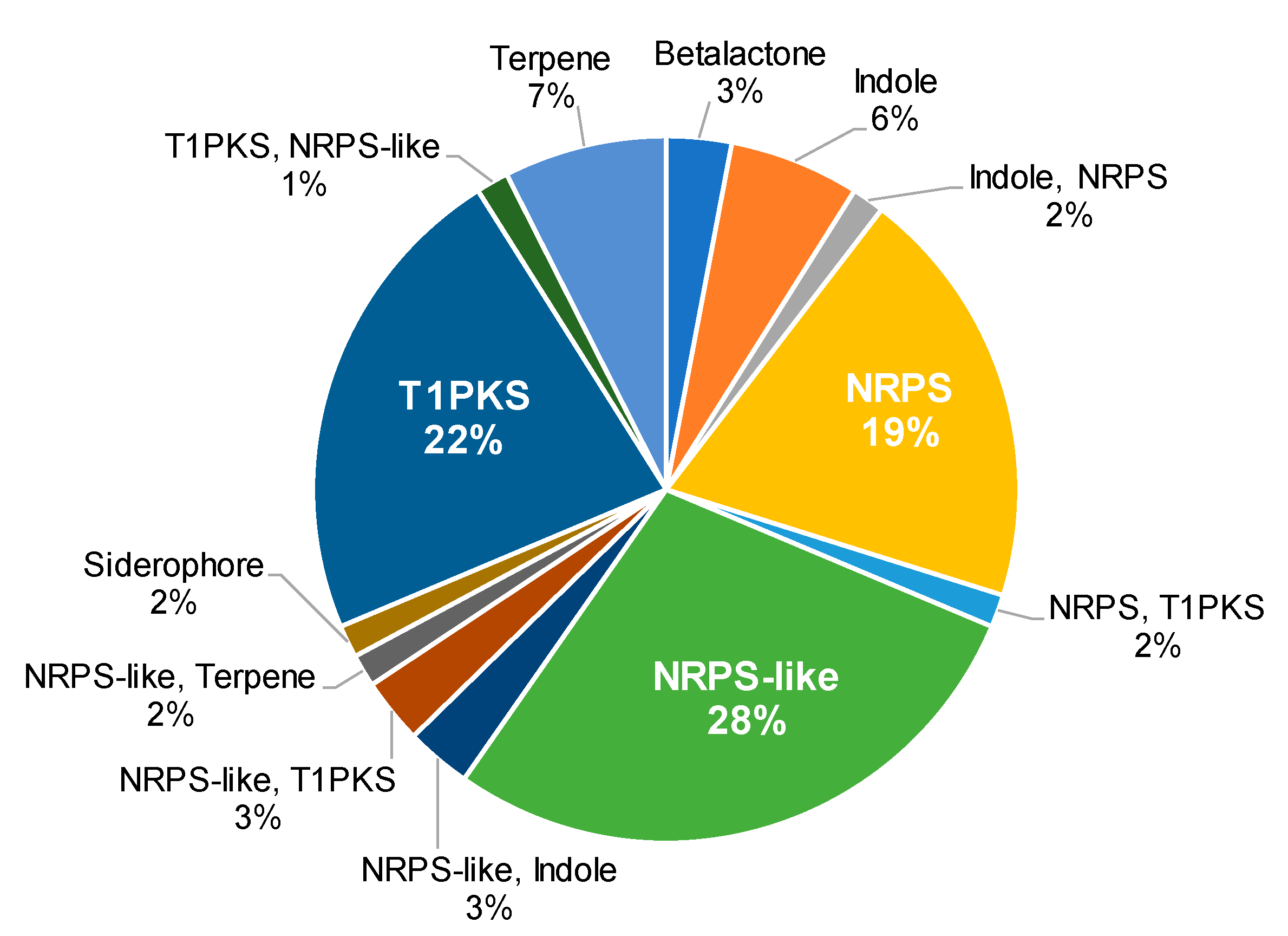

| Contig Characteristics | |
|---|---|
| Total number | 11 |
| Total length (bp) | 30,756,112 |
| N50 | 3,946,678 |
| L50 | 4 |
| Max. length (bp) | 5,241,077 |
| GC (%) | 53 |
| Genome Characteristics | |
| Genome assembly (MB) | 30.75 |
| Predicted protein coding sequences | 10,822 |
| % GO terms | 75 |
| Average gene length (bp) | 2934 |
| Cluster | Contig/Location/Similarity (%) | |||||
|---|---|---|---|---|---|---|
| Aspergillus sp. MEXU 27854 | A. terreus NIH2624 | |||||
| Asperfuranone | 4 | 2,673,561–2,721,442 | 18 | 11 | 634,985–694,556 | 81 |
| Asperphenamate | 3 | 271,678–331,344 | 75 | 14 | 139,864–204,269 | 100 |
| Azanigerone A | 8 | 1,592,427–1,640,144 | 20 | 5 | 1,778,394–1,863,542 | 20 |
| Burnettramic acid A | 3 | 390,502–438,846 | 22 | 14 | 209,359–297,831 | 22 |
| Citreoviridin | 1 | 862,457–908,022 | 80 | 15 | 353,636–401,160 | 100 |
| Clavaric acid | 6 | 1,529,334–1,551,729 | 100 | 12 | 102,884–121,870 | 100 |
| Clavaric acid | 6 | 1,529,334–1,551,729 | 100 | 12 | 102,884–121,870 | 100 |
| Dimethylcoprogen | 4 | 2,608,491–2,654,299 | 100 | 11 | 195,335–278,566 | 100 |
| Dimethylcoprogen | 4 | 2,608,491–2,654,299 | 100 | 11 | 195,335–278,566 | 100 |
| Duclauxin | 5 | 102,877–149,557 | 21 | 11 | 195,335–278,566 | 14 |
| Monascorubrin | 4 | 2,673,561–2,721,442 | 100 | 4 | 1,778,394–1,863,542 | 100 |
| Naphthopyrone | 4 | 2,500,933–2,543,731 | 100 | 8 | 1,584,471–1,627,478 | 100 |
| Nidulanin A | 6 | 2,333,314–2,390,373 | 50 | 1 | 620,230–676,423 | 100 |
| Ochrindole A | 6 | 3,324,939–3,362,374 | 29 | 1 | 1,948,860–1,985,238 | 11 |
| Pyranonigrin E | 6 | 2,504,128–2,546,577 | 100 | 1 | 786,879–829,500 | 100 |
| Squalestatin S1 | 8 | 1,544,948–1,587,995 | 60 | 12 | 501,832–541,227 | 60 |
| Terrequinone A | 6 | 3,324,939–3,362,374 | 100 | 1 | 1,948,860–1,985,238 | 60 |
| Adox Proteins (AA) | Function (% Identity to Corresponding Adx/Not Proteins) a | Adx Proteins (AA) | Function (% Identity to Corresponding Not Proteins) a |
|---|---|---|---|
| g7012/AdoxG | FAD monooxygenase | adxA | NRPS [A-T-C-A-T-C] |
| (426) | (30%, NotB) | (2413) | (24%, NotE) |
| g7440/AdoxA | CYP450 monooxygenase | adxB | Reductase |
| (490) | (28%, NotG) | (378) | (−) |
| g7441/AdoxB | Glucosyltransferase | adxC | Prenyltransferase |
| (155) | (−) | (422) | (35%, NoF) |
| g7442 | CYP450 monooxygenase | ||
| (261) | (−) | ||
| g7443/AdoxC | Prenyltransferase | ||
| (385) | (32%, AdxC/43%, NotF) | ||
| g7444/AdoxD | CYP450 monooxygenase | ||
| (248) | (−) | ||
| g7445/AdoxE | NRPS [A-T-C-A-T-C] | ||
| (2008) | (50%, AdxA/24%, NotE) | ||
| g7446 | Unknown | ||
| (455) | (−) | ||
| g7447 | Unknown | ||
| (252) | (−) | ||
| g7448 | Unknown | ||
| (191) | (−) | ||
| g7449 | Unknown | ||
| (915) | (−) | ||
| g7450 | Transcriptional factor | ||
| (2343) | (−) | ||
| g7531/AdoxF | NADPH-Reductase | ||
| (295) | (22%, AdxB) | ||
| g2501/AdoxH | Acetyltransferase | ||
| (531) | (−) |
| Proteins (AA) | GenBank Accession No. | Closest GenBank Homolog | Amino Acid Identity (%) |
|---|---|---|---|
| g572 (930) | MZ503789 | ATEG_02815 Non-ribosomal peptide synthetase btyA Butyrolactone IIa synthetase (A. terreus NIH2624) Q0CU19.2 | 746/931 (80%) |
| Putative non-ribosomal peptide synthetase (Cladonia uncialis subsp. Uncialis) ANM86632.1 | 431/935 (46%) | ||
| Non-ribosomal peptide synthetase (A. tanneri) XP_033423325.1 | 406/934 (43%) | ||
| g573 (271) | ATEG_02816 Methyltransferase btyB (A. terreus NIH2624) XP_001211994.1 | 203/269 (75%) | |
| Hypothetical protein ATETN484_0005003700 (A. terreus) GES60578.1 | 223/271 (82%) |
| Compound | Observed Ion a | Adduct | Molecular Formula | Exact Mass c | Mass Accuracy (ppm) |
|---|---|---|---|---|---|
| 9-deoxy-PF1233 B (1) | 419.195 | [M + H]+ | C25H26N2O4 | 418.1893 | −3.7 |
| 9-deoxy-PF1233 A (2) | 461.206 | [M + H]+ | C27H28N2O5 | 460.1998 | −2.4 |
| seco-PF1233 B carboxylic acid (3) | 453.202 | [M + H]+ | C25H28N2O6 | 452.1947 | 0.0 |
| 9-deoxy-seco-PF1233 B carboxylic acid (4) | 437.206 | [M + H]+ | C25H28N2O5 | 436.1998 | −2.5 |
| 4,9-dideoxy-seco-PF1233 B carboxylic acid (5) | 421.211 | [M + H]+ | C25H28N2O4 | 420.2049 | −2.4 |
| PF1233 B (6) | 435.191 | [M + H]+ | C25H26N2O5 | 434.1842 | −1.0 |
| PF1233 A (7) | 477.202 | [M + H]+ | C27H28N2O6 | 476.1941 | 0.0 |
| Caletasin (8) | 634.359 | [M + H]+ | C35H47N5O6 | 633.3526 | −1.4 |
| 3-O-methylbutyrolactone II (9) | 371.112 | [M + H]+ | C20H18O7 | 370.1052 | +2.7 |
| Butyrolactone II (10) | 357.097 | [M + H]+ | C19H16O7 | 356.0896 | +2.7 |
| Asperphenamate b | 507.228 | [M + H]+ | C32H30N2O4 | 506.2206 | +2.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Cárdenas, A.; Cruz-Zamora, Y.; Fajardo-Hernández, C.A.; Villanueva-Silva, R.; Cruz-García, F.; Raja, H.A.; Figueroa, M. Genome Mining and Molecular Networking-Based Metabolomics of the Marine Facultative Aspergillus sp. MEXU 27854. Molecules 2021, 26, 5362. https://doi.org/10.3390/molecules26175362
Martínez-Cárdenas A, Cruz-Zamora Y, Fajardo-Hernández CA, Villanueva-Silva R, Cruz-García F, Raja HA, Figueroa M. Genome Mining and Molecular Networking-Based Metabolomics of the Marine Facultative Aspergillus sp. MEXU 27854. Molecules. 2021; 26(17):5362. https://doi.org/10.3390/molecules26175362
Chicago/Turabian StyleMartínez-Cárdenas, Anahí, Yuridia Cruz-Zamora, Carlos A. Fajardo-Hernández, Rodrigo Villanueva-Silva, Felipe Cruz-García, Huzefa A. Raja, and Mario Figueroa. 2021. "Genome Mining and Molecular Networking-Based Metabolomics of the Marine Facultative Aspergillus sp. MEXU 27854" Molecules 26, no. 17: 5362. https://doi.org/10.3390/molecules26175362
APA StyleMartínez-Cárdenas, A., Cruz-Zamora, Y., Fajardo-Hernández, C. A., Villanueva-Silva, R., Cruz-García, F., Raja, H. A., & Figueroa, M. (2021). Genome Mining and Molecular Networking-Based Metabolomics of the Marine Facultative Aspergillus sp. MEXU 27854. Molecules, 26(17), 5362. https://doi.org/10.3390/molecules26175362