The Impacts of Different Biological Treatments on the Transformation of Explosives Waste Contaminated Sludge
Abstract
:1. Introduction
2. Materials and Methods
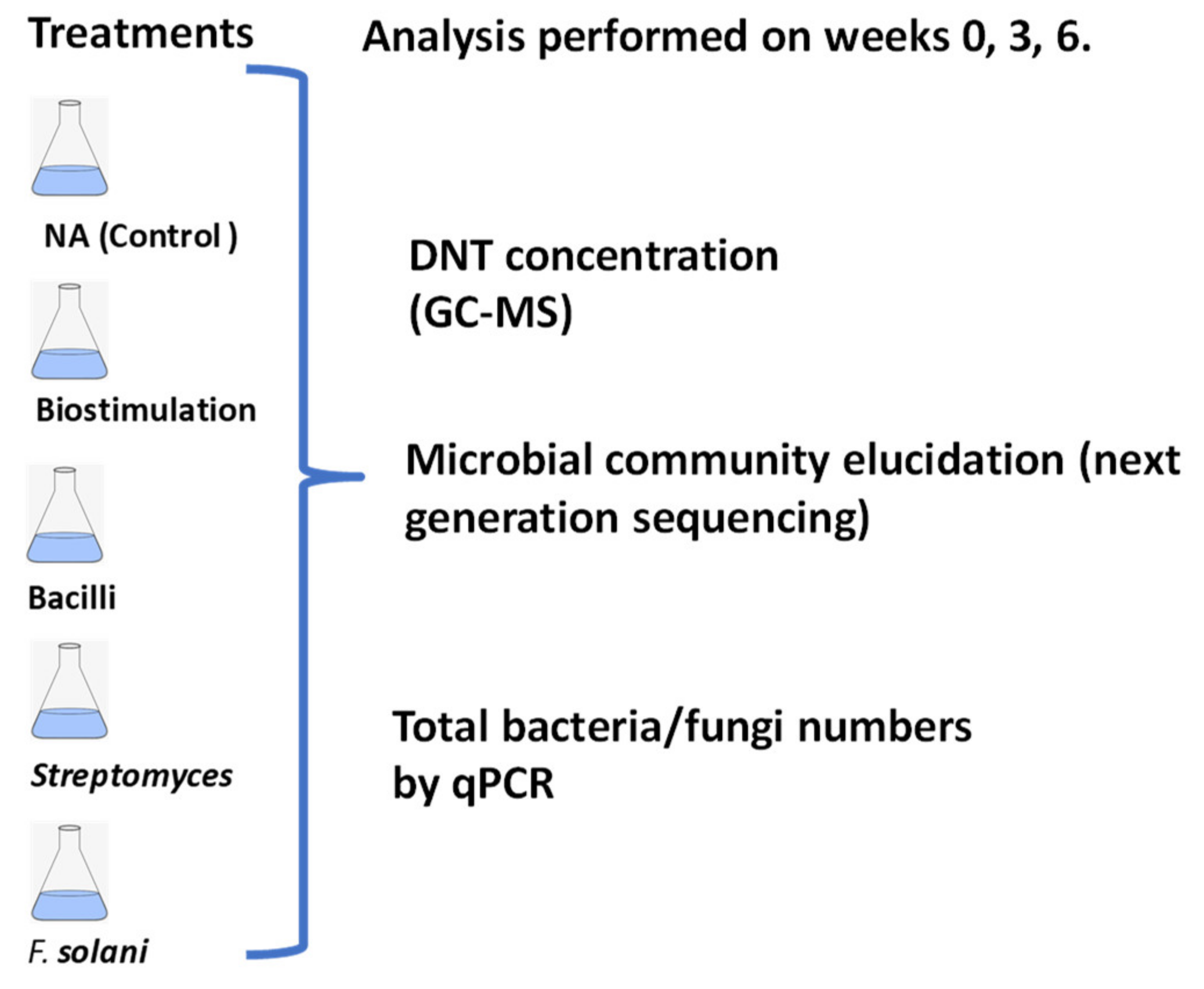
2.1. Mesocosms
2.2. DNT Concentration
2.3. Microbial Community Analysis (16S DNA Illumina MiSeq Sequencing)
2.4. Quantification of Total Fungi, Bacteria (qPCR)
2.5. Data Analysis
3. Results and Discussion
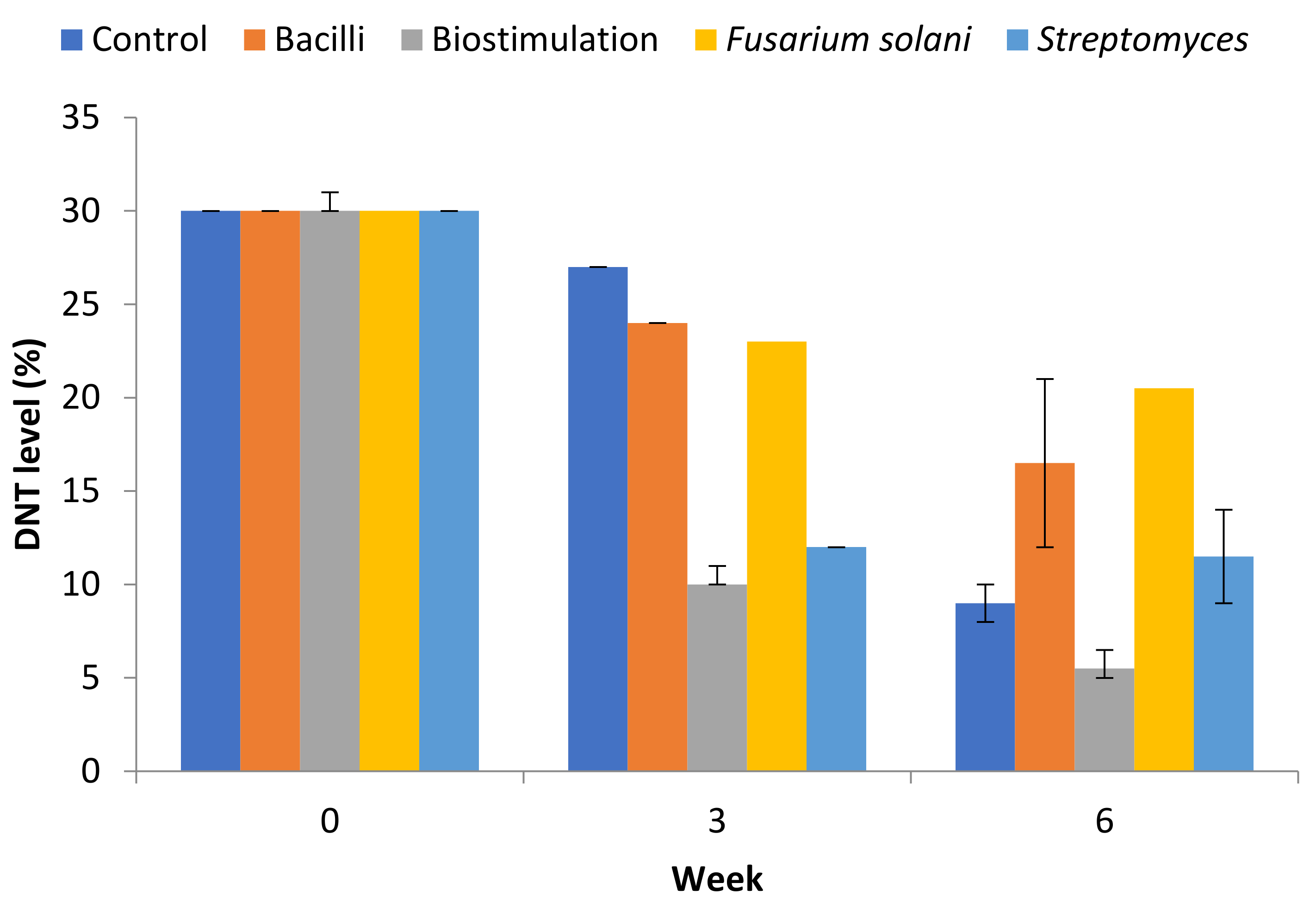
3.1. DNT Remediation
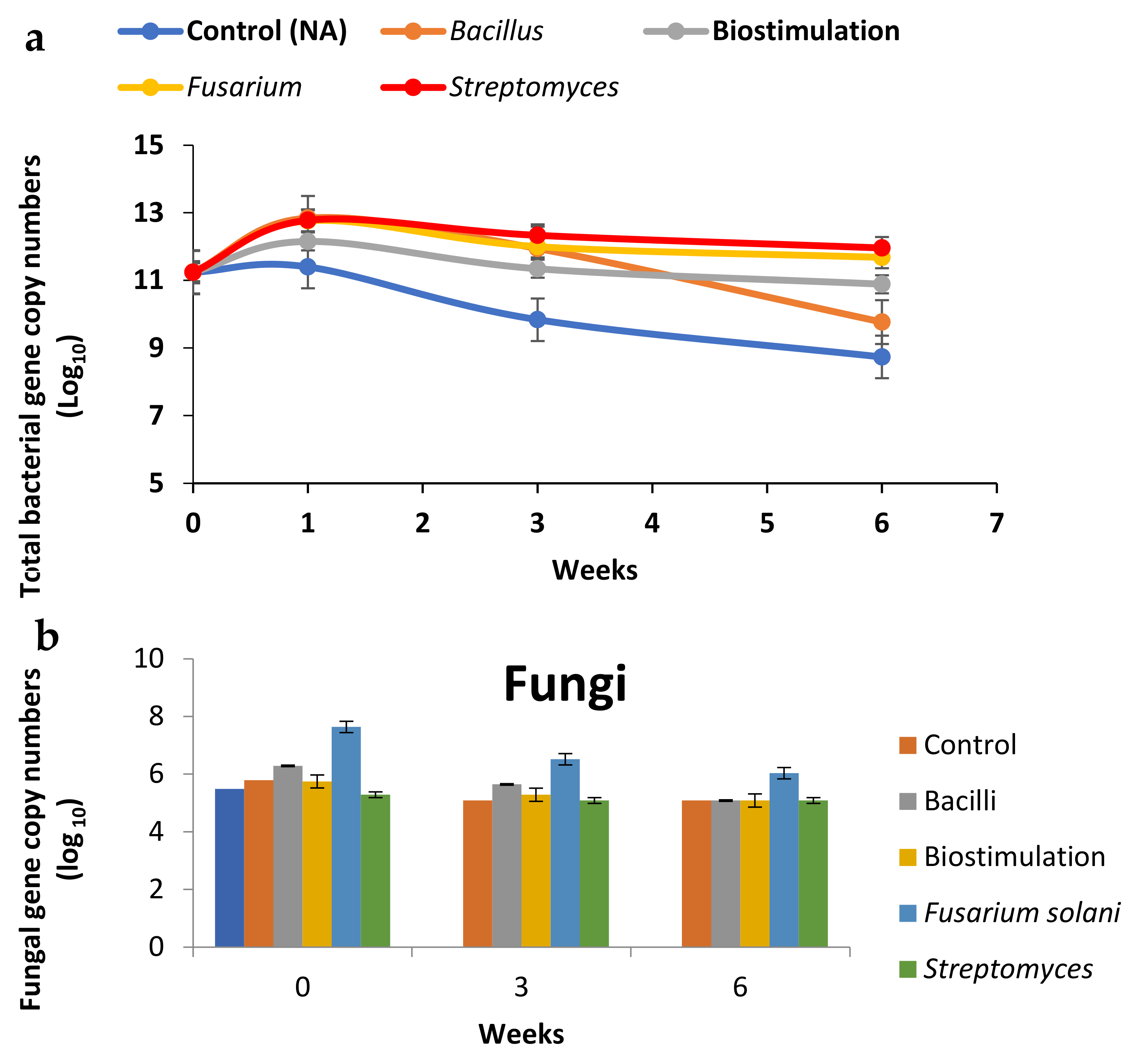
3.2. Total Microbial Community Analysis Quantitative PCR
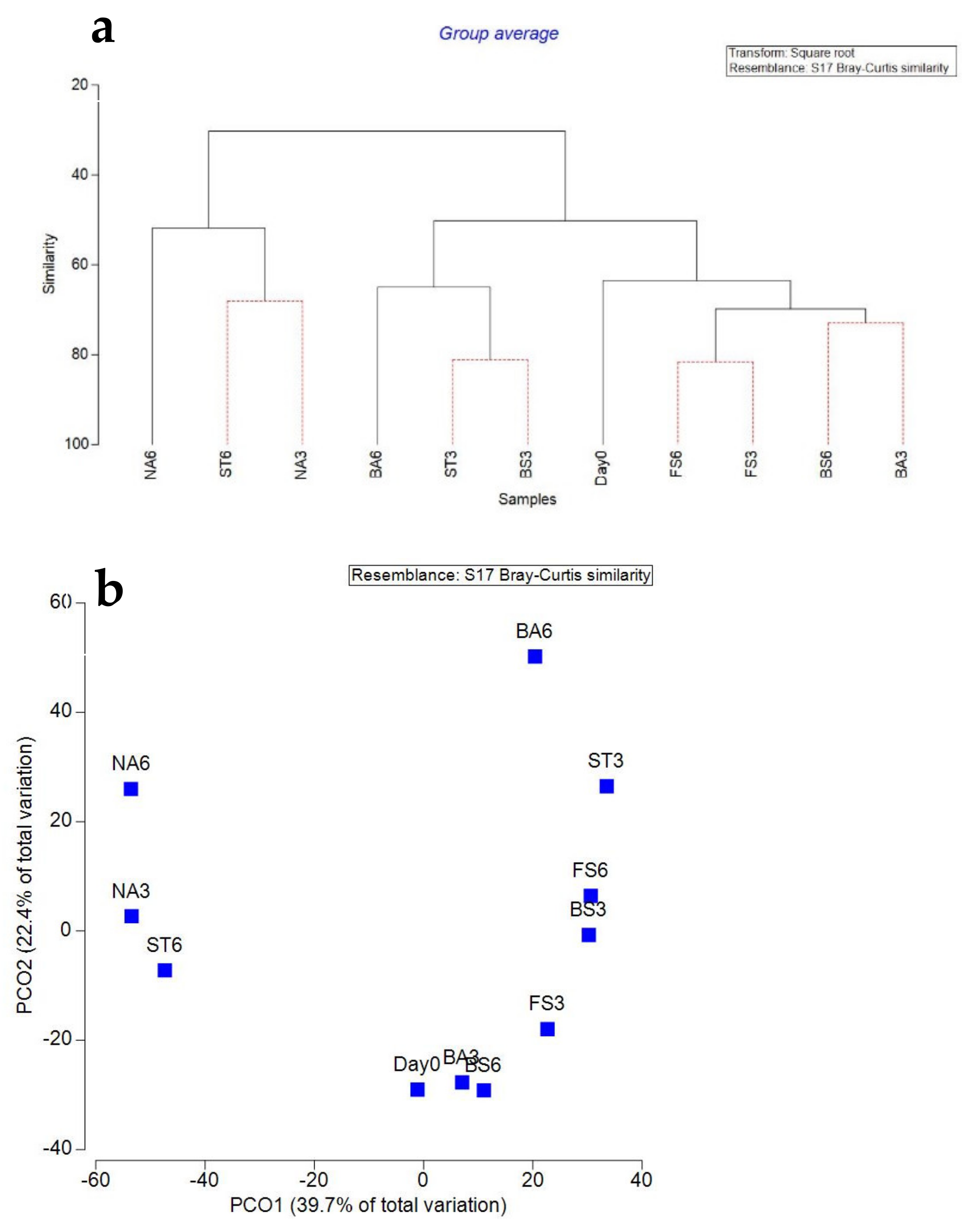
3.3. Bacterial Community Diversity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Chatterjee, S.; Deb, U.; Datta, S.; Walther, C.; Gupta, D.K. Common explosives (TNT, RDX, HMX) and their fate in the environment: Emphasizing bioremediation. Chemosphere 2017, 184, 438–451. [Google Scholar] [CrossRef]
- Bruns-Nagel, D.; Steinbach, K.; Gemsa, D.; von Löw, E. Composting (Humification) of Nitroaromatic Compounds. Biodegradation of Nitroaromatic Compounds and Explosives; Lewis Publishers: Boca Raton, FL, USA, 2000; pp. 357–393. [Google Scholar]
- Hudcova, T.; Halecky, M.; Kozliak, E.; Stiborova, M.; Paca, J. Aerobic degradation of 2,4-dinitrotoluene by individual bacterial strains and defined mixed population in submerged cultures. J. Hazard. Mater. 2011, 192, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Thijs, S.; Weyens, N.; Sillen, W.; Gkorezis, P.; Carleer, R.; Vangronsveld, J. Potential for plant growth promotion by a consortium of stress-tolerant 2,4-dinitrotoluene-degrading bacteria: Isolation and characterization of a military soil. Microb. Biotechnol. 2014, 7, 294–306. [Google Scholar] [CrossRef]
- Dodard, S.; Renoux, A.; Hawari, J.; Ampleman, G.; Thiboutot, S.; Sunahara, G. Ecotoxicity characterization of dinitrotoluenes and some of their reduced metabolites. Chemosphere 1999, 38, 2071–2079. [Google Scholar] [CrossRef]
- Ellis, H.V., III; Hagensen, J.H.; Hodgson, J.R.; Minor, J.L.; Hong, C.B. Mammalian Toxicity of Munition Compounds. Phase III. Effects of Lifetime Exposure. Part I. 2,4-Dinitrotoluene; Midwest Research Institute: Kansas, MO, USA, 1979. [Google Scholar]
- Gong, P.; Kuperman, R.G.; Sunahara, G.I. Genotoxicity of 2,4- and 2,6-dinitrotoluene as measured by the Tradescantia micronucleus (Trad-MCN) bioassay. Mutat. Res. Genet. Toxicol. Environ. Mutagenesis 2003, 538, 13–18. [Google Scholar] [CrossRef]
- Rickert, D.E.; Butterworth, B.E.; Popp, J.A.; Krahn, D.F. Dinitrotoluene: Acute Toxicity, Oncogenicity, Genotoxicity, and Metabolism. CRC Crit. Rev. Toxicol. 1984, 13, 217–234. [Google Scholar] [CrossRef]
- Habineza, A.; Zhai, J.; Mai, T.; Mmereki, D.; Ntakirutimana, T. Biodegradation of 2, 4, 6-Trinitrotoluene (TNT) in Contaminated Soil and Microbial Remediation Options for Treatment. Periodica Polytechnica. Chem. Eng. 2017, 61, 171. [Google Scholar]
- Johnson, G.R.; Jain, R.K.; Spain, J.C. Properties of the trihydroxytoluene oxygenase from Burkholderia cepacia R34: An extradiol dioxygenase from the 2,4-dinitrotoluene pathway. Arch. Microbiol. 1999, 173, 86–90. [Google Scholar] [CrossRef]
- Küce, P.; Coral, G.; Kantar, C. Biodegradation of 2,4-dinitrotoluene (DNT) by Arthrobacter sp. K1 isolated from a crude oil contaminated soil. Ann. Microbiol. 2014, 65, 467–476. [Google Scholar] [CrossRef]
- Kundu, D.; Hazra, C.; Chaudhari, A. Isolation, screening and assessment of microbial isolates for biodegradation of 2, 4-and 2, 6-dinitrotoluene. Int. J. Curr. Microbiol. App. Sci. 2015, 4, 564–574. [Google Scholar]
- Nishino, S.F.; Paoli, G.C.; Spain, J.C. Aerobic Degradation of Dinitrotoluenes and Pathway for Bacterial Degradation of 2,6-Dinitrotoluene. Appl. Environ. Microbiol. 2000, 66, 2139–2147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, J.; Kanjo, Y.; Suidan, M.T.; Venosa, A.D. Anaerobic biotransformation of 2,4-dinitrotoluene with ethanol as primary substrate: Mutual effect of the substrates on their biotransformation. Water Res. 1996, 30, 307–314. [Google Scholar] [CrossRef]
- Cheng, J.; Suidan, M.T.; Venosa, A.D. Anaerobic biotransformation of 2,4-dinitrotoluene with ethanol, methanol, acetic acid and hydrogen as primary substrates. Water Res. 1998, 32, 2921–2930. [Google Scholar] [CrossRef]
- Huang, J.; Ning, G.; Li, F.; Sheng, G.D. Biotransformation of 2,4-dinitrotoluene by obligate marine Shewanella marisflavi EP1 under anaerobic conditions. Bioresour. Technol. 2015, 180, 200–206. [Google Scholar] [CrossRef]
- Hughes, J.B.; Wang, C.Y.; Zhang, C. Anaerobic Biotransformation of 2,4-Dinitrotoluene and 2,6-Dinitrotoluene by Clostridium acetobutylicum: A Pathway through Dihydroxylamino Intermediates. Environ. Sci. Technol. 1999, 33, 1065–1070. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Ye, Z.-F.; Zhang, M.-H. Bioremediation of 2,4-dinitrotoluene (2,4-DNT) in immobilized micro-organism biological filter. J. Appl. Microbiol. 2011, 110, 1476–1484. [Google Scholar] [CrossRef] [PubMed]
- Spanggord, R.J.; Spain, J.C.; Nishino, S.F.; Mortelmans, K.E. Biodegradation of 2,4-dinitrotoluene by a Pseudomonas sp. Appl. Environ. Microbiol. 1991, 57, 3200–3205. [Google Scholar] [CrossRef] [Green Version]
- Aburto-Medina, A.; Taha, M.; Shahsavari, E.; Ball, A.S. Degradation of the dinitrotoluene isomers 2,4- and 2,6-DNT: Appraising the role of microorganisms. In Enhancing Cleanup of Environmental Pollutants: Biological Approaches; Anjum, N.A., Gill, S.S., Tuteja, N., Eds.; Springer International Publishing: Cham, Switzerland, 2017; Volume 1, pp. 5–20. [Google Scholar]
- Erkelens, M.; Adetutu, E.M.; Taha, M.; Tudararo-Aherobo, L.; Antiabong, J.; Provatas, A.; Ball, A. Sustainable remediation—The application of bioremediated soil for use in the degradation of TNT chips. J. Environ. Manag. 2012, 110, 69–76. [Google Scholar] [CrossRef]
- Almeida, C.M.R.; Reis, I.; Couto, N.; Bordalo, A.; Mucha, A.P. Potential of the microbial community present in an unimpacted beach sediment to remediate petroleum hydrocarbons. Environ. Sci. Pollut. Res. 2012, 20, 3176–3184. [Google Scholar] [CrossRef]
- Nikolopoulou, M.; Pasadakis, N.; Kalogerakis, N. Evaluation of autochthonous bioaugmentation and biostimulation during microcosm-simulated oil spills. Mar. Pollut. Bull. 2013, 72, 165–173. [Google Scholar] [CrossRef]
- Paca, J.; Halecky, M.; Bárta, J.; Bajpai, R. Aerobic biodegradation of 2,4-DNT and 2,6-DNT: Performance characteristics and biofilm composition changes in continuous packed-bed bioreactors. J. Hazard. Mater. 2009, 163, 848–854. [Google Scholar] [CrossRef] [PubMed]
- Poi, G.; Aburto-Medina, A.; Mok, P.C.; Ball, A.S.; Shahsavari, E. Bioremediation of Phenol-Contaminated Industrial Wastewater Using a Bacterial Consortium—from Laboratory to Field. Water Air Soil Pollut. 2017, 228, 89. [Google Scholar] [CrossRef]
- Poi, G.; Aburto-Medina, A.; Mok, P.C.; Ball, A.S.; Shahsavari, E. Large scale bioaugmentation of soil contaminated with petroleum hydrocarbons using a mixed microbial consortium. Ecol. Eng. 2017, 102, 64–71. [Google Scholar] [CrossRef]
- Poi, G.; Shahsavari, E.; Aburto-Medina, A.; Mok, P.C.; Ball, A.S. Large scale treatment of total petroleum-hydrocarbon contaminated groundwater using bioaugmentation. J. Environ. Manag. 2018, 214, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Holmgren, E.; Ek, S.; Colmsjö, A. Extraction of explosives from soil followed by gas chromatography–mass spectrometry analysis with negative chemical ionization. J. Chromatogr. A 2012, 1222, 109–115. [Google Scholar] [CrossRef]
- Shahsavari, E.; Adetutu, E.M.; Anderson, P.; Ball, A. Plant residues—A low cost, effective bioremediation treatment for petrogenic hydrocarbon-contaminated soil. Sci. Total Environ. 2013, 443, 766–774. [Google Scholar] [CrossRef]
- Schäfer, H.; Muyzer, G. Denaturing gradient gel electrophoresis in marine microbial ecology. Methods Microbiol. 2001, 425–468. [Google Scholar]
- Fierer, N.; Jackson, J.A.; Vilgalys, R.; Jackson, R.B. Assessment of Soil Microbial Community Structure by Use of Taxon-Specific Quantitative PCR Assays. Appl. Environ. Microbiol. 2005, 71, 4117–4120. [Google Scholar] [CrossRef] [Green Version]
- Cébron, A.; Norini, M.P.; Beguiristain, T.; Leyval, C. Real-time PCR quantification of PAH-ring hydroxylating dioxygenase (PAH-RHD α) genes from Gram positive and Gram negative bacteria in soil and sediment samples. J. Microbiol. Methods 2008, 73, 148–159. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Huson, D.H.; Beier, S.; Flade, I.; Górska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.-J.; Tappu, R. MEGAN Community Edition-Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef] [Green Version]
- Clarke, K.; Gorley, R.J.P. PRIMER v7: User Manual/Tutorial, 3rd ed.; Primer-E Ltd.: Devon, UK, 2015. [Google Scholar]
- Snellinx, Z.; Taghavi, S.; Vangronsveld, J.; Van Der Lelie, D. Microbial consortia that degrade 2,4-DNT by interspecies metabolism: Isolation and characterisation. Biodegradation 2003, 14, 19–29. [Google Scholar] [CrossRef]
- Johnson, G.R.; Spain, J.C. Evolution of catabolic pathways for synthetic compounds: Bacterial pathways for degradation of 2,4-dinitrotoluene and nitrobenzene. Appl. Microbiol. Biotechnol. 2003, 62, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Suen, W.C.; Haigler, B.E.; Spain, J.C. 2,4-Dinitrotoluene dioxygenase from Burkholderia sp. strain DNT: Similarity to naphthalene dioxygenase. J. Bacteriol. 1996, 178, 4926–4934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suen, W.C.; Spain, J.C. Cloning and characterization of Pseudomonas sp. strain DNT genes for 2,4-dinitrotoluene degradation. J. Bacteriol. 1993, 175, 1831–1837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
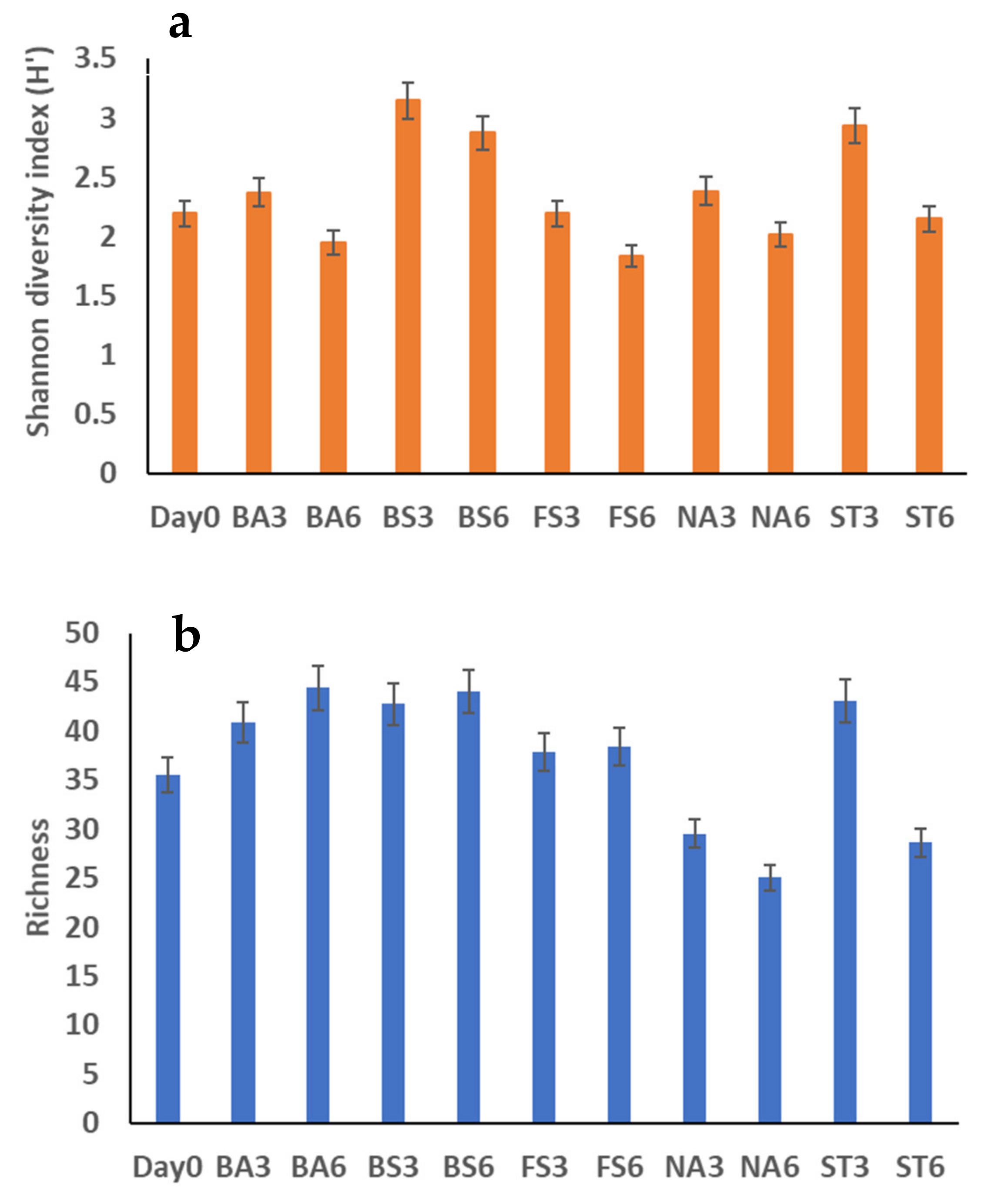
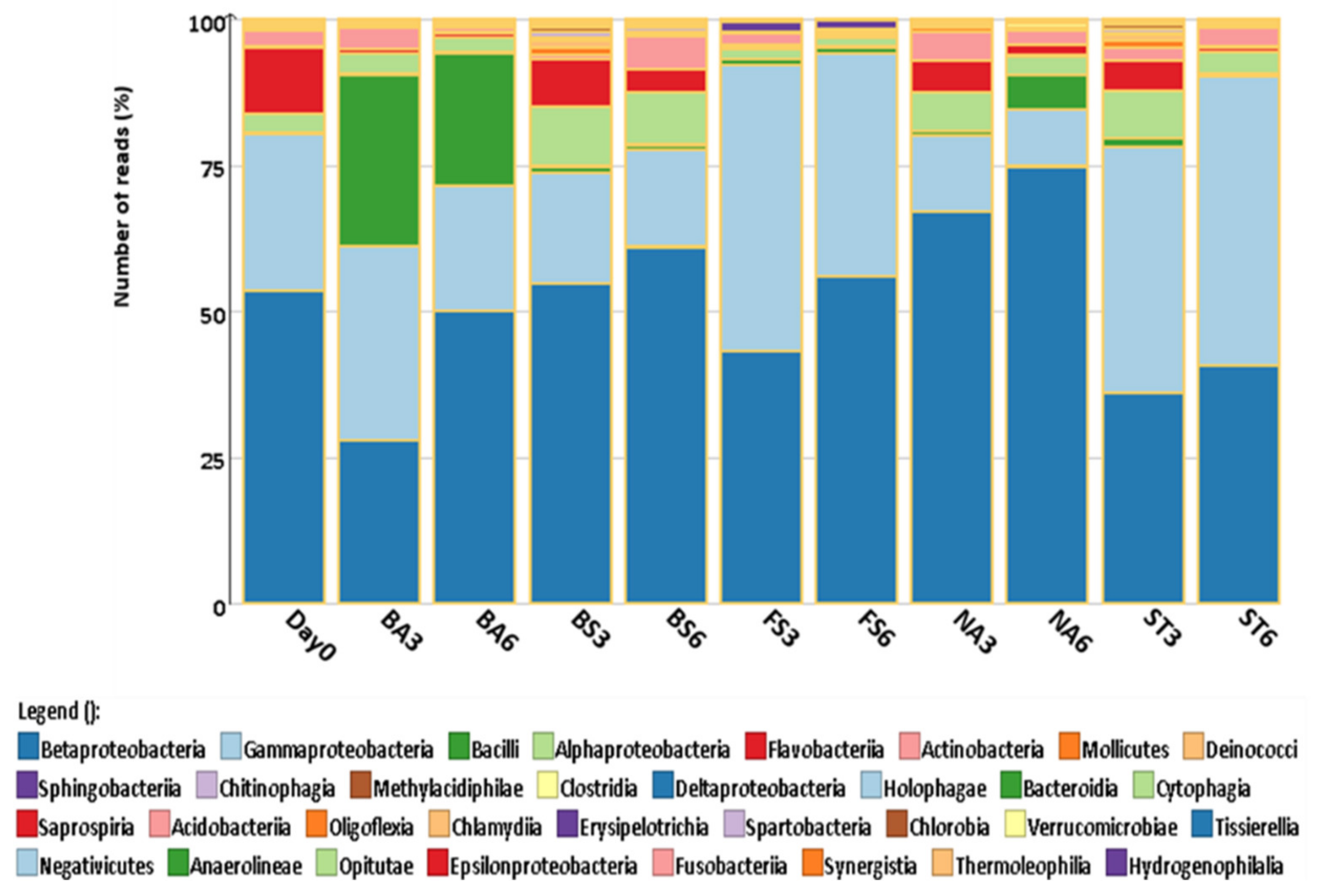
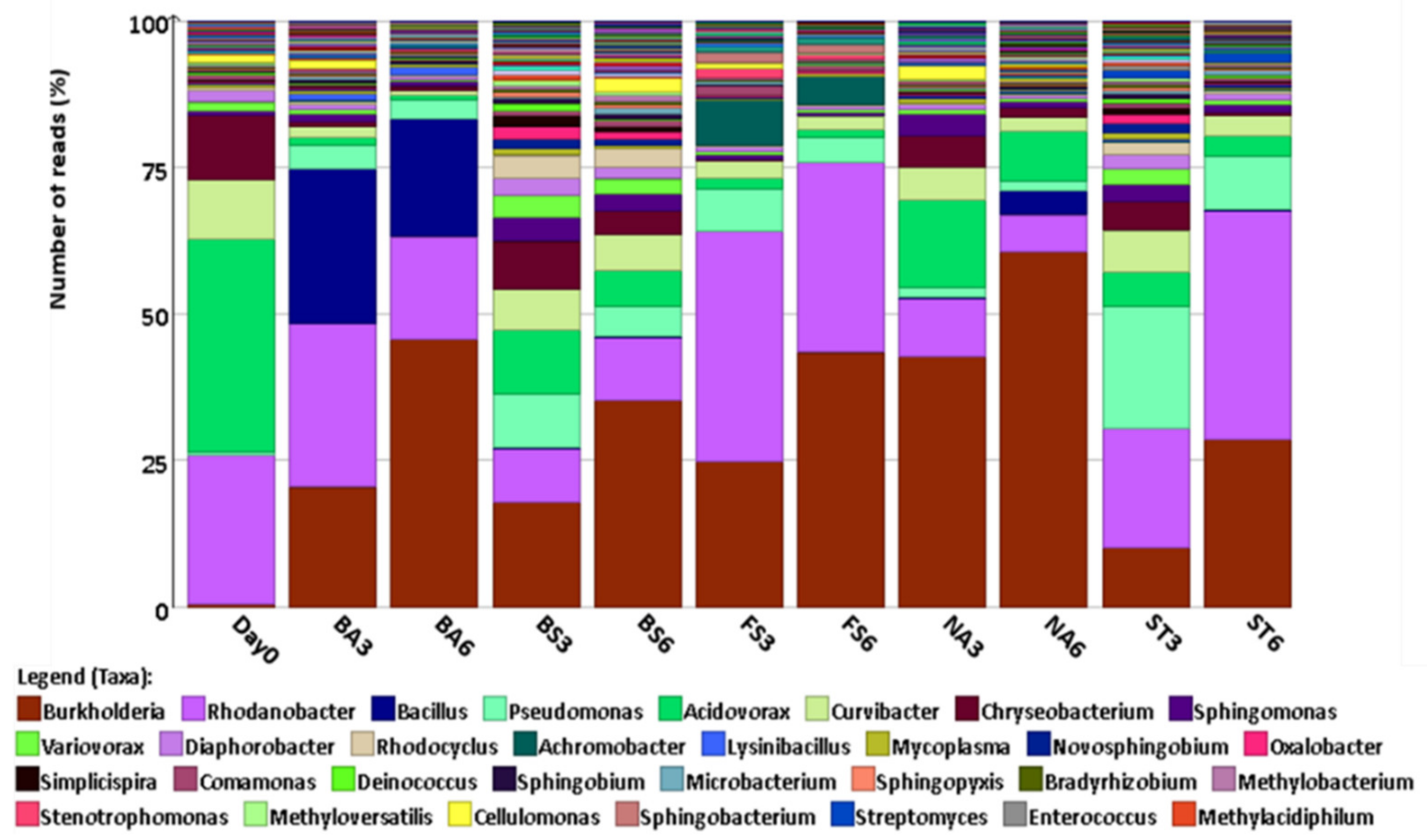
| Sludge Elements/Parameters | Units | Value |
|---|---|---|
| pH | 9.2 | |
| Organic C | % | 7.7 |
| Colour | Yellow | |
| Nitrite | mg/kg | 1900 |
| Nitrate | mg/kg | 7200 |
| Sulphate | mg/kg | 6700 |
| Moisture | % | 41 |
| 2,4,6 TNT | mg/kg | <50 |
| 2,4 DNT | g/L | 300 |
| 2,6 DNT | g/L | 0.056 |
| RDX | <50 |
| Treatments | Symbol | Nutrient Formulation | Hydrocarbonoclastic Bacteria |
|---|---|---|---|
| Natural attenuation (control) | NA | - | None |
| Bioaugmentation 1 | BA | - | Bacilli consortium |
| Bioaugmentation 2 | ST | - | Streptomyces sp. |
| Bioaugmentation 3 | FS | - | Fusarium solani |
| Biostimulation | BS | + | None |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aburto-Medina, A.; Shahsavari, E.; Taha, M.; Bates, A.; Van Ieperen, L.; Ball, A.S. The Impacts of Different Biological Treatments on the Transformation of Explosives Waste Contaminated Sludge. Molecules 2021, 26, 4814. https://doi.org/10.3390/molecules26164814
Aburto-Medina A, Shahsavari E, Taha M, Bates A, Van Ieperen L, Ball AS. The Impacts of Different Biological Treatments on the Transformation of Explosives Waste Contaminated Sludge. Molecules. 2021; 26(16):4814. https://doi.org/10.3390/molecules26164814
Chicago/Turabian StyleAburto-Medina, Arturo, Esmaeil Shahsavari, Mohamed Taha, Andrew Bates, Leon Van Ieperen, and Andrew S. Ball. 2021. "The Impacts of Different Biological Treatments on the Transformation of Explosives Waste Contaminated Sludge" Molecules 26, no. 16: 4814. https://doi.org/10.3390/molecules26164814
APA StyleAburto-Medina, A., Shahsavari, E., Taha, M., Bates, A., Van Ieperen, L., & Ball, A. S. (2021). The Impacts of Different Biological Treatments on the Transformation of Explosives Waste Contaminated Sludge. Molecules, 26(16), 4814. https://doi.org/10.3390/molecules26164814






