Abstract
α-Glucosidase inhibitors (AGIs) are used as medicines for the treatment of diabetes mellitus. The α-Glucosidase enzyme is present in the small intestine and is responsible for the breakdown of carbohydrates into sugars. The process results in an increase in blood sugar levels. AGIs slow down the digestion of carbohydrates that is helpful in controlling the sugar levels in the blood after meals. Among heterocyclic compounds, benzimidazole moiety is recognized as a potent bioactive scaffold for its wide range of biologically active derivatives. The aim of this study is to explore the α-glucosidase inhibition ability of benzimidazolium salts. In this study, two novel series of benzimidazolium salts, i.e., 1-benzyl-3-{2-(substituted) amino-2-oxoethyl}-1H-benzo[d]imidazol-3-ium bromide 9a–m and 1-benzyl-3-{2-substituted) amino-2-oxoethyl}-2-methyl-1H-benzo[d] imidazol-3-ium bromide 10a–m were screened for their in vitro α-glucosidase inhibitory potential. These compounds were synthesized through a multistep procedure and were characterized by 1H-NMR, 13C-NMR, and EI-MS techniques. Compound 10d was identified as the potent α-glucosidase inhibitor among the series with an IC50 value of 14 ± 0.013 μM, which is 4-fold higher than the standard drug, acarbose. In addition, compounds 10a, 10e, 10h, 10g, 10k, 10l, and 10m also exhibited pronounced potential for α-glucosidase inhibition with IC50 value ranging from 15 ± 0.037 to 32.27 ± 0.050 µM when compared with the reference drug acarbose (IC50 = 58.8 ± 0.12 μM). A molecular docking study was performed to rationalize the binding interactions of potent inhibitors with the active site of the α-glucosidase enzyme.
1. Introduction
The benzimidazole ring system has established its reputation amongst heterocyclic templates for its biologically active derivatives. Benzimidazole derivatives have displayed exceptional clinical and biological applications, such as anticancer [1], antifungal [2], antimalarial [3], antibacterial [4], anti-HIV [5], anti-inflammatory [6], antioxidant [7], antiprotozoal [8], and enzyme inhibitory activities [9]. In addition, it serves as an essential component of various commercially available biocides [10,11].
Benzimidazolium salts (derived from benzimidazole) have been extensively explored the for design and discovery of new drugs [12,13,14]. Particularly, in organometallic chemistry, benzimidazolium salts have emerged as an influential class of ligands due to strong σ-donor and weak π-accepting ability. Besides multiple synthetic and catalytic applications, benzimidazolium salts are also proven as effective antibacterial 1 [15], anticancer 2 [16], antimalarial, and anti-leishmanial agents 3 [17,18] (Figure 1).
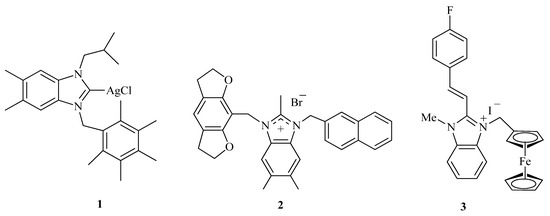
Figure 1.
The chemical structures of biologically active benzimidazolium salts.
Diabetes mellitus is a prevailing disorder, arisen due to a high level of blood glucose. The α-Glucosidase enzyme is responsible for the lysis of α-glucopyranoside bonds in disaccharides and oligosaccharides to form monosaccharides, ultimately regulating the blood glucose availability and postprandial hyperglycemia in type-2 diabetic patients [19]. The role of α-glucosidase inhibitors is to reduce hyperglycemia, thus considered as first-line sugar-lowering agents [20]. At present, a number of α-glucosidase inhibitors such as acarbose, voglibose, and miglitol have been discovered [21]. These α-glucosidase inhibitors are capable to interrupt the hydrolysis of carbohydrates and thus, reduce postprandial blood glucose levels [22,23]. However, continuous intake of these drugs may cause severe side effects such as diarrhea, body pain, and gastrointestinal disorders [24,25]. Subsequently, the development of new α-glucosidase inhibitors associated with minimum side effects is the need of the time.
Over the recent years, heterocyclic compounds have been extensively studied as α-glucosidase inhibitors [26,27,28]. More specifically, the benzimidazole ring system has emerged as an efficient template for new and potential anti-diabetic agents [29,30,31]. Benzimidazolium salts having α-glucosidase inhibitory potential have appeared in a few literature reports [32]. To the best of our knowledge, this research work is among the few successful efforts to explore benzimidazolium salts as α-glucosidase inhibitors. Keeping in view with evidence from the literature for the biological activities of benzimidazolium salts, we have synthesized a library of novel 1,3-disubstituted benzimidazolium salts and have screened them for α-glucosidase inhibition studies. Furthermore, the structure-activity relationship has been established with the help of molecular docking studies.
2. Results and Discussion
2.1. Chemistry
Twenty-six benzimidazolium salts 9(a–m) and 10(a–m) bearing amide functionality have been synthesized by following the methodology as illustrated in Scheme 1. It involves the reaction of o-phenylenediamine and respective carboxylic acid for 3 to 5 h to give 1H-benzimidazole and 2-methyl-1H-benzimidazole. These precursor benzimidazoles were treated with benzyl chloride in the presence of a base and using acetonitrile as a solvent to form corresponding N-benzylatedbenzimidazoles in 65–85% yields. Quaternization of the synthesized N-benzylbenzimidazole precursors with a range of substituted α-bromoacetamides resulted in the formation of targeted benzimidazolium salts 9(a–m) and 10(a–m). The chemical reaction was monitored by TLC using chloroform: methanol (4:1) mixture as a mobile phase. After completion of the reaction, products were filtered, washed, and purified using column chromatography.
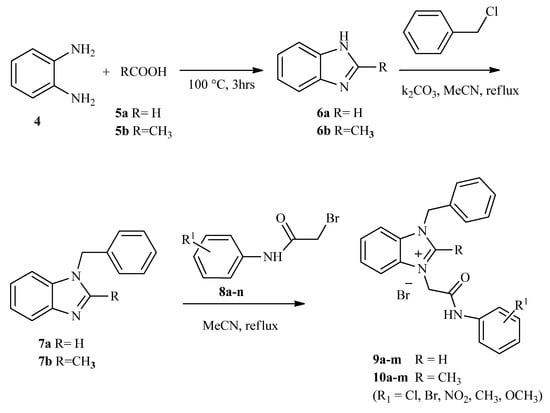
Scheme 1.
The lay-out for the synthesis of targeted benzimidazolium salts.
2.2. Characterization
The formation of benzimidazolium salts was confirmed by 1H-NMR, 13C-NMR, and mass spectrometry (Supplementary Materials File). A characteristic sharp singlet between 9.91 and 10.3 ppm is observed for NCHN protons (imidazole ring) of compounds 9a–m. For all benzimidazolium salts, protons of the methylene group attached to phenyl (–CH2–Ph) resonated at 5.51–5.86 ppm as a sharp singlet whereas the occurrence of another slightly downfield singlet in the range of 5.66–6.00 ppm is integrated for two protons of methylene linker of acetamide moiety. The acetanilide NH in benzimidazolium bromides appeared as an isolated singlet at 10.0–11.78 ppm. In 1H-NMR spectra of 9i, 9j, 9k, 10i, 10j, and 10k, a singlet peak in the range of 2.25–2.30 ppm confirmed the presence of methyl substituent. Similarly, the appearance of a singlet at 3.71–3.91ppm corresponded to the methoxy group of compounds 9l, 9m, 10l, and 10m. Aromatic protons of benzimidazolium salts 9(a–m) and 10(a–m) were detected in the range of 6.90–8.61ppm. In the case of 13C-NMR spectra, signals of methylene carbons and carbonyl carbons were observed in the range of 49–52 ppm and 162.6–166 ppm respectively. Furthermore, mass spectrometry data also confirmed the formation of title compounds. The NMR data of similar functionalities in the literature [33,34] was found supportive of our findings.
2.3. In Silico Studies (Molecular Docking)
The in silico techniques have established their reputation in the field of drug discovery. The study is helpful in identifying the potential biologically active compounds as the targets for synthesis, thus minimizing the hardships of laborious synthetic work. Considering the effectiveness of this tool, we executed the molecular docking studies of a library of benzimidazolium salts. Molecular docking was performed with the help of the MOE software to screen out the compounds having the best residual interactions with the receptor protein. Compounds were ranked on the basis of docking scores, rmsd values, and interactions with the selected pocket of the receptor enzyme. Results of molecular docking are presented in Figure 2 and Figure 3 and Table 1.
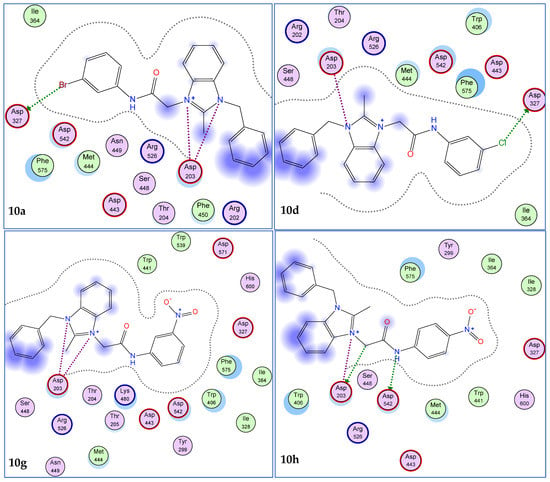
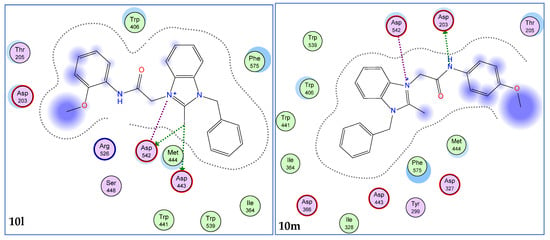
Figure 2.
2D interaction modes of active compounds (10a, 10d, 10g, 10h, 10l, and 10m). The blue color indicates ligand exposure and the green color dotted lines indicates the interactions of receptor enzyme with ligands.
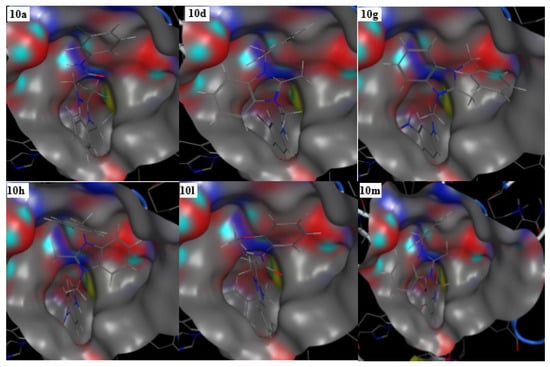
Figure 3.
3D interaction modes of active compounds (10a, 10d, 10g, 10h, 10l, and 10m). Color portion shows the pocket. Selected and red portions indicate the ligand exposure points.

Table 1.
In silico screening results of benzimidazolium salts 9a–m and 10a–m.
Among the screened derivatives, the benzimidazolium salts 10a–m showed good docking scores and significant interactions with the selected pocket of the receptor protein in comparison to the benzimidazolium salts 9a–m (Table 1, Figure 2). The binding energy values of most of these derivatives were found close to the standard drug, acarbose (−16.18 Kcal/mol). These ligands also occupied the selected pocket of the receptor enzyme effectively, as shown in 3D maps (Figure 3).
Most of the ligands interacted with Asp203, Asp327, and Asp542 residues among the reference/selected residues (Asp203, Asp542, Asp327, His600, Arg526) as revealed by the 2D maps and were considered responsible for the inhibition of the α-glucosidase enzyme (Figure 2). Nakamura and group had also observed the interactions of Asp203, Asp327, and Asp542 residues with the salacinol and its derivatives during in silico screening against α-glucosidase enzyme [35]. Similarly, the aglycone of curculigoside A, and derivatives, blocked the activity of α-glucosidase enzyme via bindings with Asp203, Asp327, and Asp542 residues [36]. In silico analysis of anthocyanidins and anthocyanins proved them good inhibitors of the α-glucosidase enzyme. These compounds also exhibited inhibitory action by interacting with the selected residues, Asp203, Asp327, and Asp542 [37].
Prominently, the derivatives 10a, 10d, 10g, 10h, 10l, and 10m exhibited best docking scores (−10.06 to −11.15 Kcal/mol) and rmsd values less than 2Å (Table 1). Regarding the interaction modes, the compounds 10a and 10d showed interactions with the common residues, Asp203 and Asp327 (Figure. 2). The docking scores for these ligands were found to be −10.87 and −10.85 Kcal/mol, respectively. Similarly, the ligands 10h and 10m also exhibited good docking scores (−10.94 and −13.43 Kcal/mol) and bindings with common residues, Asp203 and Asp542 (Figure 2). The ligands 10g and 10m showed interactions with Asp203 and Asp542, respectively. In order to check the validity of docking experiments, acarbose was also docked into the selected pocket of the enzyme and it showed binding interactions with selected residues, His600, Asp542, Arg526, and Asp327 (Figure 4). It is important to note that the presence of methyl, bromo, chloro, methylene, acetamide groups, and imidazole ring were found indispensable for the in silico inhibition of the α-glucosidase enzyme.
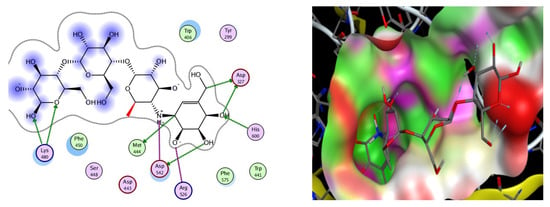
Figure 4.
2D (Left) and 3D (Right) interaction modes of the standard drug, acarbose.
2.4. In Vitro α-Glucosidase Inhibitory Activity
Benzimidazole derivatives have been reported as excellent inhibitors for α-glucosidase, dipeptidyl Peptidase IV, aldose reductase, and diacylglycerol acyltransferase [38,39]. In this study, two series of benzimidazolium salts that are quaternary derivatives of benzimidazole have been synthesized in search of potent α-glucosidase inhibitors. The compounds 9(a–m) and 10(a–m) were screened for in vitro α-glucosidase inhibitory activity according to reported protocol by using acrobose as a reference drug. The synthesized benzimidazolium salts exhibited a varying degree of α-glucosidase inhibitory potential with IC50 values ranging from 14 to 279.28 µM presented in Table 2. The structure–activity relationship established the role of various substituents on phenyl rings and their positions with biological activities. First of all, compounds 9a–m were found to be inactive or weakly active against the enzyme, while compounds having C2-substituted benzimidazolium salts 10a–m demonstrated excellent α-glucosidase inhibitory profile (Table 2). These results strengthen the fact that C2-substituted benzimidazole variants are more susceptible to α-glucosidase inhibition. Recently, Mentes and his coworkers synthesized 4-(5-fluoro-2-substituted-1H-benzimidazol-6-yl)morpholine derivatives and indicated that C2 substitution of benzimidazole has a significant effect on α-glucosidase inhibitory activities [40]. The strong impact of C2 substituted benzimidazole on α-glucosidase and urease inhibitory activities is observed by other researchers too [41]. Predominately, compound 10d (1-benzyl-3-(2-((3-bromophenyl)amino)-2-oxoethyl)-2-methyl-1H-benzo[d]imidazole-3-ium bromide) exhibited the highest α-glucosidase inhibitory potential with IC50 value 14 + 0.013 µM, which is 4-fold higher than reference drug. Compounds 10m, 10h, 10g, 10a, 10l, 10e, and 10k exhibited better activity than acarbose with an IC50 value of 16.16 ± 0.026, 17 ± 0.067, 19 ± 0.086, 22 ± 0.040, 25 ± 0.011, 30 ± 0.049 and 32.27 ± 0.050 µM, respectively.

Table 2.
In vitro α-glucosidase inhibitory (IC50) activities of synthesized benzimidazolium salts.
Compounds 10c, 10d, 10e having ortho, meta, and para-substituted bromo group on phenyl rings, showed higher activity than acarbose (standard drug). The shifting of bromo from position 2 to position 3 led to a dramatic increase in activity. Among all of the halo substituted derivatives, 10d was the most active with the highest α-glucosidase inhibitory potential. Replacing of the bromo substituent with the chloro substituent caused a slight decline in the α-glucosidase inhibitory potential for 10a and 10b, as the compound appeared as a moderate to poor inhibitor. It is reflected that nature and change in position of the halo-group have a notable effect on the α-glucosidase inhibitory activity.
Among nitro-substituted compounds, 10g and 10h, having a substitution at the meta and para positions, demonstrated promising α-glucosidase inhibitory results due to a strong electron-withdrawing effect. However, 10f, having nitro-substitution at the ortho position, showed considerable decline in activity.
The bioactivity results of 10l and 10m benzimidazolium salts with methoxy substitution at the ortho and para positions, also showed their effectiveness for α-glucosidase inhibition. Higher activities of these compounds containing an electron-donating methoxy group are due to resonance stabilization of the ortho and para positions. Contrary to these results, methyl-substituted derivatives 10i–k showed varying degrees of α-glucosidase inhibitory potential. Compounds 10i and 10j containing methyl groups at the ortho and meta positions appeared as poor inhibitors, whereas, 10k having methoxy at the para position was found to be moderately active (Figure 5).
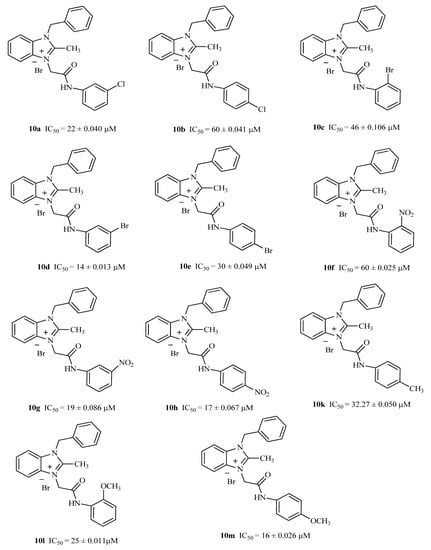
Figure 5.
The chemical structures of potent α-glucosidase inhibitors.
It is clear from direct comparison of halo and methoxy/methyl-substituted compounds that there is a slight decrease in the α-glucosidase inhibitory profile for methoxy/methyl-substituted benzimidazolium salts. The decreasing order of α-glucosidase inhibitory potential of the active compounds can be listed as 10d > 10m > 10h > 10g > 10a > 10l > 10e > 10k > 10c > acarbose. Compounds 9m in the first series, and 10i in the second series, having electron-donating substitution (methoxy and methyl), exhibited the lowest α-glucosidase inhibitory results in their respective series. It can be concluded from the above IC50 results that the position of both electron-donating and withdrawing groups on the phenyl ring have a vital role in determining the efficacy of α-glucosidase inhibition.
3. Materials and Methods
3.1. General
All chemicals and solvents used in this synthesis were purchased from Sigma-Aldrich (Munich, Germany) and Daejung Chemicals (Siheung-si, South Korea). Solvents such methanol, acetonitrile, and chloroform used in this synthesis were purified and dried by a solvents purification system. Melting points were recorded by using the Stuart SMP 40 melting point apparatus. Spectroscopic studies (1H and 13C-NMR) were carried out by using the Bruker 600 MHz spectrometer in DMSO-d6 solvent.
3.2. General Procedure for the Preparation of Benzimidazole Precursors 6(a–b)
o-Phenylenediamine 4 (5.4 g, 0.05 mol) and formic acid or acetic acid (0.075 mol) were mixed in a round bottom flask. The mixture was heated during stirring at 100 °C for 3 h. Afterwards, the contents of the flask were cooled to room temperature and an aqueous solution of sodium hydroxide (10% w/v) was added dropwise to attain a pH~8. The ppts. of the corresponding benzimidazole were formed, which were filtered, washed with ice-cold water, and dried. The products were purified by recrystallization from methanol.
- 1H-Benzimidazole (6a): Yield: 85%; m.p. 172 °C (Literature m.p. 171–173 °C [42]).
- 2-Methyl-1H-Benzimidazole (6b): Yield: 81%; m.p. 176 °C (Literature m.p. 176–178 °C [42]).
3.3. General Procedure for Preparation of N-Benzylatedbenzimidazoles 7(a–b)
A mixture of benzimidazole 6a or 6b (0.05 mol), CTAB (0.05 mol), acetonitrile (30 mL), and K2CO3 (0.1 mol) was stirred at room temperature. Benzyl chloride (0.075 mol) was added drop-wise to the reaction mixture over a period of 5 min. Afterwards, after stirring, and the reaction mixture was subjected to reflux for 8 h. The progress of the reaction was monitored by using TLC in a mixture of chloroform and methanol in a 4:1 ratio as a mobile phase. After the completion of the reaction, the contents of the flask were cooled to room temperature and poured into crushed ice. The crude product thus obtained was purified by column chromatography.
- 1-Benzyl-1H-benzimidazole (7a): White crystalline solid; Yield: 85%; m.p. 118 °C (Literature m.p. 115–118 °C [43,44]); 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 5.60 (s, 2H, CH2-Ph), 7.10–7.22 (m, 3H, Ar-H), 7.30–7.35 (m, 5H, Ar-H), 7.75 (d, J = 7.8 Hz, 1H, Ar-H), 7.86 (s, 1H, CH=N). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 48.8, 111.0, 121.5 (2C), 122.5, 124, 128 (2C), 128.5, 130, 134, 135.7, 144.1, 144.8 (C = N).
- 1-Benzyl-2-methyl-1H-benzimidazole (7b): Waxy solid; Yield: 65%; 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 2.45 (s, 3H, CH3), 5.30 (s, 2H, CH2-Ph), 7.10 (t, 3H, Ar-H), 7.35–7.42 (m, 5H, Ar-H), 7.70 (d, J = 7.2 Hz, 1H, Ar-H). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 14.3, 46.4, 110.0, 121.5, 122.5 (2C), 124, 126 (2C), 127.5, 129, 133, 135.7, 141.2, 143.0 (C = N).
3.4. General Procedure for the Preparation of Benzimidazolium Salts 9(a–m)
1-Benzyl-1H-benzimidazole 7a (0.22 g, 1 mmol) was dissolved in acetonitrile (30 mL) in a round bottom flask. The solution of 2-bromo-N-arylacetamides 8(a–m) (1 mmol) in acetonitrile (10 mL) was added dropwise in the round bottom flask and the reaction mixture was refluxed for 12–14 h. The completion of the reaction was observed by thin-layer chromatography. Finally, the contents were cooled in an ice bath to obtain the crude solid salts which were purified by column chromatography.
- 1-Benzyl-3-[2-{(3-chlorophenyl)amino}-2-oxoethyl]-1H-benzo[d]imidazol-3-ium bromide (9a): White crystalline solid, Yield: 80%; m.p. 205–207 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 5.63 (s, 2H, CH2-Ph), 5.89 (s, 2H, CH2-CO), 7.19 (d, 1H, J = 7.98 Hz, Ar-H), 7.41 (t, 2H, J = 8.07 Hz, Ar-H), 7.45 (t, 2H, J = 7.38 Hz, Ar-H), 7.51 (d, 1H, J = 8.28 Hz, Ar-H), 7.54 (d, 2H, J = 7.32 Hz, Ar-H), 7.67–7.72 (m, 2H, Ar-H), 7.81 (s, 1H, Ar-H), 8.01 (d, 1H, J = 7.68 Hz, Ar-H), 8.07 (d, 1H, J = 7.86 Hz, Ar-H), 9.97 (s, 1H, NCHN), 11.02 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 49.2, 50.0, 113.8, 114.0, 117.7, 118.8, 123.8, 126.8, 127.0, 128.2 (2C), 128.4, 129.1(3C), 130.4, 130.7, 133.2, 133.7, 139.5, 143.5, 164.0 (C=O). MS (ESI+): m/z calcd for C22H19ClN3O+ [M − Br]+ 376; found 377 [M − Br + H]+.
- 1-Benzyl-3-[2-{(4-chlorophenyl)amino}-2-oxoethyl]-1H-benzo[d]imidazol-3-ium bromide (9b): White crystalline solid; Yield: 78%; m.p. 197–198 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 5.62 (s, 2H, CH2-Ph), 5.89 (s, 2H, CH2-CO), 7.39- 7.42 (m, 2H, Ar-H), 7.43–7.46 (m, 3H, Ar-H), 7.54 (d, 2H, J = 7.2 Hz, Ar-H), 7.65–7.67 (m, 3H, Ar-H), 7.68–7.71 (m, 1H, Ar-H), 8.01 (d, 1H, J = 6.72 Hz, Ar-H), 8.07 (d, 1H, J = 6.96 Hz, Ar-H), 9.98 (s, 1H, NCHN), 10.96 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 49.2, 50.0, 113.8, 114.0, 121.0 (2C), 126.8, 127.0, 127.6, 128.2 (2C), 128.8 (2C), 129.0 (2C), 130.4, 132.0, 132.0, 133.7, 137.1, 143.6, 163.6 (C=O): MS (ESI+): m/z calcd for C22H19ClN3O+ [M − Br]+ 376; found 376 [M − Br]+.
- 1-Benzyl-3-[2-{(2-bromophenyl)amino}-2-oxoethyl]-1H-benzo[d]imidazol-3-ium bromide (9c): White crystalline solid; Yield: 78%; m.p. 192–194 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 5.68 (s, 2H, CH2-Ph), 5.87 (s, 2H, CH2-CO), 7.19 (t, 1H, J = 7.2 Hz, Ar-H), 7.39 (t, 2H, J = 7.2 Hz, Ar-H), 7.42 (q, 2H, J = 12.6 Hz, J = 14.4 Hz, Ar-H), 7.2 (t, 2H, J = 8.75 Hz, Ar-H) 7.60 (d, 1H, J = 7.26 Hz, Ar-H), 7.66–7.72 (m, 3H, Ar-H), 8.00 (d, 1H, J = 8.00 Hz, Ar-H), 8.06 (d, 1H, J = 8.06 Hz, Ar-H), 10.04 (s, 1H, NCHN), 10.50 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 49.0, 50.0, 113.9 (2C), 126.8, 127.0, 127.5, 128.2 (5C), 128.8 (4C), 129.0, 131.0, 132.9, 133.8, 143.7, 164.1 (C=O). MS (ESI+): m/z calcd for C22H19BrN3O+ [M − Br]+ 420.0706; found 420.0714 [M − Br]+.
- 1-Benzyl-3-[2-{(3-bromophenyl)amino}-2-oxoethyl]-1H-benzo[d]imidazol-3-ium bromide (9d): White crystalline solid; Yield: 85%; m.p. 176–178 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 5.62 (s, 2H, CH2-Ph), 5.88 (s, 2H, CH2-CO), 7.31–7.36 (m, 2H, Ar-H), 7.42 (t, 1H, J = 8.46 Hz, Ar-H), 7.41–7.46 (m, 2H, Ar-H), 7.53–7.55 (m, 3H, Ar-H), 7.67–7.72 (m, 2H, Ar-H), 7.95 (t, 1H, J = 3.6 Hz, Ar-H), 8.01 (d, 1H, J = 6.93 Hz, Ar-H), 8.07 (d, 1H, J = 6.93 Hz, Ar-H), 9.96 (s, 1H, NCHN), 10.99 (s, 1H, NH).13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 49.2, 50.0, 113.8, 114.0, 118.1, 121.6, 121.7, 126.7, 126.8, 127.0, 128.8, 128.2 (2C), 129.1 (2C), 130.4, 131.0, 132.0, 133.7, 139.7, 143.6, 163.9 (C=O). MS (ESI+): m/z calcd for C22H19BrN3O+ [M − Br]+ 420; found 420 [M − Br]+.
- 1-Benzyl-3-[2-{(4-bromophenyl)amino)}-2-oxoethyl]-1H-benzo[d]imidazol-3-ium bromide (9e): White crystalline solid; Yield: 75%; m.p. 150–152 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 5.60 (s, 2H, CH2-Ph), 5.88 (s, 2H, CH2-CO), 7.40 (t, 1H, J = 7.26 Hz, Ar-H), 7.45 (t, 2H, J = 7.35 Hz, Ar-H), 7.52–7.55 (m, 3H, Ar-H), 7.55 (t, 1H, J = 3.4 Hz, Ar-H), 7.6 (d, 2H, J =8.8 Hz, Ar-H), 7.76–7.71 (m, 2H, Ar-H), 8.0 (d, 1H, J =8.3 Hz, Ar-H), 8.06 (d, 1H, J =7.9 Hz, Ar-H), 9.95 (s, 1H, NCHN), 10.92 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 49.2, 50.0, 113.8, 114.0, 115.7, 121.3, 126.8, 127.0, 128.2 (3C), 128.8, 129.1 (3C), 130.4, 131.8 (2C), 132.0, 133.7, 143.6, 144.20, 163.6 (C=O). MS (ESI+): m/z calcd for C22H19BrN3O+ [M − Br]+ 420; found 420 [M − Br]+.
- 1-Benzyl-3-[2-{(2-nitrophenyl)amino}-2-oxoethyl]-1H-benzo[d]imidazol-3-ium bromide (9f): White crystalline solid; Yield: 74%; m.p. 142–143 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 5.69 (s, 2H, CH2-Ph), 5.8 (s, 2H, CH2-CO), 7.38–7.41 (m, 1H, Ar-H), 7.44 (q, 3H, J = 7.4 Hz, Ar-H), 7.52 (d, 2H, J = 7.2 Hz, Ar-H), 7.67–7.73 (m, 3H, Ar-H), 7.77 (t, 1H, J = 7.8 Hz, Ar-H), 8.0 (t, 3H, J = 7.35 Hz, Ar-H), 9.99 (s, 1H, NCHN), 11.2 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 49.0, 50.0, 113.8, 114.0, 121.0, 125.0 (2C), 126.8, 127.0, 128.2 (3C), 128.8, 129.0 (3C), 130.0, 132.0, 134.1, 142.8, 143.5, 164.2 (C=O). MS (ESI+): m/z calcd for C22H19N4O3+ [M − Br]+ 387; found 388 [M − Br + H]+.
- 1-Benzyl-3-[2-{(3-nitrophenyl)amino}-2-oxoethyl]-1H-benzo[d]imidazol-3-ium bromide (9g): White crystalline solid; Yield: 75%; m.p. 162–163 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 5.63 (s, 2H, CH2-Ph), 5.86 (s, 2H, CH2-CO), 7.38–7.41 (m, 1H, Ar-H), 7.44 (t, 2H, J = 14.7 Hz, Ar-H), 7.52 (d, 2H, J = 7.86 Hz, Ar- H), 7.66–7.71 (m, 3H, Ar-H), 7.90 (d, 1H, J = 7.68 Hz, Ar-H), 7.97–8.00 (m, 2H, Ar-H), 8.05 (d, 1H, J = 7.68 Hz, Ar-H), 8.61 (s, 1H, Ar-H), 9.93 (s, 1H, NCHN), 11.27 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 49.2, 50.0, 113.4, 113.8, 114.0, 118.6, 125.3, 126.8, 127.0, 128.2 (2C), 128.87, 129.1 (2C), 130.4 (2C), 130.5, 132.0, 139.2, 143.5, 148.0, 164.3 (C=O). MS (ESI+): m/z calcd for C22H19N4O3+ [M − Br]+ 387; found 387 [M − Br]+.
- 1-Benzyl-3-[2{(4-nitrophenyl)amino}-2-oxoethyl]-1H-benzo[d]imidazol-3-ium bromide (9h): White crystalline solid; Yield: 70%; m.p. 152–153 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 5.64 (s, 2H, CH2-Ph), 5.87 (s, 2H, CH2-CO), 7.41 (t, 1H, J = 7.2 Hz, Ar-H), 7.45 (t, 2H, J = 7.35 Hz, Ar-H), 7.52 (d, 2H, J = 7.26 Hz, Ar-H), 7.68–7.71 (m, 2H, Ar-H), 7.85 (d, 2H, J = 9.1 Hz, Ar-H), 8.01 (t, 1H, J = 9.2 Hz, Ar-H), 8.07 (d, 1H, J = 7.8 Hz, Ar-H), 8.28 (d, 2H, J = 9.18 Hz, Ar-H), 9.91 (s, 1H, NCHN), 11.34 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 49.3, 50.0, 113.8, 114.0, 119.15 (2C), 125.1 (2C), 126.8, 127, 128.2 (2C), 128.9, 129.1 (2C), 130.4, 132.0, 133.7, 142.7, 143.6, 144.2, 164.6 (C=O). MS (ESI+): m/z calcd for C22H19N4O3+ [M − Br]+ 387; found 387 [M − Br]+.
- 1-Benzyl-3-{2-oxo-2-(o-tolylamino)ethyl}-1H-benzo[d]imidazol-3-ium bromide (9i): White crystalline solid; Yield: 90%; m.p. 162–163 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 2.3 (s, 3H, CH3), 5.67 (s, 2H, CH2-Ph), 5.9 (s, 2H, CH2-CO), 7.13 (t, 1H, J = 7.14 Hz, Ar-H), 7.19 (t, 1H, J = 7.11 Hz, Ar-H), 7.2 (d, 1H, J = 7.32 Hz, Ar-H), 7.39 (t, 1H, J = 7.2 Hz, Ar-H), 7.44 (t, 3H, J = 7.26 Hz, Ar-H), 7.53 (d, 2H, J = 7.26 Hz, Ar-H), 7.66–7.73 (m, 2H, Ar-H), 8.0 (d, 1H, J =8.2 Hz, Ar-H), 8.07 (d, 1H, J = 8.28 Hz, Ar-H), 10.0 (s, 1H, NCHN), 10.14 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 17.9, 49.0, 50.0, 113.7, 113.9, 124.9, 126.1, 128.2 (4C), 128.8 (4C), 129.0 (3C), 130.5 (2C), 131.8, 143.6, 163.6 (C=O). MS (ESI+): m/z calcd for C23H22N3O+ [M − Br]+ 356; found 357 [M − Br + H]+.
- 1-Benzyl-3-{2-oxo-2-(m-tolylamino)ethyl}-1H-benzo[d]imidazol-3-ium bromide (9j): White crystalline solid; Yield: 88%; m.p. 132–134 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 2.3 (s, 3H, CH3), 5.60 (s, 2H, CH2-Ph), 5.88 (s, 2H, CH2-CO), 6.92 (d, 1H, J = 7.6 Hz, Ar-H), 7.22 (t, 1H, J = 7.8 Hz, Ar-H), 7.39–7.40 (m, 2H, Ar-H), 7.42–7.46 (m, 3H, Ar-H), 7.53 (d, 2H, J = 7.3 Hz, Ar-H), 7.65–7.71 (m, 2H, Ar-H), 7.99 (d, 1H, J = 7.98 Hz, Ar-H), 8.04 (d, 1H, J = 8.04 Hz, Ar-H), 9.99 (s, 1H, NCHN), 10.74 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 21.1, 49.2, 50.0, 113.8, 116.5, 119.8, 124.7, 126.8, 127.0, 128.2 (3C), 129.0 (3C), 130.4, 132.0, 133.7, 137.1, 138.0, 138.2, 143.5, 163.3 (C=O). MS (ESI+): m/z calcd for C23H22N3O+ [M − Br]+ 356; found 356 [M − Br]+.
- 1-Benzyl-3-{2-oxo-2-(p-tolylamino)ethyl}-1H-benzo[d]imidazol-3-ium bromide (9k): White crystalline solid; Yield: 90%; m.p. 148–149 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 2.26 (3H, s, CH3), 5.59 (s, 2H, CH2-Ph), 5.88 (s, 2H, CH2-CO), 7.16 (d, 2H, J = 8.34 Hz, Ar-H), 7.39–7.40 (m, 1H, Ar-H), 7.45 (t, 2H, J = 6 Hz, Ar-H), 7.52 (dd, 4H, J = 7.8, 16.1 Hz, Ar-H), 7.66–7.71 (m, 2H, Ar-H), 8.03 (d, 1H, J = 7.32 Hz, Ar-H), 8.06 (d, 1H, J = 7.38 Hz, Ar-H), 9.99 (s, 1H, NCHN), 10.72 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 20.4, 49.1, 49.9, 113.8 (2C), 119.3 (3C); 126.9, 128.2 (4C); 128.8 (5C), 129.0, 129.3, 133.7, 143.5, 163.1 (C=O). MS (ESI+): m/z calcd for C23H22N3O+ [M − Br]+ 356; found 357 [M − Br + H]+.
- 1-Benzyl-3-[2-{(2-methoxyphenyl)amino}-2-oxoethyl]-1H-benzo[d]imidazol-3-ium bromide (9l): White crystalline solid; Yield: 92%; m.p. 188–189 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 3.88 (s, 3H, OCH3), 5.67 (s, 2H, CH2-Ph), 5.87 (s, 2H, CH2-CO), 6.90 (t, 1H, J = 7.20 Hz, Ar-H), 7.09 (d, 1H, J = 8.04 Hz, Ar-H), 7.13 (t, 1H, J = 7.68 Hz, Ar-H), 7.38 (t, 1H, J = 7.2 Hz, Ar-H), 7.48 (t, 2H, J = 7.2 Hz, Ar-H), 7.52 (d, 2H, J = 7.38 Hz, Ar-H), 7.64–7.70 (m, 2H, Ar-H), 7.88 (d, 1H, J = 7.90 Hz, Ar-H), 8.00 (dd, 2H, J = 8.2, 15.99 Hz, Ar-H), 10.03 (s, 1H, NCHN), 10.07 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 49.2, 49.9, 55.8, 111.4, 113.7, 113.8, 120.3, 121.9, 125.4, 126.2, 126.8, 127.0, 128.2 (2C), 128.8, 129.0 (2C), 130.4, 131.8, 133.7, 143.5, 149.8, 163.6 (C=O). HRMS (ESI+): m/z calcd for C23H22N3O+ [M − Br]+ 372.1707; found 372.1714 [M − Br]+.
- 1-Benzyl-3-[2-{(4-methoxyphenyl)amino}-2-oxoethyl]-1H-benzo[d]imidazol-3-ium bromide (9m): White crystalline solid; Yield: 90%; m.p. 171–173 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 3.71 (s, 3H, OCH3), 5.58 (s, 2H, CH2-Ph), 5.87 (s, 2H, CH2-CO), 6.90–6.93 (m, 2H, Ar-H), 7.37–7.39 (m, 1H, Ar-H), 7.43 (t, 2H, J = 14.7 Hz, Ar-H), 7.53 (dd, 4H, J = 3.4, 8.3 Hz, Ar-H), 7.64–7.69 (m, 2H, Ar-H), 7.99 (d, 1H, J = 7.92 Hz, Ar-H), 8.08 (d, 1H, J = 7.98 Hz, Ar-H), 10.00 (s, 1H, NCHN), 10.69 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 49.1, 50.0, 55.2, 113.8, 114.0, 121.0 (2C), 126.8, 127, 128.2 (3C), 128.8, 129.0 (3C), 130.4, 131.2, 132.0, 133.7, 143.5, 155.70, 162.8 (C=O). MS (ESI+): m/z calcd for C23H22N3O+ [M − Br]+ 372; found 373 [M − Br + H]+.
3.5. General Procedure for the Preparation of Benzimidazolium Salts 10(a–m)
1-Benzyl-2-methyl-1H-benzimidazole 7b (0.23 g, 1 mmol) was dissolved in acetonitrile (30 mL) in a round bottom flask. The solution of 2-bromo-N-arylacetamides 8(a–m) (1 mmol) in acetonitrile (10 mL) was added dropwise in the round bottom flask and the reaction mixture was refluxed for 12–14 h. The completion of the reaction was observed by thin-layer chromatography. Finally, the contents were cooled in an ice bath to obtain the crude solid salt which was purified by column chromatography.
- 1-Benzyl-3-[2-{(3-chlorophenyl)amino}-2-oxoethyl]-2-methyl-1H-benzo[d]imidazol-3-ium bromide (10a): White crystalline solid; Yield: 74%; m.p. 210–213 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 2.94 (s, 3H, Benzimidazole-CH3), 5.52 (s, 2H, CH2-Ph), 5.83 (s, 2H, CH2-CO), 7.17 (d, 1H, J = 9.52 Hz, Ar-H), 7.31 (d, 2H, J = 9.2 Hz, Ar-H), 7.35–7.41 (m, 4H, Ar-H), 7.44 (t, 1H, J = 9.6 Hz, Ar-H), 7.61–7.67 (m, 2H, Ar-H), 7.74 (s, 1H, Ar-H), 7.94 (d, 1H, J = 9.4 Hz, Ar-H), 7.89 (d, 1H, J = 8.84 Hz, Ar-H), 10.94 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 11.3, 48.2, 48.7, 113.3, 113.7, 118.5, 119.6, 124.5, 127.1, 127.2, 127.6 (2C), 129.0, 129.6 (2C), 131.1, 131.2, 131.8, 133.7, 134.3, 139.8, 153.6, 164.0 (C=O). MS (ESI+): m/z calcd for C23H21ClN3O+ [M − Br]+ 390; found 391 [M − Br + H]+.
- 1-Benzyl-3-[2-{(4-chlorophenyl)amino}-2-oxoethyl]-2-methyl-1H-benzo[d]imidazol-3-ium bromide (10b): White crystalline solid; Yield: 74%; m.p. 178–180 °C. 1H-NMR: (DMSO-d6, 600 MHz) δ (ppm): 2.95 (s, 3H, Benzimidazole-CH3), 5.52 (s, 2H, CH2-Ph), 5.66 (s, 2H, CH2-CO), 7.33(d, 2H, J =17.4 Hz, Ar-H), 7.36 (t, 2H, J = 4.9 Hz, Ar-H), 7.39–7.42 (m, 3H, Ar-H), 7.60 (d, 2H, J = 8.94 Hz, Ar-H), 7.65 (q, 2H, J1 = J2 = 18 Hz, Ar-H), 7.95 (d, 1H, J =7.85 Hz, Ar-H), 7.99 (d, 1H, J =7.74 Hz, Ar-H), 10.88 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 10.8, 47.7, 48.2, 112.8, 113.2, 121.1 (2C), 126.5, 127.1 (2C), 127.8, 128.5, 128.8 (2C), 129.1 (3C), 130.5, 131.3, 133.8, 136.8, 153.2, 163.3 (C=O). HRMS (ESI+): m/z calcd for C23H21ClN3O+ [M − Br]+ 390.1368; found 390.1374 [M − Br]+.
- 1-Benzyl-3-[2-{(2-bromophenyl)amino)}-2-oxoethyl]-2-methyl-1H-benzo[d]imidazol-3-ium bromide (10c): White crystalline solid; Yield: 76%; m.p. 252 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 3.0 (s, 3H, Benzimidazole-CH3), 5.64 (s, 2H, CH2-Ph), 6.0 (s, 2H, CH2-CO), 7.20 (t, 1H, J = 7.5 Hz, Ar-H), 7.32–7.42 (m, 6H, Ar-H), 7.63 (q, 2H, J1 = J2 = 7.5 Hz, Ar-H), 7.7 (q, 2H, J1 = J 2= 8.2 Hz, Ar-H), 7.98 (d, 1H, J = 8.2 Hz, Ar-H), 8.03 (d, 1H, J = 8.2 Hz, Ar-H), 10.3 (s, 1H, NH), 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 11.0, 47.7, 48.2, 113.0, 113.2, 113.6, 118.7, 125.5, 126.5, 126.6, 127.1 (2C), 128.5, 129.1 (2C), 130.5, 130.6, 131.4, 133.8, 139.1, 148.0, 153.2, 164.0 (C=O). MS (ESI+): m/z calcd for C23H21BrN3O+ [M − Br]+ 434; found 434 [M − Br]+.
- 1-Benzyl-3-[2-{(3-bromophenyl)amino}-2-oxoethyl]-2-methyl-1H-benzo[d]imidazol-3-ium bromide (10d): White crystalline solid; Yield: 80%; m.p. 242 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 3.0 (s, 3H, Benzimidazole-CH3), 5.60 (s, 2H, CH2-Ph), 5.90 (s, 2H, CH2-CO), 7.31–7.35 (m, 4H, Ar-H), 7.3 (t, 1H, J = 7.3 Hz, Ar-H), 7.4 (t, 2H, J = 7.3 Hz, Ar-H), 7.5 (d, 1H, J = 7.3 Hz, Ar-H), 7.6–7.7 (m, 2H, Ar-H), 8.0 (s, 1H, Ar-H), 8.1 (d, 1H, J = 8.05 Hz, Ar-H), 8.04 (d, 1H, J = 7.75 Hz, Ar-H), 11.0 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 11.0, 47.8, 48.2, 113.0, 113.3, 118.2, 121.6, 121.8, 126.5, 126.6, 126.7, 127.1 (2C), 128.4, 129.1 (2C), 130.6, 131.0, 131.4, 134.0, 139.6, 153.2, 163.6 (C=O). MS (ESI+): m/z calcd for C23H21BrN3O+ [M − Br]+ 434; found 434 [M − Br + H]+.
- 1-Benzyl-3-[2-{(4-bromophenyl)amino}-2-oxoethyl]-2-methyl-1H-benzo[d]imidazol-3-ium bromide (10e): White crystalline solid; Yield: 82%; m.p. 212 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 2.99 (s, 3H, Benzimidazole-CH3), 5.66 (s, 2H, CH2-Ph), 5.91 (s, 2H, CH2-CO), 7.33–7.43 (m, 5H, Ar-H), 7.52–7.54 (m, 2H, Ar-H), 7.68–7.61 (m, 4H, Ar-H), 7.98 (d, 1H, J = 7.4 Hz, Ar-H), 8.11 (d, 1H, J = 7.3 Hz, Ar-H), 11.54 (s, 1H, NH). MS (ESI+): m/z calcd for C23H21BrN3O+ [M − Br]+ 434; found 435 [M − Br + H]+.
- 1-Benzyl-2-methyl-3-[2-{(2-nitrophenyl)amino}-2-oxoethyl]-1H-benzo[d]imidazol-3-ium bromide 10f): White crystalline solid; Yield: 72%; m.p. 160–161 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): (500 MHz, DMSO-d6) δ (ppm): 2.98 (s, 3H, Benzimidazole-CH3), 5.73 (s, 2H, CH2-Ph), 5.91 (s, 2H, CH2-CO), 7.31–7.34 (m, 2H, Ar-H), 7.37–7.43 (m, 3H, Ar-H), 7.62–7.66 (m, 4H, Ar-H), 7.74–7.77 (m, 1H, Ar-H), 7.97 (t, J = 6.7 Hz, 2H, Ar-H), 8.09 (d, J = 8.0 Hz, 1H, Ar-H), 11.78 (s, 1H, NH). MS (ESI+): m/z calcd for C23H21N4O3+ [M − Br]+ 401; found 402 [M − Br + H]+.
- 1-Benzyl-2-methyl-3-[2-{(3-nitrophenyl)amino}-2-oxoethyl]-1H-benzo[d]imidazol-3-ium bromide (10g): White crystalline solid; Yield: 73%; m.p. 180–182 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 2.98 (s, 3H, Benzimidazole-CH3), 5.61 (s, 2H, CH2-Ph), 5.90 (s, 2H, CH2-CO), 7.34 (d, 2H, J = 7.26 Hz, Ar-H), 7.37 (t, 1H, J = 7.2 Hz, Ar-H), 7.42 (t, 2H, J = 7.5 Hz, Ar-H), 7.63 (dd, 1H, J = 1.1, 7.32 Hz, Ar-H), 7.65–7.68 (m, 3H, Ar-H), 7.91 (d, 1H, J = 8.16 Hz, Ar-H), 7.95–8.0 (m, 2H, Ar-H), 8.04 (d, 1H, J = 7.78 Hz, Ar-H), 11.29 (s, 1H, NH). 13C-NMR: (DMSO-d6, 150 MHz) δ (ppm): 11.0, 47.8, 48.2, 113.0, 113.2, 113.6, 118.7, 125.5, 126.5, 126.6, 127.1 (2C), 128.5, 129.1 (2C), 130.5, 130.6, 131.4, 133.8, 139.1, 148.0, 153.2, 164.0 (C=O). MS (ESI+): m/z calcd for C23H21N4O3+ [M − Br]+ 401; found 401 [M − Br]+.
- 1-Benzyl-2-methyl-3-[2-{(4-nitrophenyl)amino)}-2-oxoethyl]-1H-benzo[d]imidazol-3-ium bromide (10h): White crystalline solid; Yield: 76%; m.p. 222–224 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 2.96 (s, 3H, Benzimidazole-CH3), 5.60 (s, 2H, CH2-Ph), 5.87 (s, 2H, CH2-CO), 7.32 (d, 2H, J = 7.4 Hz, Ar-H), 7.37 (t, 1H, J = 7.2 Hz, Ar-H), 7.41 (t, 2H, J = 7.2 Hz, Ar-H), 7.62–7.67 (m, 2H, Ar-H), 7.84 (d, 2H, J = 8.8 Hz, Ar-H), 7.96 (d, 1H, J = 7.9 Hz, Ar-H), 8.02 (d, 1H, J = 7.53 Hz, Ar-H), 8.25 (d, 2H, J = 9.2 Hz, Ar-H), 10.88 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm):10.8, 47.9, 48.2, 113.0, 113.2, 119.4 (2C), 125.0 (2C), 126.6, 126.7, 127.1 (2C), 128.5, 129.1 (2C), 130.5, 131.3, 133.8, 143.0, 144.0, 153.2, 164.2 (C=O). MS (ESI+): m/z calcd for C23H21N4O3+ [M − Br]+ 401; found 402 [M − Br + H]+.
- 1-Benzyl-2-methyl-3-{2-oxo-2-(o-tolylamino)ethyl}-1H-benzo[d]imidazol-3-ium bromide (10i): White crystalline solid; Yield: 84%; m.p. 168–170 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 2.25 (s, 3H, CH3), 2.51 (s, 3H, Benzimidazole-CH3), 5.59 (s, 2H, CH2-Ph), 5.87 (s, 2H, CH2-CO), 7.11–7.19 (m, 2H, Ar-H), 7.24 (d, 1H, J = 8.60 Hz, Ar-H), 7.30–7.41 (m, 6H, Ar-H), 7.63.7–68 (m, 2H, Ar-H), 7.99 (q, 2H, J1 = J2 = 9.6 Hz, Ar-H),10.08 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 11.3, 18.2, 48.0, 48.7, 113.2, 113.8, 125.5, 126.5, 126.6, 127.0, 127.1, 127.6 (2C), 129.0, 129.6 (2C), 131.0, 131.1, 132.0, 132.4, 134.4, 135.6, 153.6, 164.0 (C=O). MS (ESI+): HR m/z calcd for C24H24N3O+ [M − Br]+ 370.1914; found 370.1917 [M − Br]+.
- 1-Benzyl-2-methyl-3-{2-oxo-2-(m-tolylamino)ethyl}-1H-benzo[d]imidazol-3-ium bromide (10j): White crystalline solid; Yield: 82%; m.p. 192–194 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 2.26 (s, 3H, CH3), 2.95 (s, 3H, Benzimidazole-CH3), 5.51 (s, 2H, CH2-Ph), 5.87 (s, 2H, CH2-CO), 6.94 (d, 1H, J = 7.5 Hz, Ar-H), 7.22 (t, 1H, J = 7.8 Hz, Ar-H), 7.32 (d, 2H, J = 7.2 Hz, Ar-H), 7.36 (q, 2H, J1 = J2 = 7.2 Hz, Ar-H), 7.41 (t, 3H, J = 7.39 Hz, Ar-H), 7.62–7.67 (m, 2H, Ar-H), 7.95 (d, 1H, J = 8.01 Hz, Ar-H), 7.98 (d, 1H, J =8.01 Hz, Ar-H), 10.68 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 10.8, 21.0, 47.7, 48.1, 112.8, 113.2, 116.6, 120.0, 125.0, 126.5, 126.6, 127.1(2C), 128.5, 128.8, 129.1 (2C), 130.6, 131.4, 133.8, 137.8, 138.3, 153.1, 163.0 (C=O). MS (ESI+): m/z calcd for C24H24N3O+ [M − Br]+ 370; found 370 [M − Br]+.
- 1-Benzyl-2-methyl-3-{2-oxo-2-(p-tolylamino)ethyl}-1H-benzo[d]imidazol-3-ium bromide (10k): White crystalline solid; Yield: 82%; m.p. 220–221 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 2.26 (s, 3H, CH3), 2.96 (s, 3H, Benzimidazole-CH3), 5.51 (s, 2H, CH2-Ph), 5.90 (s, 2H, CH2-CO), 7.15 (d, 2H, J = 8.3 Hz, Ar-H), 7.33 (d, 2H, J = 7.17 Hz, Ar-H), 7.36 (t, 1H, J = 7.4 Hz, Ar-H), 7.41 (t, 2H, J = 7.3 Hz, Ar-H), 7.48 (d, 2H, J = 8.41 Hz, Ar-H), 7.62–7.67 (m, 2H, Ar-H), 7.98 (d, 1H, J = 7.5 Hz, Ar-H), 8.03 (d, 1H, J = 7.5 Hz, Ar-H), 10.64 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 10.8, 20.4, 47.8, 48.1, 112.8, 113.0, 113.3, 119.4 (2C), 127.1(2C), 128.4, 129.0 (3C), 129.3 (2C), 130.6, 131.5, 133.1, 134.0, 135.6, 153.1, 163.0 (C=O). MS (ESI+): m/z calcd for C24H24N3O+ [M − Br]+ 370; found 370 [M − Br]+.
- 1-Benzyl-3-[2-{(2-methoxyphenyl)amino}-2-oxoethyl]-2-methyl-1H-benzo[d]imidazol-3-ium bromide (10l): White crystalline solid; Yield: 84%; m.p. 244–246 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 2.97 (s, 3H, Benzimidazole-CH3), 3.91 (s, 3H, OCH3), 5.67 (s, 2H, CH2-Ph), 5.91 (s, 2H, CH2-CO), 6.90–7.14 (m, 3H, Ar-H), 7.35–7.42 (m, 5H, Ar-H), 7.64–7.67 (m, 2H, Ar-H), 7.92 (d, 1H, J = 9.05 Hz, Ar-H), 8.0 (dd, 2H, J = 9.27, 9.61 Hz, Ar-H) 10.0 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 11.4, 48.4, 48.7, 56.3, 112.0, 113.3, 113.8, 120.8, 122.3, 125.8, 127.0, 127.1, 127.6 (3C), 129.0, 131.1 (3C), 132.0, 134.5, 150.2, 153.8, 164.0 (C=O). MS (ESI+): m/z calcd for C24H24N3O2+ [M − Br]+ 386; found 386 [M − Br]+.
- 1-Benzyl-3-[2-{(4-methoxyphenyl)amino}-2-oxoethyl]-2-methyl-1H-benzo[d]imidazole-3-ium bromide (10m): White crystalline solid; Yield: 82%; m.p. 235–238 °C. 1H-NMR (DMSO-d6, 600 MHz) δ (ppm): 2.96 (s, 3H, Benzimidazole-CH3), 3.72 (s, 3H, OCH3), 5.51 (s, 2H, CH2-Ph), 5.88 (s, 2H, CH2-CO), 6.92 (d, 2H, J = 9.06 Hz, Ar-H), 7.33 (d, 2H, J = 7.2 Hz, Ar-H), 7.37 (t,1H, J = 7.2 Hz, Ar-H), 7.41 (t, 2H, J = 7.3 Hz, Ar-H), 7.50 (d, 2H, J = 9.03 Hz, Ar-H), 7.62–7.67 (m, 2H, Ar-H), 7.96 (d, 1H, J = 7.93 Hz, Ar-H), 8.01 (d, 1H, J = 7.93 Hz, Ar-H) 10.63 (s, 1H, NH). 13C-NMR (DMSO-d6, 150 MHz) δ (ppm): 10.8, 47.7, 48.2, 55.2, 112.8, 113.2, 114.0 (2C), 121.1 (2C), 126.5, 126.6, 127.1 (2C), 128.5, 129.1 (2C), 130.6, 131.0, 131.4, 134.0, 153.2, 155.8, 162.6 (C=O). MS (ESI+): m/z calcd for C24H24N3O2+ [M − Br]+ 386; found 387 [M − Br + H]+.
3.6. In Silico α-Glucosidase Inhibition Studies
The in silico analysis of all the benzimidazolium salts along with the reference acarbose was carried out by using the MOE Ver.2014.0901 (Molecular Operating Environment, Chemical Computing Group ULC., Montreal, QC, Canada).
3.6.1. Ligand Preparation
The software (ChemDraw Ultra 12 + Serial, Perkin Elmer, MA, USA) was utilized to draw the structures of all these compounds. In order to display these files in MOE, structures of these compounds were saved in the MDL file (“.sdf”). The 2D structure of reference acarbose in the SDF file was retrieved from NCBI Pubchem. Afterwards, 3D protonation and energy minimizations of compounds as well as acarbose were executed under default parameters in MOE.
3.6.2. Protein Preparation
The 3D structure of Saccharomyces cerevisiae α-glucosidase (PDB ID: 2QMJ) was downloaded from the Protein Data Bank (http://www.rcsb.org/pdb, accessed on 10 June 2021). The binding pocket with the protein residues (Asp203, Asp542, Asp327, His600, Arg526) was selected and the interactions of the ligand (acarbose) were checked. Afterwards ligand, solvent molecules, and unknown atoms from the receptor protein were removed. The active sites from the 3D structure of the protein were determined using MOE-Site Finder Module. The active sites of the ligand consist of Asp203, Tyr299, Asp327, Ile328, Asp366, Trp406, Trp441, Asp443, Met444, Arg526, Trp539, Gly541, Asp542, Asp571, Phe575, Arg598, and His600 residues. These sites were occupied by dummy atoms and the protein was 3D-protonated and energy-minimized using default parameters in the MOE software.
3.6.3. Molecular Docking
Ten conformations of all the ligands were docked into the selected site of the enzyme. The MOE-Dock module was used to access binding modes of ligands with the α-glucosidase enzyme. The parameters, placement: triangle matcher, rescoring: London dG, refinement: forcefield, retain: 10 were used. The root mean square deviation rmsd values and docking scores of their top-ranked conformations were used to study the binding modes of ligands. Results of the molecular docking are depicted in Table 1.
3.7. In Vitro α-Glucosidase Inhibition
The literature protocol was used to assess α-glucosidase inhibition of synthesized compounds [45]. The solution of the α-glucosidase enzyme (obtained from Saccharomyces cerevisiae (Sigma-Aldrich, Munich, Germany)) was prepared in phosphate buffer (pH 6.8, 100 mM). The synthesized compounds were dissolved in dimethyl sulfoxide (DMSO). A mixture of 40 μL of the α-glucosidase enzyme (0.5 U/mL), 120 μL of phosphate buffer, and 12.5 μL of each sample was prepared in 96-well microtiter plates and incubated for 5 min at 37 °C. Afterwards, 40 μL of 5 mM p-nitrophenyl-α-d-glucopyranoside (Sigma-Aldrich, Munich, Germany) was poured into each well and further incubated for an additional half-hour. The absorbance at 405 nm and 37 °C was noted using a microplate reader. Acarbose (Sigma-Aldrich, Munich, Germany) and DMSO were used as reference inhibitors and negative control, respectively. The tests were conducted in triplicate and percent inhibitions were calculated using the following formula:
where ‘A’ stands for absorbance.
% Inhibition = [(A control − A sample)/A control] × 100
The IC50 values of the synthesized compounds and acarbose were also determined.
4. Conclusions
In conclusion, two novel series of benzimidazolium salts bearing amide functionality, 9(a–m) and 10(a–m), were synthesized and screened for α-glucosidase inhibitory potential. The C2-substituted benzimidazolium derivatives 10(a–m) emerged as excellent α-glucosidase inhibitors. The bromo-substituted compound 10d exhibited the best activity among the series, with an IC50 value of 14 + 0.013 µM which is 4-fold higher than the standard drug. Moreover, molecular docking studies of the most active compounds were carried out to find the interaction mode and structure-activity relationship. This study, being pioneering in its nature, will possibly be a landmark for further research for the discovery of safer drugs for the treatment of diabetic mellitus.
Supplementary Materials
The following are available online: 1H-NMR, 13C-NMR, and MS spectra of the synthesized compounds having potent enzyme inhibition activities.
Author Contributions
Conceptualization, M.A. and U.A.A.; methodology, I.A.K., M.A. and U.A.A.; synthesis, I.A.K. and M.A.; characterization, I.A.K., M.A., S.S. and M.E.A.Z.; enzyme inhibition studies, I.A.K., M.A. and U.A.A.; writing—original draft preparation, I.A.K., M.A., S.S., U.A.A. and M.E.A.Z.; writing—review and editing, I.A.K., M.A., S.S., U.A.A. and M.E.A.Z.; supervision, M.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Higher Education Commission of Pakistan, grant number NRPU-5614.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the Supplementary Materials.
Acknowledgments
The authors (I.A.K., M.A. and U.A.A.) greatly acknowledge the Government College University, Faisalabad for support.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are available from the authors.
References
- Elsayed, S.A.; Harrypersad, S.; Sahyon, H.A.; El-Magd, M.A.; Walsby, C.J. Ruthenium(II)/(III) DMSO-Based Complexes of 2-Aminophenyl Benzimidazole with in Vitro and in Vivo Anticancer Activity. Molecules 2020, 25, 4284. [Google Scholar] [CrossRef]
- Kankate, R.S.; Gide, P.S.; Belsare, D.P. Design, Synthesis and Antifungal Evaluation of Novel Benzimidazole Tertiary Amine Type of Fluconazole Analogues. Arab. J. Chem. 2019, 12, 2224–2235. [Google Scholar] [CrossRef] [Green Version]
- Okombo, J.; Brunschwig, C.; Singh, K.; Dziwornu, G.A.; Barnard, L.; Njoroge, M.; Wittlin, S.; Chibale, K. Antimalarial Pyrido[1,2-a]benzimidazole Derivatives with Mannich Base Side Chains: Synthesis, Pharmacological Evaluation, and Reactive Metabolite Trapping Studies. ACS Infect. Dis. 2019, 5, 372–384. [Google Scholar] [CrossRef] [PubMed]
- Alasmary, F.A.S.; Snelling, A.M.; Zain, M.E.; Alafeefy, A.M.; Awaad, A.S.; Karodia, N. Synthesis and Evaluation of Selected Benzimidazole Derivatives as Potential Antimicrobial Agents. Molecules 2015, 20, 15206–15223. [Google Scholar] [CrossRef]
- Srivastava, R.; Gupta, S.K.; Naaz, F.; Gupta, P.S.S.; Yadav, M.; Singh, V.K.; Singh, A.; Rana, M.K.; Gupta, S.K.; Schol, D.; et al. Alkylated benzimidazoles: Design, Synthesis, Docking, DFT Analysis, ADMET Property, Molecular Dynamics and Activity Against HIV and YFV. Comput. Biol. Chem. 2020, 89, 107400. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Hang, N.; Zhang, C.; Chen, Y.; Li, S.; Sun, Y.; Li, Z.; Meng, X. Synthesis and Biological Evaluation of Novel Benzimidazole Derivatives and Analogs Targeting the NLRP3 Inflammasome. Molecules 2017, 22, 213. [Google Scholar] [CrossRef] [Green Version]
- Taha, M.; Mosaddik, A.; Rahim, F.; Ali, S.; Ibrahim, M.; Almandil, N.B. Synthesis, Antiglycation and Antioxidant Potentials of Benzimidazole Derivatives. J. King Saud Univ. Sci. 2020, 32, 191–194. [Google Scholar] [CrossRef]
- Flores-Carrillo, P.; Velazquez-Lopez, K.M.; Aguayo-Ortiz, R.; Hernández-Campos, A.; Trejo-Soto, P.J.; Yépez-Mulia, L.; Castillo, R. Synthesis, Antiprotozoal Activity, and Chemoinformatic Analysis of 2-(Methylthio)-1 H-Benzimidazole- 5-Carboxamide Derivatives: Identification of New Selective Giardicidal and Trichomonicidal Compounds. Eur. J. Med. Chem. 2017, 137, 211–220. [Google Scholar] [CrossRef]
- Can, O.D.; Osmaniye, D.; Ozkay, U.D.; Saglik, B.N.; Levent, S.; Igin, S.; Baysal, M.; Ozkay, Y.; Kaplancikli, Z.A. MAO Enzymes Inhibitory Activity of New Eenzimidazole Derivatives Including Hydrazone and Propargyl Side Chains. Eur. J. Med. Chem. 2017, 131, 92–106. [Google Scholar] [CrossRef]
- Zikos, C.; Evangelou, A.; Karachaliou, C.E.; Gourma, G.; Blouchos, P.; Moschopoulou, G.; Yialouris, C.; Griffiths, J.; Johnson, G.; Petrou, P.; et al. Commercially Available Chemicals as Immunizing Haptens for the Development of a Polyclonal Antibody Recognizing Carbendazim and Other Benzimidazole-type Fungicides. Chemosphere 2015, 119, 16–20. [Google Scholar] [CrossRef]
- Kamil, A.; Akhter, S.; Ahmed, M.; Rizwani, G.H.; Hassan, S.; Naeem, S.; Jahan, S.; Khursheed, R.; Zahid, H. Antimalarial and Insecticidal Activities of Newly Synthesized Derivatives of Benzimidazole. Pak. J. Pharm. Sci. 2015, 28, 2179–2184. [Google Scholar]
- Karatas, M.O.; Uslu, H.; Alici, B.; Gokce, B.; Gencer, N.; Arslan, O.; Arslan, B.N.; Ozdemir, N. Functionalized Imidazolium and Benzimidazolium Salts as Paraoxonase 1 Inhibitors: Synthesis, Characterization and Molecular Docking Studies. Bioorg. Med. Chem. Lett. 2016, 24, 1392–1401. [Google Scholar] [CrossRef]
- Rodriguez-Sanz, A.; Sanchez-Alonso, P.; Bellon, T.; Alajarin, R.; Martínez-Cabeza, V.; Selgas, R.; Vaquero, J.J.; Alvarez-Buill, J. Synthesis and Biological Evaluation of Pyridazino[10,60:1,2] Pyrido [3,4-b]Indolinium and Pyridazino[1,6-a] Benzimidazolium Salts as Anti-Inflammatory Agents. Eur. J. Med. Chem. 2015, 93, 83–92. [Google Scholar] [CrossRef]
- Habib, A.; Nazari, M.; Iqbal, M.A.; Bhatti, H.N.; Ahmed, M.B.A.; Majid, A.M.S.A. Unsymmetrically Substituted Benzimidazolium Based Silver(I)-N-Heterocyclic Carbene Complexes: Synthesis, Characterization and in Vitro Anticancer Study Against Human Breast Cancer and Colon Cancer. J. Saudi Chem. Soc. 2019, 23, 795–808. [Google Scholar] [CrossRef]
- Sharhan, O.; Heidelberg, T.; Hashim, N.M.; Al-Madhagi, W.M.; Ali, H.M. Benzimidazolium-Acridine-Based Silver N-HeteroCyclic Carbene Complexes as Potential Anti-Bacterial and Anti-Cancer Drug. Inorganica Chim. Acta 2020, 504, 119462. [Google Scholar] [CrossRef]
- Shelton, K.L.; DeBord, M.A.; Wagers, P.O.; Southerland, M.R.; Williams, T.M.; Robishaw, N.K.; Shriver, L.P.; Tessier, C.A.; Panzner, M.J.; Youngs, W.J. Synthesis, Anti-Proliferative Activity, SAR Study, and Preliminary in Vivo Toxicity Study of SubStituted N,N′-Bis(Arylmethyl) Benzimidazolium Salts Against a Panel of Non-Small Cell Lung Cancer Cell Lines. Bioorg. Med Chem. 2017, 25, 421–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howarth, J.; Hanlon, K. Novel N-Ferrocenylmethyl, N’-Methyl-2-Substituted Benzimidazolium Iodide Salts with in Vitro Activity Against the P. falciparum Malarial Parasite Strain NF54. Tetrahedron Lett. 2001, 42, 751–754. [Google Scholar] [CrossRef]
- Joshua Howarth, J.; Keith Hanlon, K. N-Ferrocenylmethyl, N’-Methyl-2-Substituted Benzimidazolium Iodide Salts with in Vitro Activity Against the Leishmania infantum Parasite Strain L1. Bioorg. Med. Chem. Lett. 2003, 13, 2017–2020. [Google Scholar] [CrossRef]
- Dhameja, M.; Gupta, P. Synthetic Heterocyclic Candidates as Promising α-Glucosidase Inhibitors: An Overview. Eur. J. Med. Chem. 2019, 176, 343–373. [Google Scholar] [CrossRef] [PubMed]
- Vieira, R.; Souto, S.B.; Sanchez-Lopez, E.; Machado, A.L.; Severino, P.; Jose, S.; Santini, A.; Fortuna, A.; Garcia, M.L.; Silva, A.M.; et al. Sugar-Lowering Drugs for Type 2 Diabetes Mellitus and Metabolic Syndrome-Review of Classical and New Compounds: Part-I. Pharmaceuticals 2019, 12, 152. [Google Scholar] [CrossRef] [Green Version]
- Rahim, F.; Zaman, K.; Taha, M.; Ullah, H.; Ghufran, M.; Wadood, A.; Rehman, W.; Uddin, N.; Shah, S.A.A.; Sajid, M.; et al. Synthesis, in Vitro Alpha-Glucosidase Inhibitory Potential of Benzimidazole Bearing Bis-Schiff Bases and Their Molecular Docking Study. Bioorg. Chem. 2020, 94, 103394. [Google Scholar] [CrossRef] [PubMed]
- Campbell, L.K.; Baker, D.E.; Campbell, R.K. Miglitol: Assessment of its Role in the Treatment of Patients with Diabetes Mellitus. Ann. Pharmacother. 2000, 34, 1291–1301. [Google Scholar] [CrossRef]
- Krentz, A.J.; Bailey, C.J. Oral Antidiabetic Agents: Current Role in Type 2 Diabetes Mellitus. Drugs 2005, 65, 385–411. [Google Scholar] [CrossRef] [PubMed]
- Chiasson, J.L.; Josse, R.G.; Gomis, R.; Hanefeld, M.; Karasik, A.; Laakso, M. Acarbose Treatment and the Risk of Cardiovascular Disease and Hypertension in Patients with Impaired Glucose Tolerance. JAMA 2003, 290, 486–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawamori, R.; Tajima, N.; Iwamoto, Y.; Kashiwagi, A.; Shimamoto, K.; Kaku, K. Voglibose for Prevention of Type 2 Diabetes Mellitus: A Randomised, Double-Blind Trial in Japanese Individuals with Impaired Glucose Tolerance. Lancet 2009, 373, 1607–1614. [Google Scholar] [CrossRef]
- Saddique, F.A.; Aslam, S.; Ahmad, M.; Ashfaq, U.A.; Muddassar, M.; Sultan, S.; Taj, S.; Hussain, M.; Sung Lee, D.; Zaki, M.E.A. Synthesis and α-Glucosidase Inhibition Activity of 2-[3-(Benzoyl/4-bromobenzoyl)-4-hydroxy-1,1-dioxido-2H-benzo[e][1,2]thiazin-2-yl]-N-arylacetamides: An in Silico and Biochemical Approach. Molecules 2021, 26, 3043. [Google Scholar] [CrossRef]
- Al-Majid, A.M.; Islam, M.S.; Atef, S.; El-Senduny, F.F.; Badria, F.A.; Elshaier, Y.A.M.M.; Ali, M.; Barakat, A.; Motiur Rahman, A.F.M. Synthesis of Pyridine-Dicarboxamide-Cyclohexanone Derivatives: Anticancer and α-Glucosidase Inhibitory Activities and in Silico Study. Molecules 2019, 24, 1332. [Google Scholar] [CrossRef] [Green Version]
- Gong, Z.; Xie, Z.; Qiu, J.; Wang, G. Synthesis, Biological Evaluation and Molecular Docking Study of 2-Substituted-4,6-DiarylpyRimidines as α-Glucosidase Inhibitors. Molecules 2017, 22, 1865. [Google Scholar] [CrossRef] [Green Version]
- Özil, M.; Emirik, M.; Beldüz, A.; Ülker, S. Molecular Docking Studies and Synthesis of Novel Bisbenzimidazole Derivatives as Inhibitors of α-Glucosidase. Bioorg. Med. Chem. 2017, 24, 5103. [Google Scholar] [CrossRef]
- Özil, M.; Emirik, M.; Etlik, S.Y.; Ülkerb, S.; Kahveci, B. A Simple and Efficient Synthesis of Novel Inhibitors of Alpha-Glucosidase Based on Benzimidazole Skeleton and Molecular Docking Studies. Bioorg. Chem. 2016, 68, 226. [Google Scholar] [CrossRef]
- Peytam, F.; Takalloobanafshi, G.; Saadattalab, T.; Norouzbahari, M.; Emamgholipour, Z.; Moghimi, S.; Firoozpour, L.; Bijanzadeh, H.R.; Faramarzi, M.A.; Mojtabavi, S.; et al. Design, Synthesis, Molecular Docking, and in Vitro α-Glucosidase Inhibitory Activities of Novel 3-Amino-2,4-diarylbenzo[4,5]imidazo[1,2-a]pyrimidines Against Yeast and Rat α-Glucosidase. Sci. Rep. 2021, 11, 11911. [Google Scholar] [CrossRef]
- Yigit, M.; Yigit, B.; Taslimi, P.; Ozdemir, I.; Karaman, M.; Gulcin, I. Novel Amine-functionalized Benzimidazolium Salts: Synthesis, Characterization, Bioactivity, and Molecular Docking Studies. J. Mol. Struct. 2020, 1207, 127802. [Google Scholar] [CrossRef]
- Akkoc, S.; Ozer, I.O.; Gok, Y.; Upadhyay, P.J.; Kayser, V. In Vitro Cytotoxic Activities of New Silver and PEPPSI Palladium N-Geterocyclic Carbene Complexes Derived from Benzimidazolium Salts. Inorganica Chim. Acta 2016, 449, 75–81. [Google Scholar] [CrossRef]
- Slimani, I.; Mansour, L.; Abutaha, N.; Harrath, A.H.; Al-Tamimi, J.; Gurbuz, N.; Ozdemir, I.; Hamdi, N. Synthesis, Structural Characterization of Silver(I)-NHC Complexes and Their Antimicrobial, Antioxidant and Antitumor Activities. J. King Saud Univ. Sci. 2020, 32, 1544–1554. [Google Scholar] [CrossRef]
- Nakamura, S.; Takahira, K.; Tanabe, G.; Morikawa, T.; Sakano, M.; Ninomiya, K.; Yoshikawa, M.; Muraoka, O.; Nakanishi, I. Docking and SAR Studies of Salacinol Derivatives as α-Glucosidase Inhibitors. Bioorg. Med. Chem. Lett. 2010, 20, 4420–4423. [Google Scholar] [CrossRef] [PubMed]
- Nursamsiar, N.; Mangande, M.M.; Awaluddin, A.; Nur, S.; Asnawi, A. In Silico Study of Aglycon Curculigoside A and Its Derivatives as α-Amylase Inhibitors. Indones. J. Pharm. Sci. Technol. 2020, 7, 29–37. [Google Scholar] [CrossRef]
- Promyos, N.; Temviriyanukul, P.; Suttisansanee, U. Investigation of Anthocyanidins and Anthocyanins for Targeting α-Glucosidase in Diabetes Mellitus. Prev. Nutr. Food Sci. 2020, 25, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Ozil, M.; Parlak, C.; Balta, N. A Simple and Efficient Synthesis of Benzimidazoles Containing Piperazine or Morpholine Skeleton at C-6 Position as Glucosidase Inhibitors with Antioxidant Activity. Bioorg. Chem. 2018, 76, 468–477. [Google Scholar] [CrossRef]
- Aboul-Enein, H.Y.; El Rashedy, A.A. Benzimidazole Derivatives as Antidiabetic Agents. Med. Chem. 2015, 5, 318–325. [Google Scholar] [CrossRef]
- Mentese, E.; Baltas, N.; Emirik, M. Synthesis, α-Glucosidase Inhibition and in Silico Studies of Some 4-(5-Fluoro-2-substituted-1H-benzimidazol-6-yl)morpholine Derivatives. Bioorg. Chem. 2020, 101, 104002. [Google Scholar] [CrossRef] [PubMed]
- Arshad, T.; Khan, K.M.; Rasool, N.; Salar, U.; Hussain, S.; Asghar, H.; Ashraf, M.; Wadood, A.; Riaz, M.; Perveen, S.; et al. 5-Bromo-2-Aryl Benzimidazole Derivatives as Non-Cytotoxic Potential Dual Inhibitors of α-Glucosidase and Urease Enzymes. Bioorg. Chem. 2017, 72, 21–31. [Google Scholar] [CrossRef]
- Ansari, K.F.; Lal, C. Synthesis, Physicochemical Properties and Antimicrobial Activity of Some New Benzimidazole Derivatives. Eur. J. Med. Chem. 2009, 44, 4028–4033. [Google Scholar] [CrossRef] [PubMed]
- Hayat, S.; Rehan, A.; Choudhary, M.I.; Khan, K.M.; Schumann, W.; Bayer, E. N-Alkylation of Anilines, Carboxamides and Several Nitrogen Heterocycles using CsF–Celite/alkyl halides/CH3CN combination. Tetrahedron 2001, 57, 9951–9957. [Google Scholar] [CrossRef]
- Milgrom, L.R.; Dempsey, P.J.F.; Yahioglu, G. 5,10,15,20-Tetrakis(N-protected-imidazol-2-yl)porphyrins. Tetrahedron 1996, 52, 9877–9890. [Google Scholar] [CrossRef]
- Mosihuzzman, M.; Naheed, S.; Hareem, S.; Talib, S.; Abbas, G.; Khan, S.N.; Israr, M. Studies on α-Glucosidase Inhibition and Anti-Glycation Potential of Iris loczyi and Iris unguicularis. Life Sci. 2013, 92, 187–192. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).