Selective Binding and Redox-Activity on Parallel G-Quadruplexes by Pegylated Naphthalene Diimide-Copper Complexes
Abstract
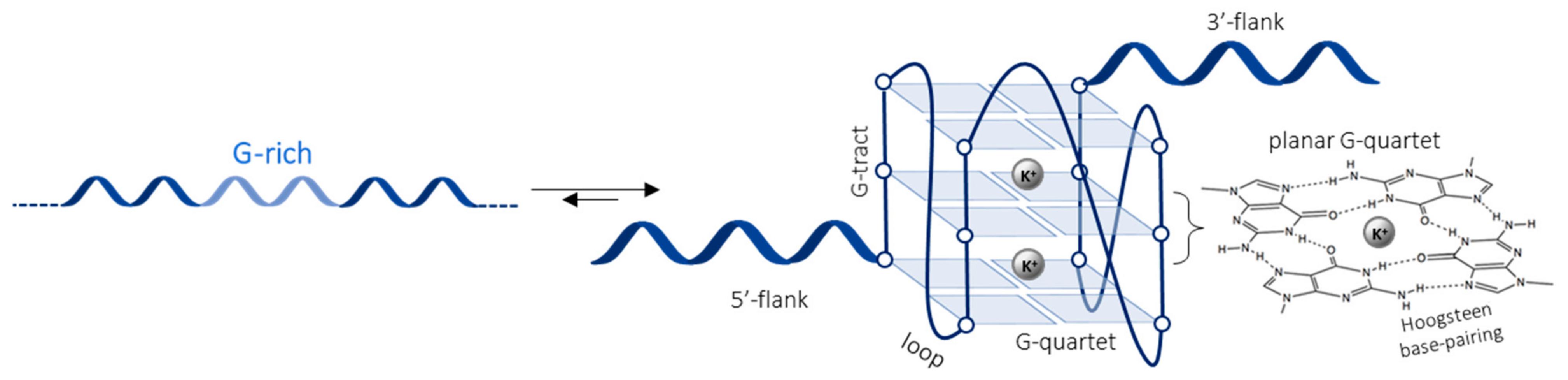
:1. Introduction
2. Results and Discussion
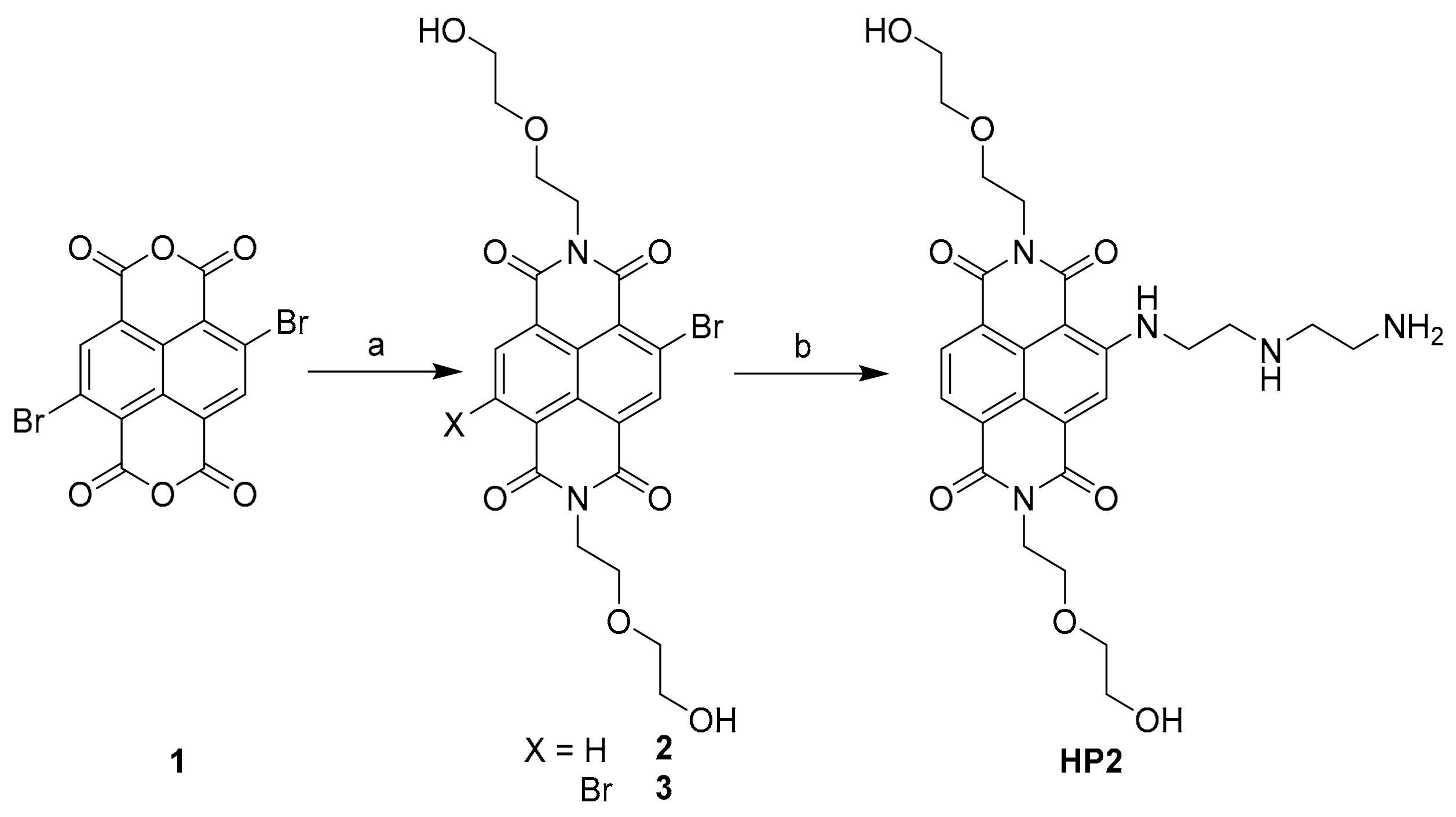
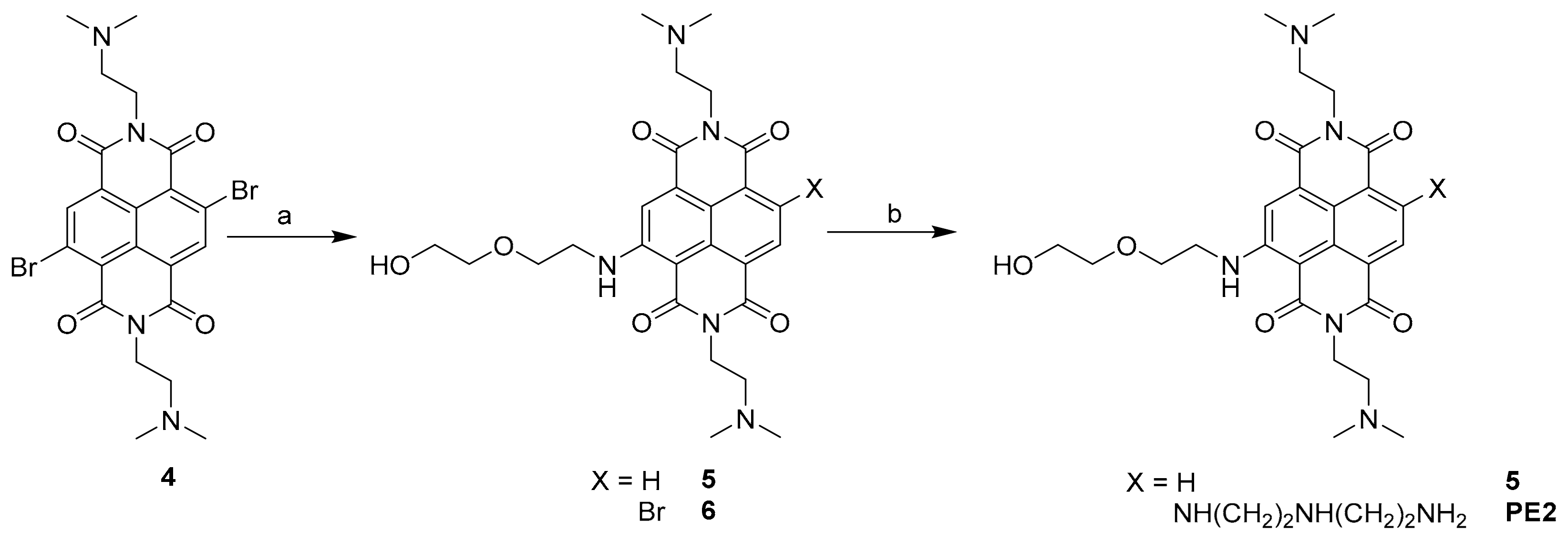
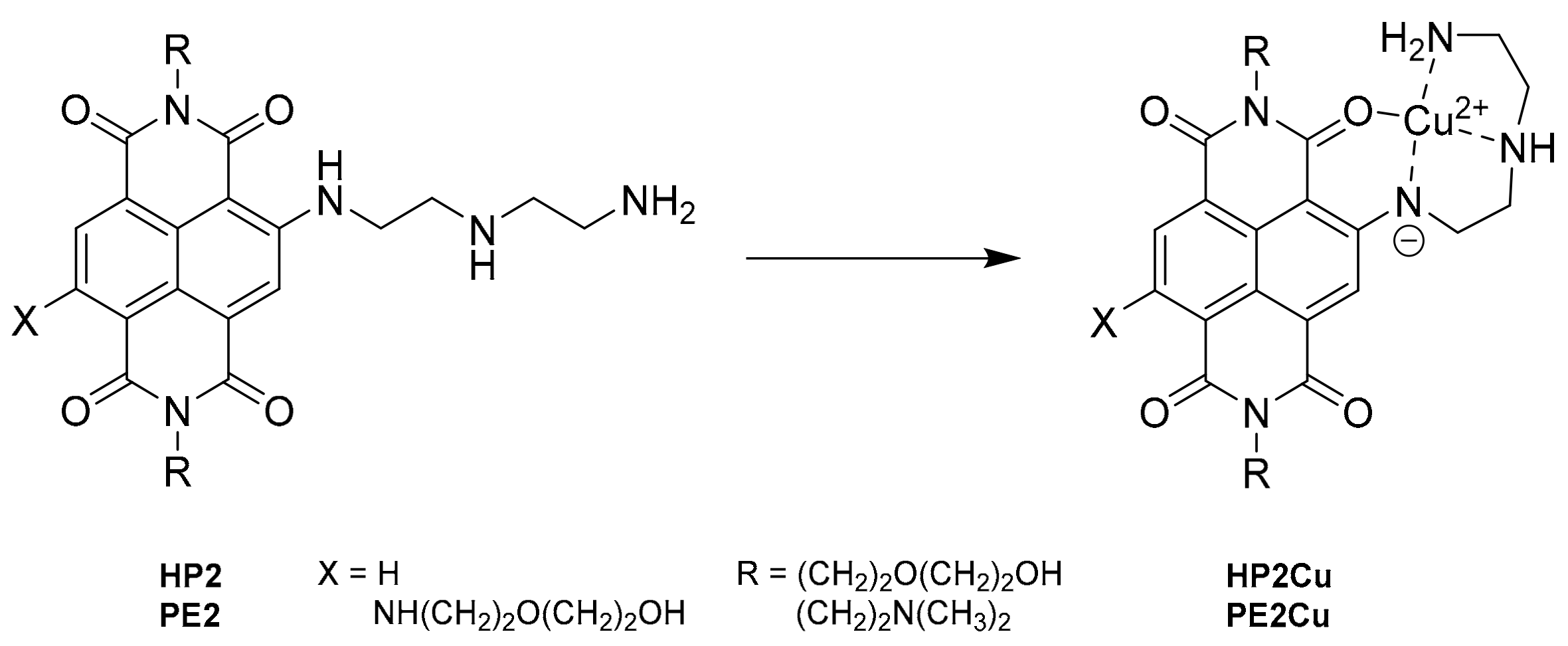
2.1. Synthesis of the New DETA-Substituted NDIs HP2 and PE2
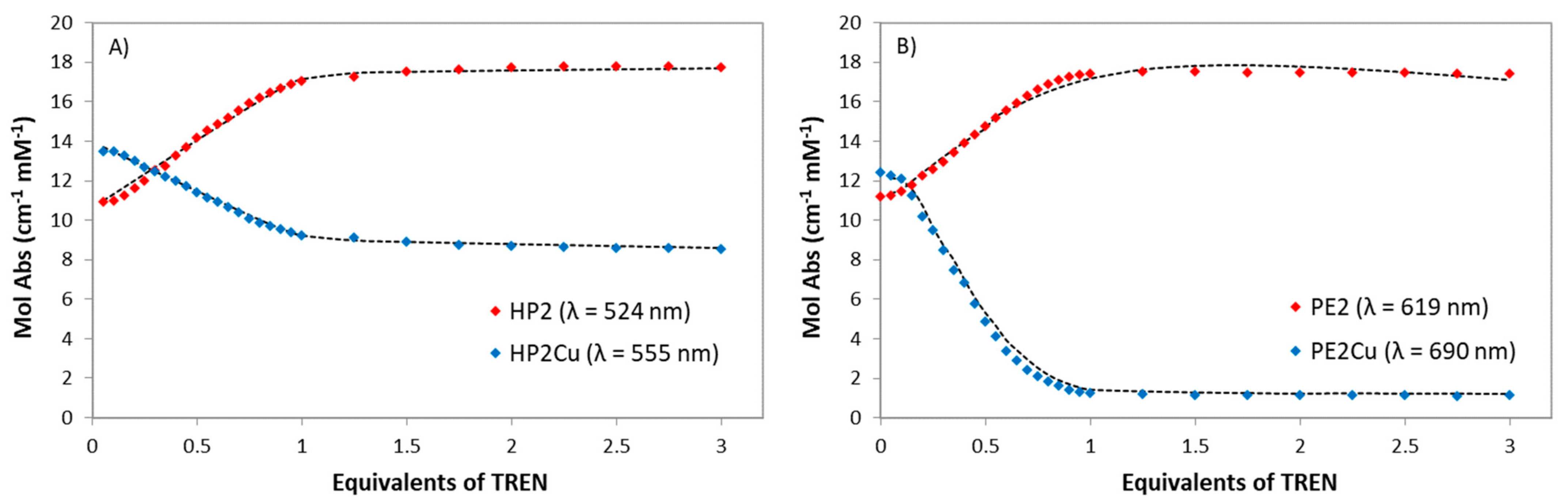
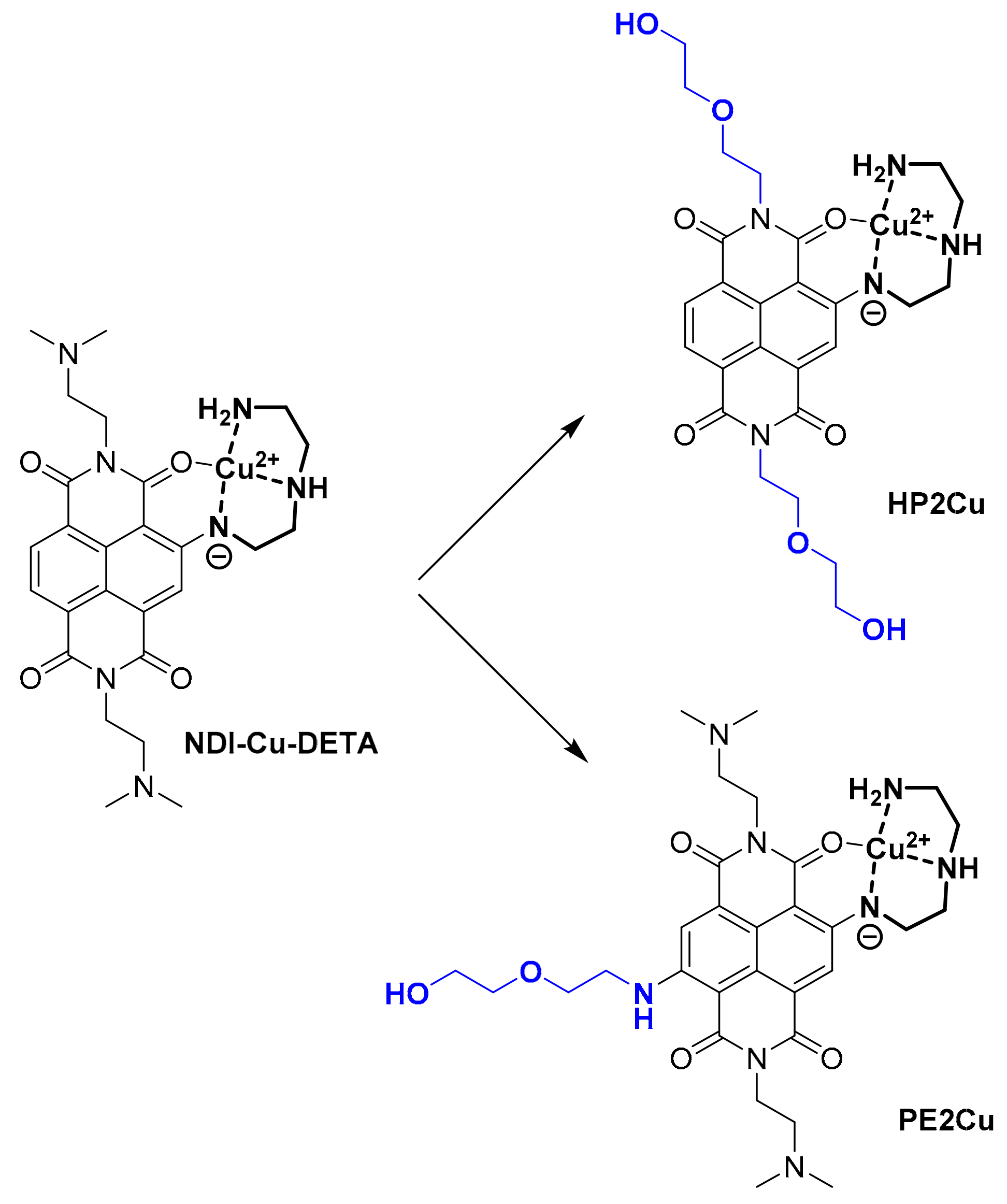
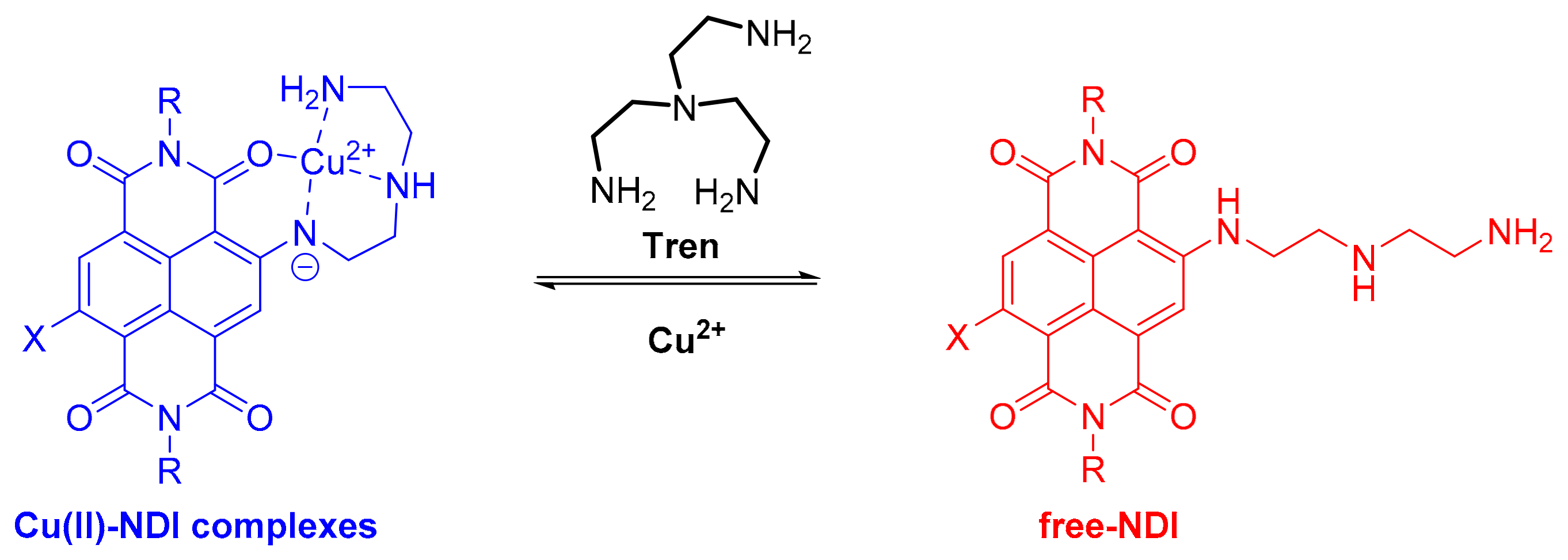
2.2. Copper Coordination to HP2 and PE2 and Complexes Synthesis
2.3. Determination of the Binding Constants
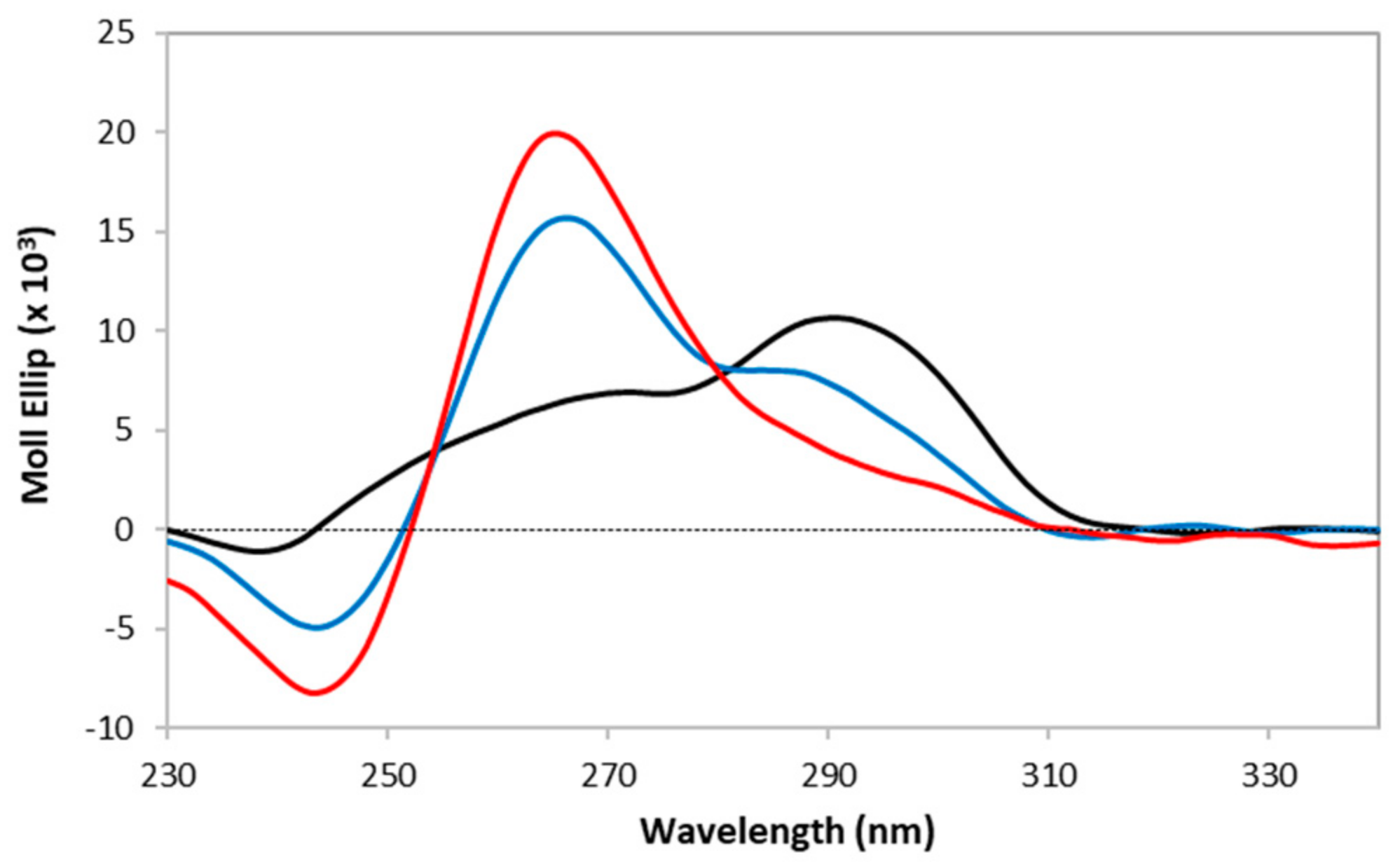
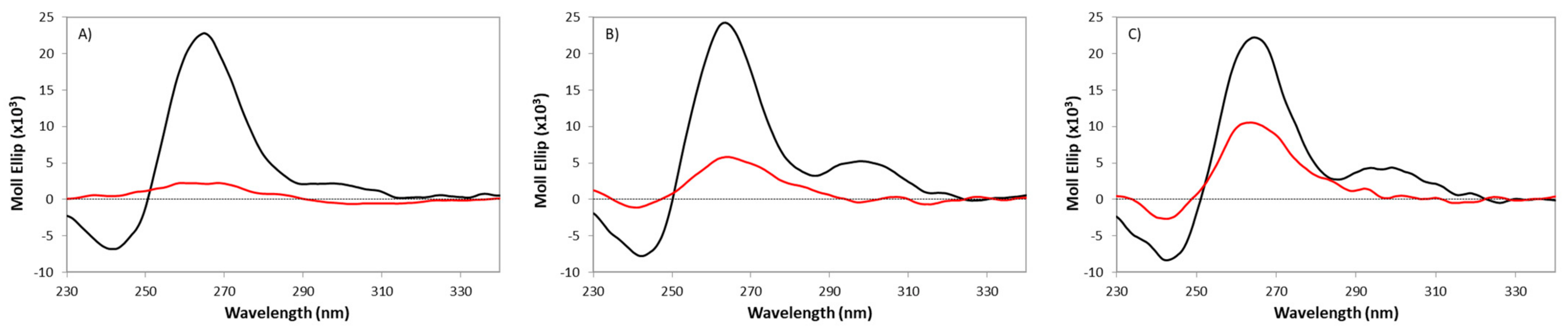
2.4. Analysis of the Effect of HP2Cu and PE2Cu on Telomeric G-Quadruplexes
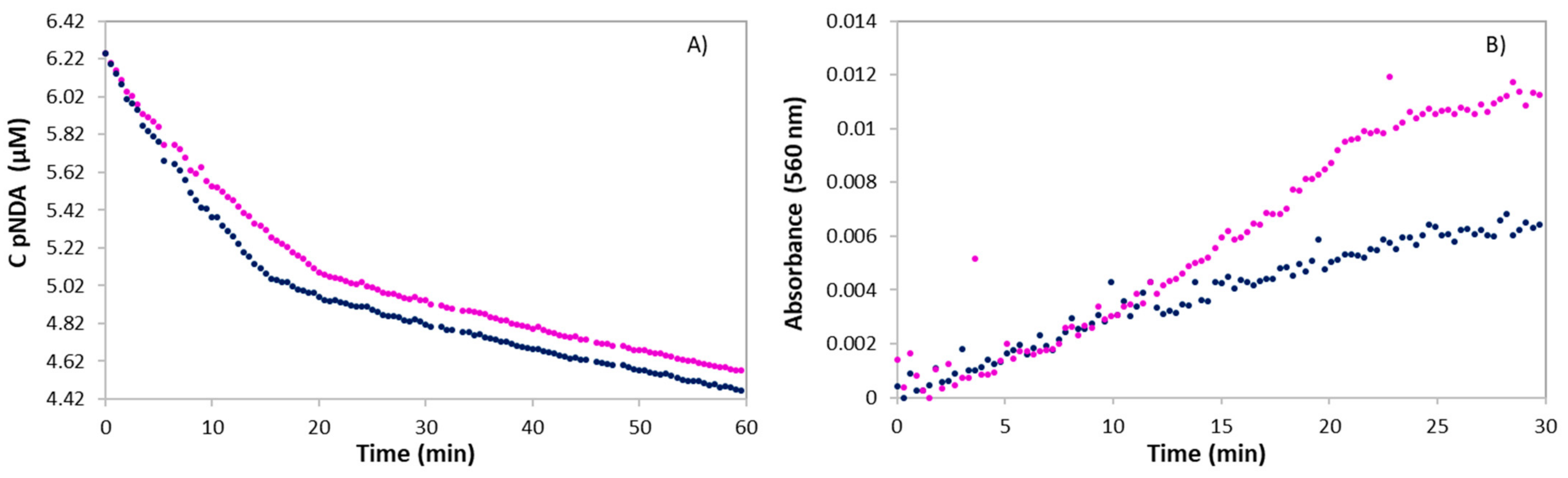
2.5. Evaluation of ROS-Mediated NA Cleavage
2.6. Determination of ROS Involved
3. Material and Methods
3.1. General Information
3.2. Synthetic Procedures
3.2.1. Synthesis of HP2
3.2.2. Synthesis of PE2
3.2.3. Synthesis of HP2Cu and PE2Cu Complexes
3.3. UV–Vis Spectrophotometric Titrations
3.4. Circular-Dichroism (CD) Analysis
3.5. Denaturing PAGE Assay
3.6. Characterization of Reactive Oxygen Species Involved
3.6.1. Bleaching of p-Nitroso-N,N-dimethylaniline (p-NDA)
3.6.2. Nitro Blue Tetrazolium (NBT) Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Kharel, P.; Becker, G.; Tsvetkov, V.; Ivanov, P. Properties and biological impact of RNA G-quadruplexes: From order to turmoil and back. Nucleic Acids Res. 2020, 48, 12534–12555. [Google Scholar] [CrossRef]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [Green Version]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Kosiol, N.; Juranek, S.; Brossart, P.; Heine, A.; Paeschke, K. G-quadruplexes: A promising target for cancer therapy. Mol. Cancer 2021, 20, 40. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Thombre, R.; Shah, Y.; Latanich, R.; Wang, J. G-Quadruplexes as pathogenic drivers in neurodegenerative disorders. Nucleic Acids Res. 2021, 49, 4816–4830. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, E.; Richter, S.N. Viral G-quadruplexes: New frontiers in virus pathogenesis and antiviral therapy. Annu. Rep. Med. Chem. 2020, 54, 101–131. [Google Scholar]
- Zhao, C.; Qin, G.; Niu, J.; Wang, Z.; Wang, C.; Ren, J.; Qu, X. Targeting RNA G-Quadruplex in SARS-CoV-2: A Promising Therapeutic Target for COVID-19? Angew. Chem. Int. Ed. 2021, 60, 432–438. [Google Scholar] [CrossRef]
- Jodoin, R.; Carrier, J.C.; Rivard, N.; Bisaillon, M.; Perreault, J.-P. G-quadruplex located in the 5′UTR of the BAG-1 mRNA affects both its cap-dependent and cap-independent translation through global secondary structure maintenance. Nucleic Acids Res. 2019, 47, 10247–10266. [Google Scholar] [CrossRef]
- Simone, R.; Balendra, R.; Moens, T.G.; Preza, E.; Wilson, K.M.; Heslegrave, A.; Woodling, N.S.; Niccoli, T.; Gilbert-Jaramillo, J.; Abdelkarim, S.; et al. G-quadruplex-binding small molecules ameliorate C9orf72 FTD/ALS pathology in vitro and in vivo. EMBO Mol. Med. 2018, 10, 22–31. [Google Scholar] [CrossRef]
- Mei, Y.; Deng, Z.; Vladimirova, O.; Gulve, N.; Johnson, F.B.; Drosopoulos, W.C.; Schildkraut, C.L.; Lieberman, P.M. TERRA G-quadruplex RNA interaction with TRF2 GAR domain is required for telomere integrity. Sci. Rep. 2021, 11, 3509. [Google Scholar] [CrossRef]
- Harpster, C.; Boyle, E.; Musier-Forsyth, K.; Kankia, B. HIV-1 genomic RNA U3 region forms a stable quadruplex-hairpin structure. Biophys. Chem. 2021, 272, 106567. [Google Scholar] [CrossRef]
- Kench, T.; Vilar, R. Chapter Fourteen—Metal complexes as G-quadruplex binders. Annu. Rep. Med. Chem. 2020, 54, 485–515. [Google Scholar]
- Nadai, M.; Doria, F.; Scalabrin, M.; Pirota, V.; Grande, V.; Bergamaschi, G.; Amendola, V.; Winnerdy, F.R.; Phan, A.T.; Richter, S.N.; et al. A Catalytic and Selective Scissoring Molecular Tool for Quadruplex Nucleic Acids. J. Am. Chem. Soc. 2018, 140, 14528–14532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirota, V.; Stasi, M.; Benassi, A.; Doria, F. Chapter Six—An overview of quadruplex ligands: Their common features and chemotype diversity. Annu. Rep. Med. Chem. 2020, 54, 163–196. [Google Scholar]
- Pirota, V.; Nadai, M.; Doria, F.; Richter, S.N. Naphthalene Diimides as Multimodal G-Quadruplex-Selective Ligands. Molecules 2019, 24, 426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, C.-F.; Shafer, R.H. Engineering the Quadruplex Fold: Nucleoside Conformation Determines Both Folding Topology and Molecularity in Guanine Quadruplexes. J. Am. Chem. Soc. 2006, 128, 5966–5973. [Google Scholar] [CrossRef] [Green Version]
- Zuffo, M.; Guédin, A.; Leriche, E.-D.; Doria, F.; Pirota, V.; Gabelica, V.; Mergny, J.-L.; Freccero, M. More is not always better: Finding the right trade-off between affinity and selectivity of a G-quadruplex ligand. Nucleic Acids Res. 2018, 46, e115. [Google Scholar] [CrossRef]
- Doria, F.; di Antonio, M.; Benotti, M.; Verga, D.; Freccero, M. Substituted heterocyclic naphthalene diimides with unexpected acidity. Synthesis, properties, and reactivity. J. Org. Chem. 2009, 74, 8616–8625. [Google Scholar] [CrossRef] [PubMed]
- Doria, F.; Manet, I.; Grande, V.; Monti, S.; Freccero, M. Water-soluble naphthalene diimides as singlet oxygen sensitizers. J. Org. Chem. 2013, 78, 8065–8073. [Google Scholar] [CrossRef] [PubMed]
- Martell, A.E.; Smith, R.M. Critical Stability Constants, 1st ed.; Springer: New York, NY, USA, 1982; Volume 5, p. 604. [Google Scholar]
- Ambrus, A.; Chen, D.; Dai, J.; Bialis, T.; Jones, R.A.; Yang, D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006, 34, 2723–2735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collie, G.W.; Haider, S.M.; Neidle, S.; Parkinson, G.N. A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex. Nucleic Acids Res. 2010, 38, 5569–5580. [Google Scholar] [CrossRef]
- Del Villar-Guerra, R.; Trent, J.O.; Chaires, J.B. G-Quadruplex Secondary Structure Obtained from Circular Dichroism Spectroscopy. Angew. Chem. Int. Ed. Engl. 2018, 57, 7171–7175. [Google Scholar] [CrossRef] [PubMed]
- Olowe, R.; Sandouka, S.; Saadi, A.; Shekh-Ahmad, T. Approaches for Reactive Oxygen Species and Oxidative Stress Quantification in Epilepsy. Antioxidants 2020, 9, 990. [Google Scholar] [CrossRef]
- McCord, J.M. The evolution of free radicals and oxidative stress. Am. J. Med. 2000, 108, 652–659. [Google Scholar] [CrossRef]
- Luo, Y.; Henle, E.S.; Linn, S. Oxidative damage to DNA constituents by iron-mediated fenton reactions. The deoxycytidine family. J. Biol. Chem. 1996, 271, 21167–21176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minotti, G.; Aust, S.D. The requirement for iron (III) in the initiation of lipid peroxidation by iron (II) and hydrogen peroxide. J. Biol. Chem. 1987, 262, 1098–1104. [Google Scholar] [CrossRef]
- Ohyashiki, T.; Nunomura, M.; Katoh, T. Detection of superoxide anion radical in phospholipid liposomal membrane by fluorescence quenching method using 1,3-diphenylisobenzofuran. Biochim. Biophys. Acta Biomembr. 1999, 1421, 131–139. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, A.S.; Gaspar, J.; Cabral, M.F.; Caneiras, C.; Guedes, R.; Rueff, J.; Castro, M.; Costa, J.; Oliveira, N.G. Macrocyclic copper(II) complexes: Superoxide scavenging activity, structural studies and cytotoxicity evaluation. J. Inorg. Biochem. 2007, 101, 849–858. [Google Scholar] [CrossRef]
- Gans, P.; Sabatini, A.; Vacca, A. Investigation of equilibria in solution. Determination of equilibrium constants with the HYPERQUAD suite of programs. Talanta 1996, 43, 1739–1753. [Google Scholar] [CrossRef]
- Del Villar-Guerra, R.; Gray, R.D.; Chaires, J.B. Characterization of Quadruplex DNA Structure by Circular Dichroism. Curr. Protoc. Nucleic Acid Chem. 2017, 68, 17.8.1–17.8.16. [Google Scholar] [CrossRef]

| Alone | HP2Cu | PE2Cu | NDI-Cu-DETA | |||||
|---|---|---|---|---|---|---|---|---|
| λ (nm) | Tm (°C) | Tm (°C) | ΔTm (°C) | Tm (°C) | ΔTm (°C) | Tm (°C) | ΔTm (°C) | |
| TERRA (K+ = 100 mM) | 264 | 81.5 ± 0.7 | >90 | >8 | >90 | >8.5 | >90 | >8.5 |
| TERRA (K+ = 10 mM) | 264 | 72.6 ± 0.3 | 84.2 ± 0.1 | +11.6 | >90 | >17.4 | 82.6 ± 0.8 | +10.0 |
| TERRA (K+ = 1 mM) | 264 | 63.7 ± 0.2 | 74.5 ± 0.4 | +10.8 | >90 | >26.3 | 79.1 ± 0.5 | +15.4 |
| TERRA (K+ = 0.1 mM) | 264 | 51.2 ± 0.2 | 58.5± 0.1 | +7.3 | 75.4 ± 0.2 | +24.2 | 66.4 ± 0.3 | +15.2 |
| hTel22 (K+ = 100 mM) | 290 | 78.24 ± 0.06 | 79.2 ± 0.6 | +1.0 | 61.3 ± 0.1 | −16.9 | 82.4 ± 0.2 | +4.16 |
| 264 | 80.3 ± 0.9 | 83.6 ± 0.4 | +3.3 | >90 | >11.6 | 89.0 ± 0.2 | +8.7 | |
| hTel22 (K+ = 10 mM) | 290 | 62.52 ± 0.05 | 66.13 ± 0.07 | +3.61 | 49.2 ± 0.1 | −13.3 | 70.34 ± 0.09 | +7.82 |
| 264 | 66.4 ± 0.3 | 80 ± 1 | +13.6 | 88.5 ± 0.3 | +22.9 | 84.9 ± 0.3 | +18.5 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pirota, V.; Lunghi, E.; Benassi, A.; Crespan, E.; Freccero, M.; Doria, F. Selective Binding and Redox-Activity on Parallel G-Quadruplexes by Pegylated Naphthalene Diimide-Copper Complexes. Molecules 2021, 26, 5025. https://doi.org/10.3390/molecules26165025
Pirota V, Lunghi E, Benassi A, Crespan E, Freccero M, Doria F. Selective Binding and Redox-Activity on Parallel G-Quadruplexes by Pegylated Naphthalene Diimide-Copper Complexes. Molecules. 2021; 26(16):5025. https://doi.org/10.3390/molecules26165025
Chicago/Turabian StylePirota, Valentina, Enrico Lunghi, Alessandra Benassi, Emmanuele Crespan, Mauro Freccero, and Filippo Doria. 2021. "Selective Binding and Redox-Activity on Parallel G-Quadruplexes by Pegylated Naphthalene Diimide-Copper Complexes" Molecules 26, no. 16: 5025. https://doi.org/10.3390/molecules26165025
APA StylePirota, V., Lunghi, E., Benassi, A., Crespan, E., Freccero, M., & Doria, F. (2021). Selective Binding and Redox-Activity on Parallel G-Quadruplexes by Pegylated Naphthalene Diimide-Copper Complexes. Molecules, 26(16), 5025. https://doi.org/10.3390/molecules26165025









