Assessment of Chemopreventive Potential of the Plant Extracts against Liver Cancer Using HepG2 Cell Line
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample Preparation and Extraction
2.3. Phytochemical Screening of Extracts and Antioxidant Activity Using DPPH Assay
2.3.1. α-. Amylase Assay
2.3.2. HPLC Analysis
2.3.3. MTT Assay for HepG2 Cell Line
2.3.4. Preparation of Cell Lines and Culture Medium for MTT Assay
2.3.5. Acridine Orange and Ethidium Bromide Dual Staining Studies Using HepG2 Cell Line
2.3.6. Apoptosis Detection Using Pi Annexin V-FITC Staining of HepG2 Cells by Flow Cytometry
2.3.7. Gene regulation of BAX and BCL 2 in HepG2 Cell Line Treated with W. somnifera Extract
3. Results and Discussion
3.1. Plant Extraction
3.2. Phytochemical Analysis
3.3. Antioxidant Activity Using DPPH Assay
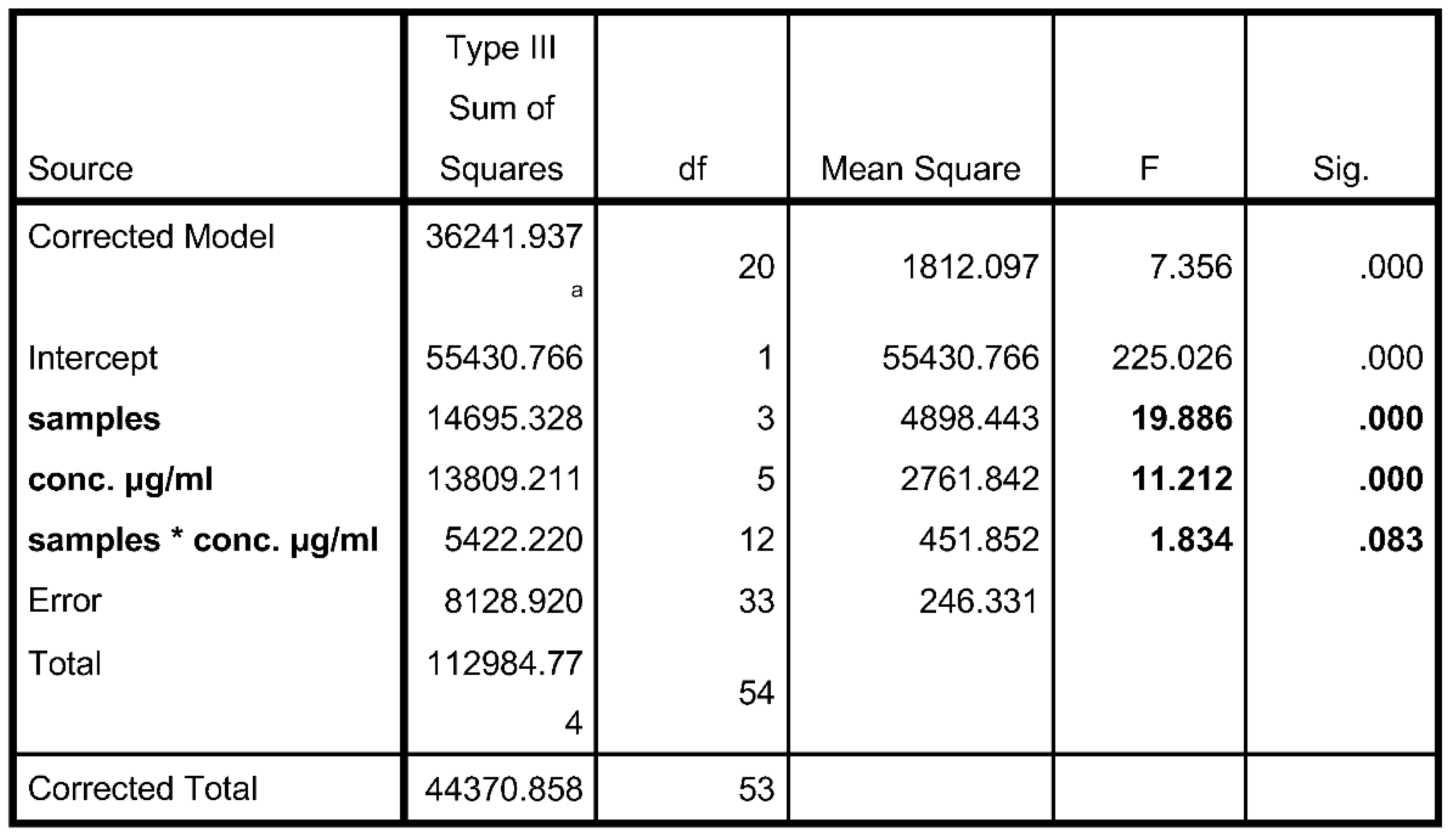
Two-Way ANOVA
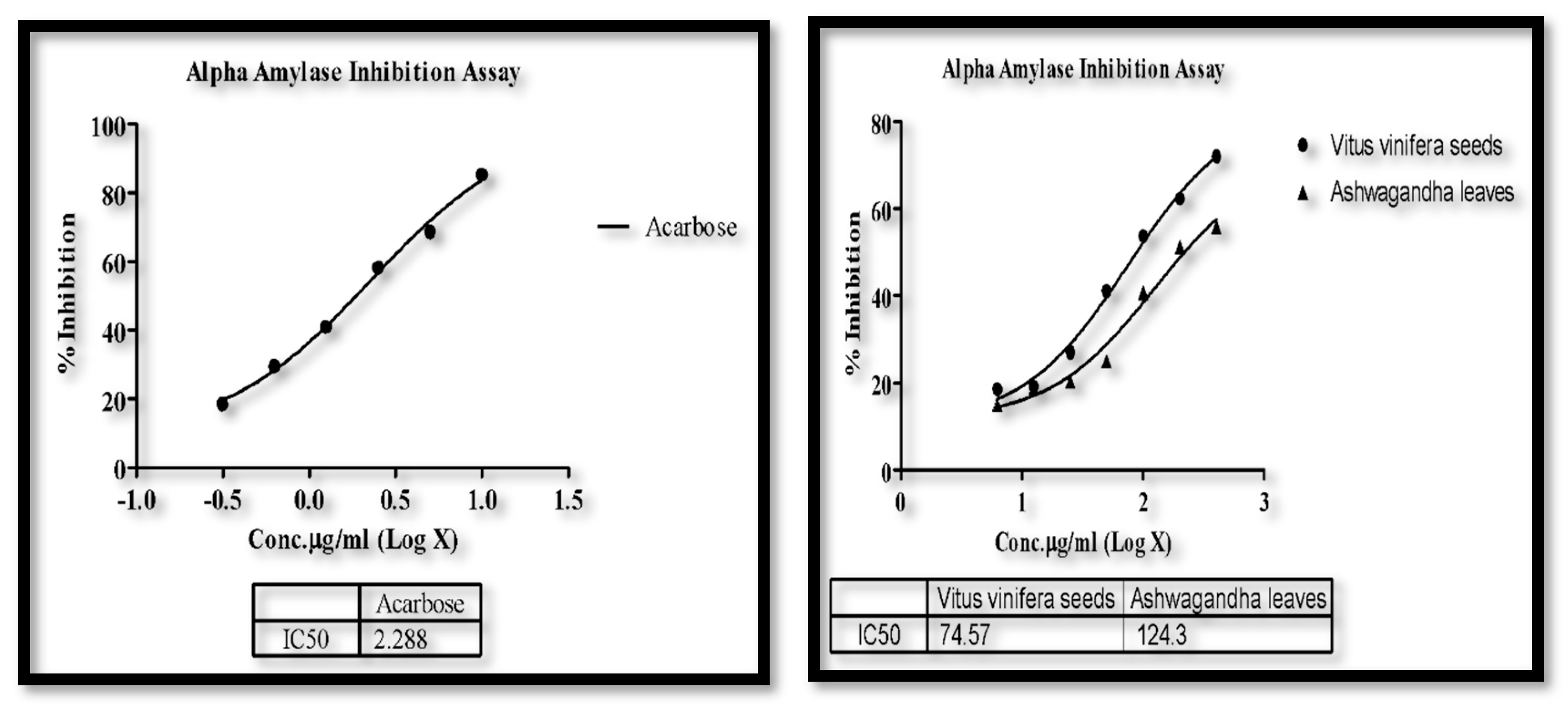
3.4. α-Amylase Assay
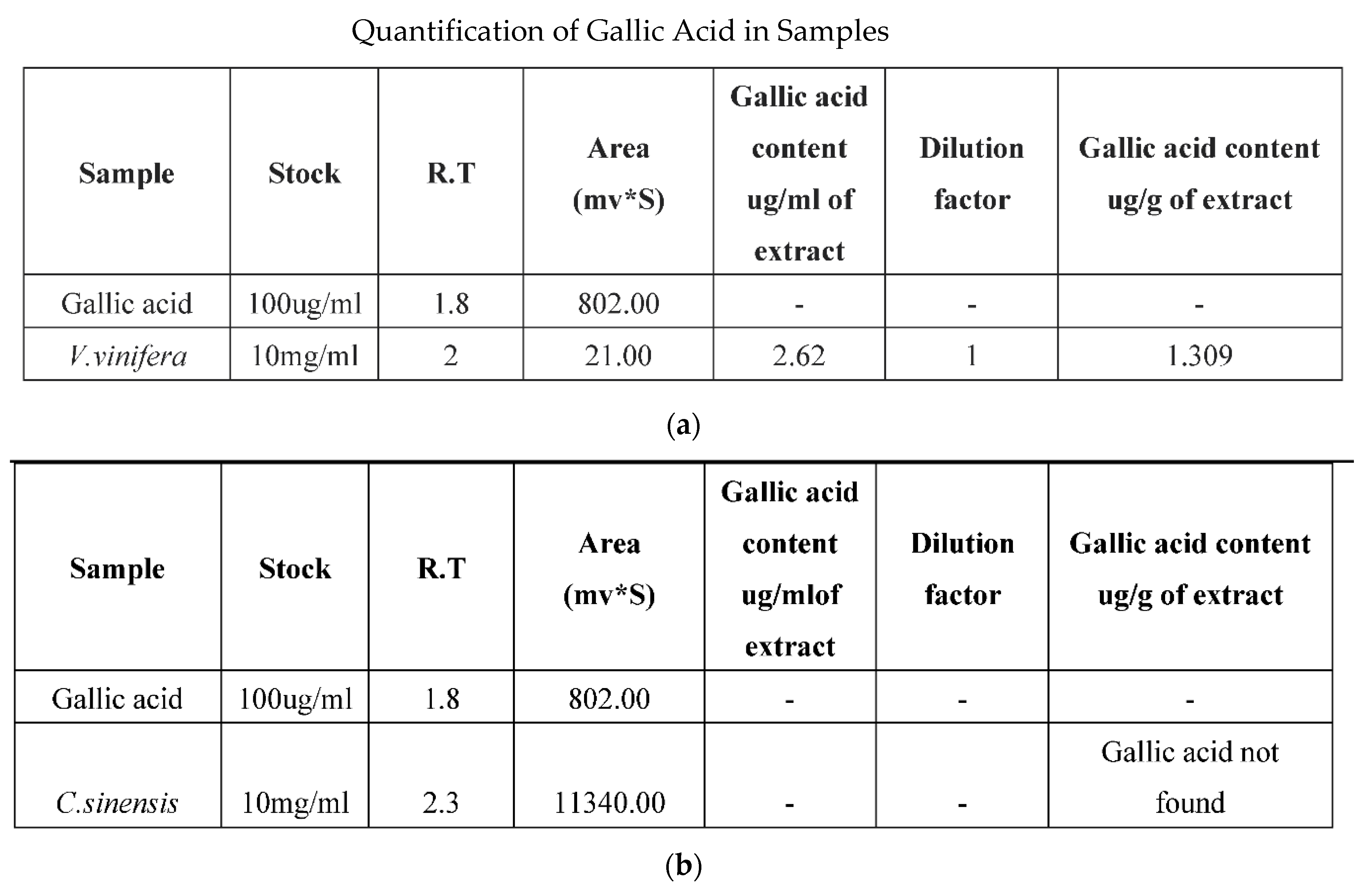
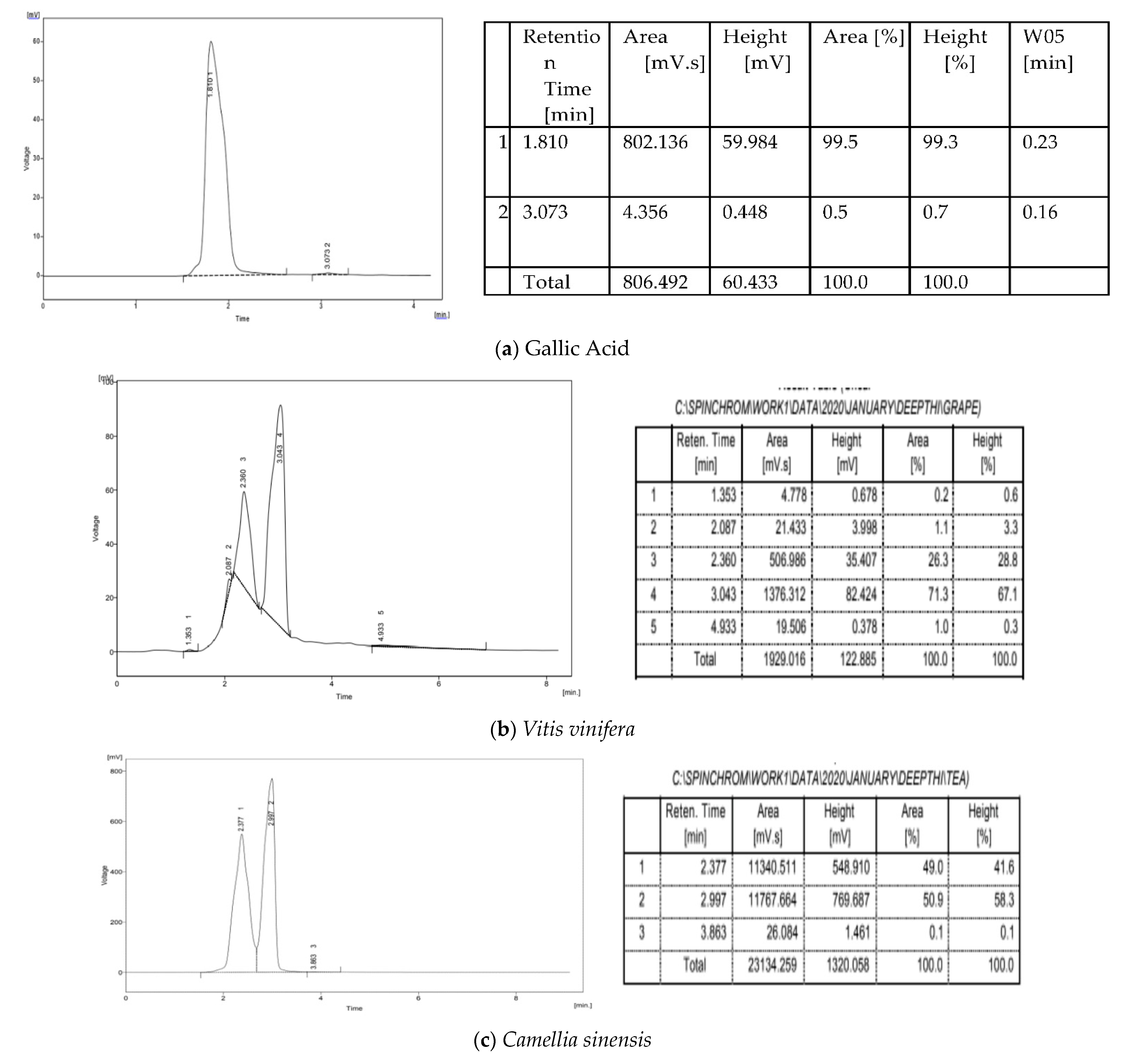
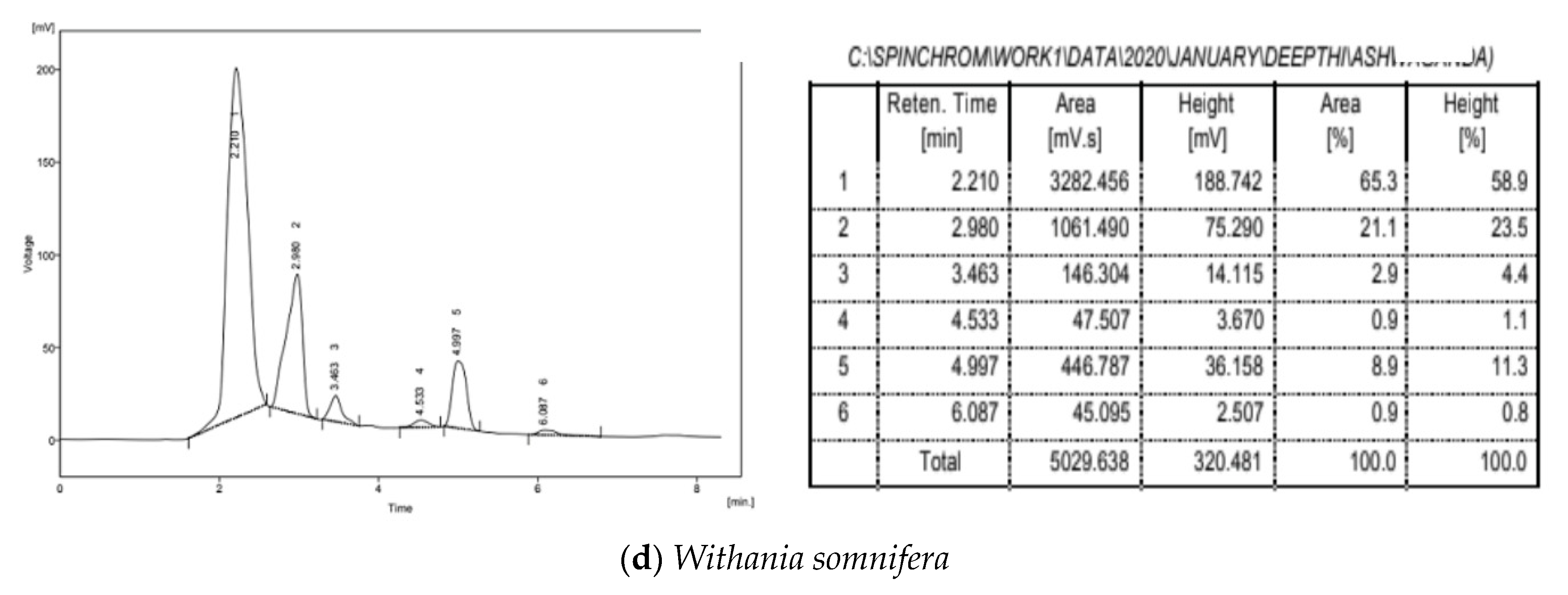
3.5. HPLC Analysis
3.6. MTT Assay for HepG2 Cell Line
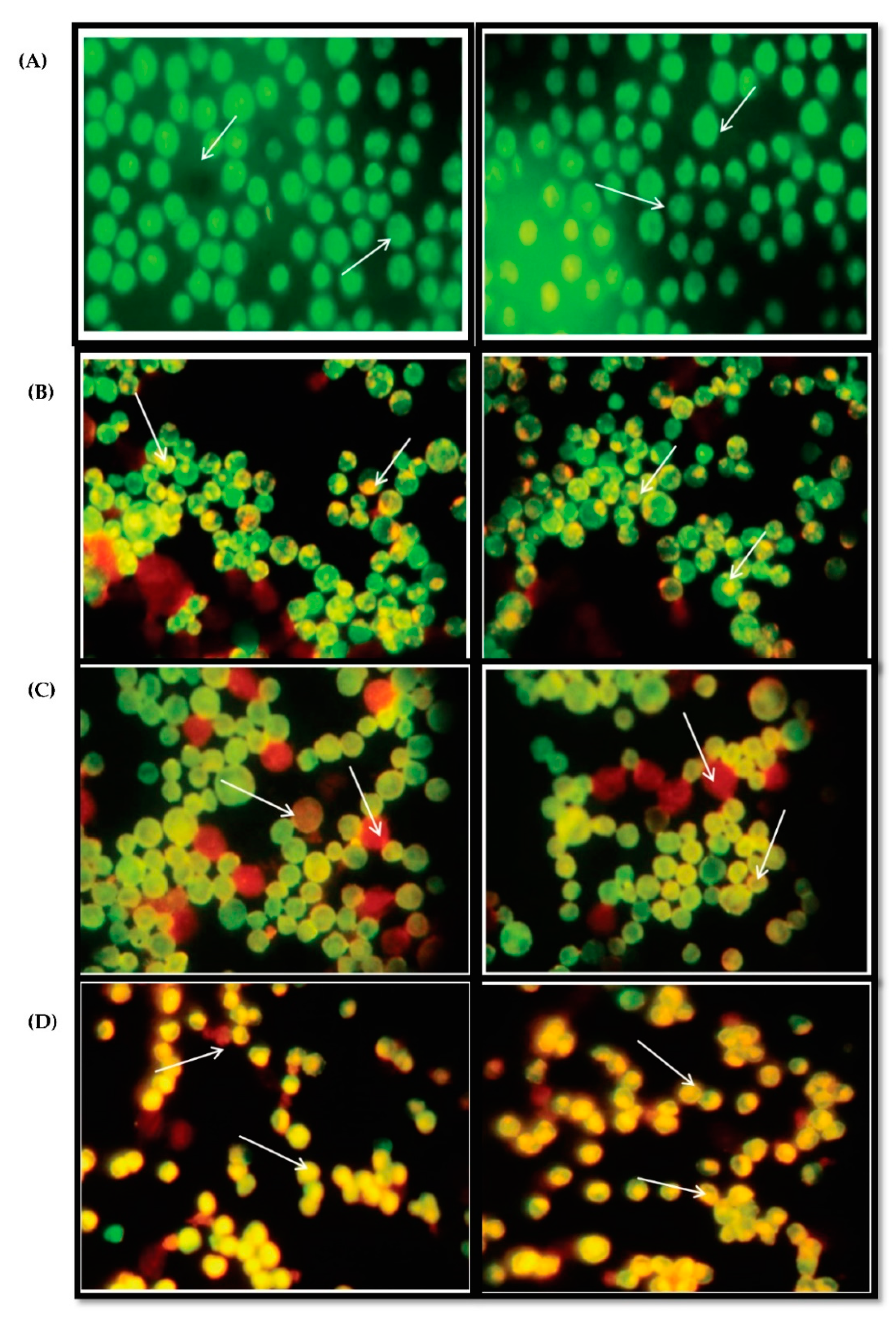
3.7. Acridine Orange and Ethidium Bromide Dual Staining Studies Using HepG2 Cell Line
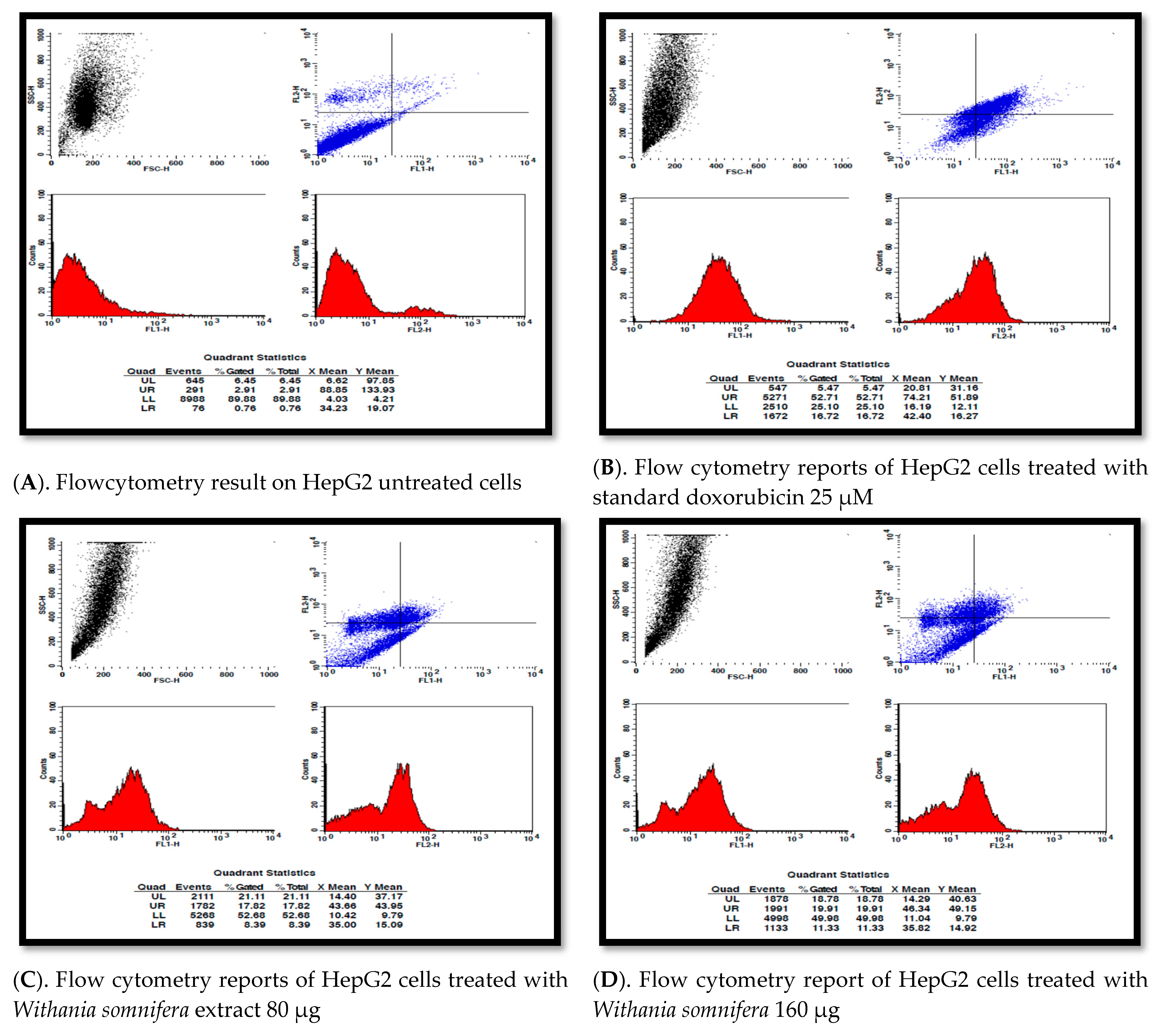
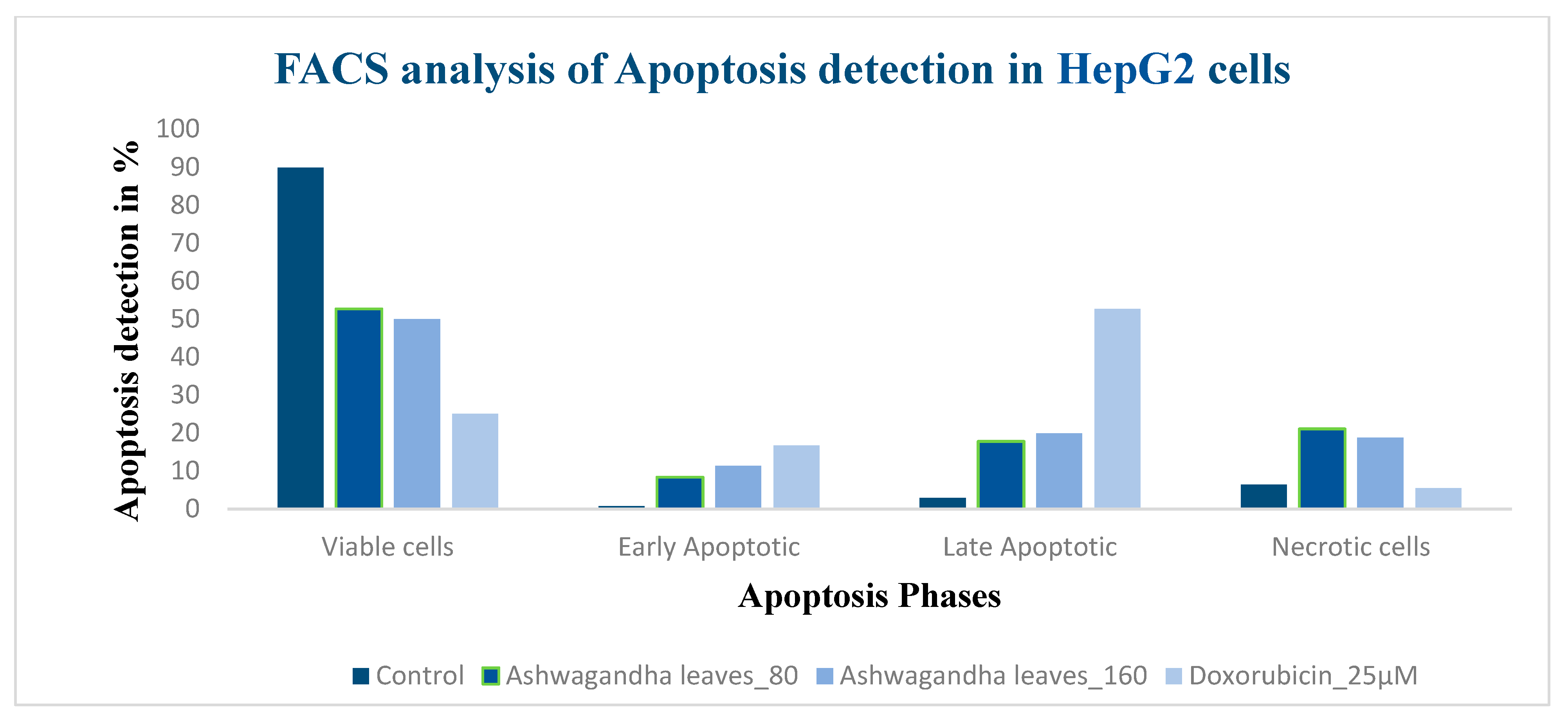
3.8. Apoptosis Detection Using Pi Annexin V-Fitc Staining of Hepg2 Cells by Flow Cytometry
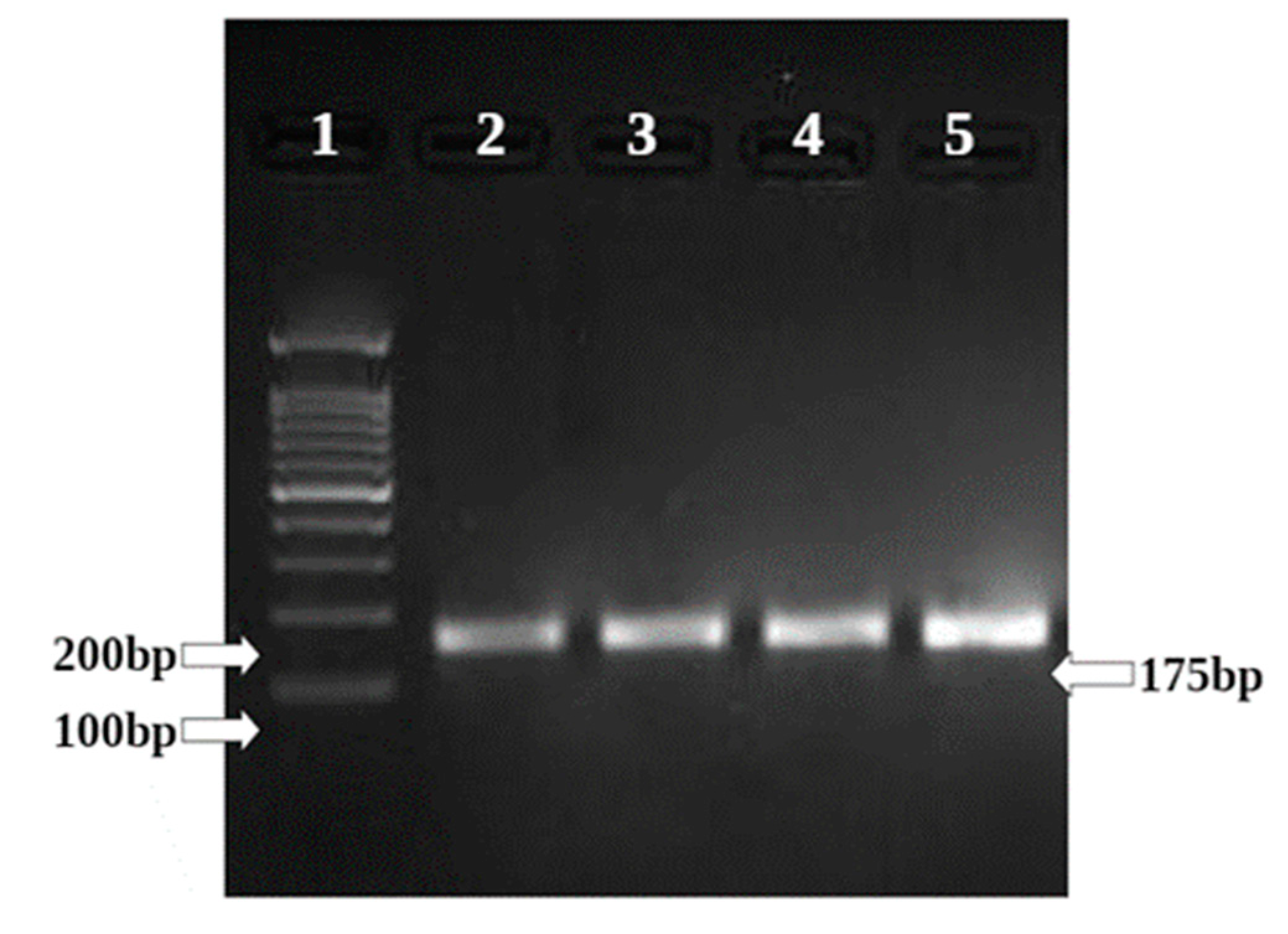
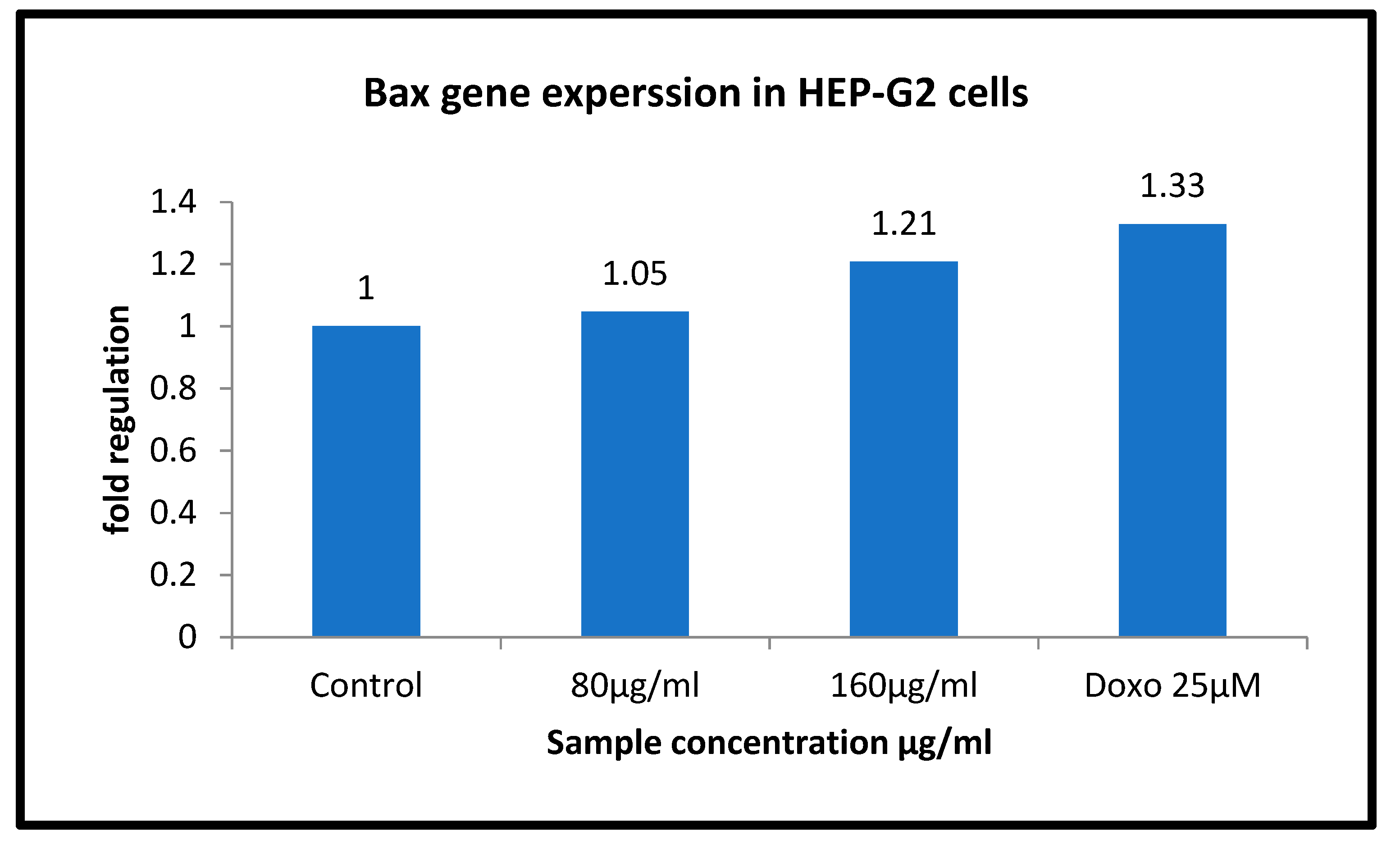
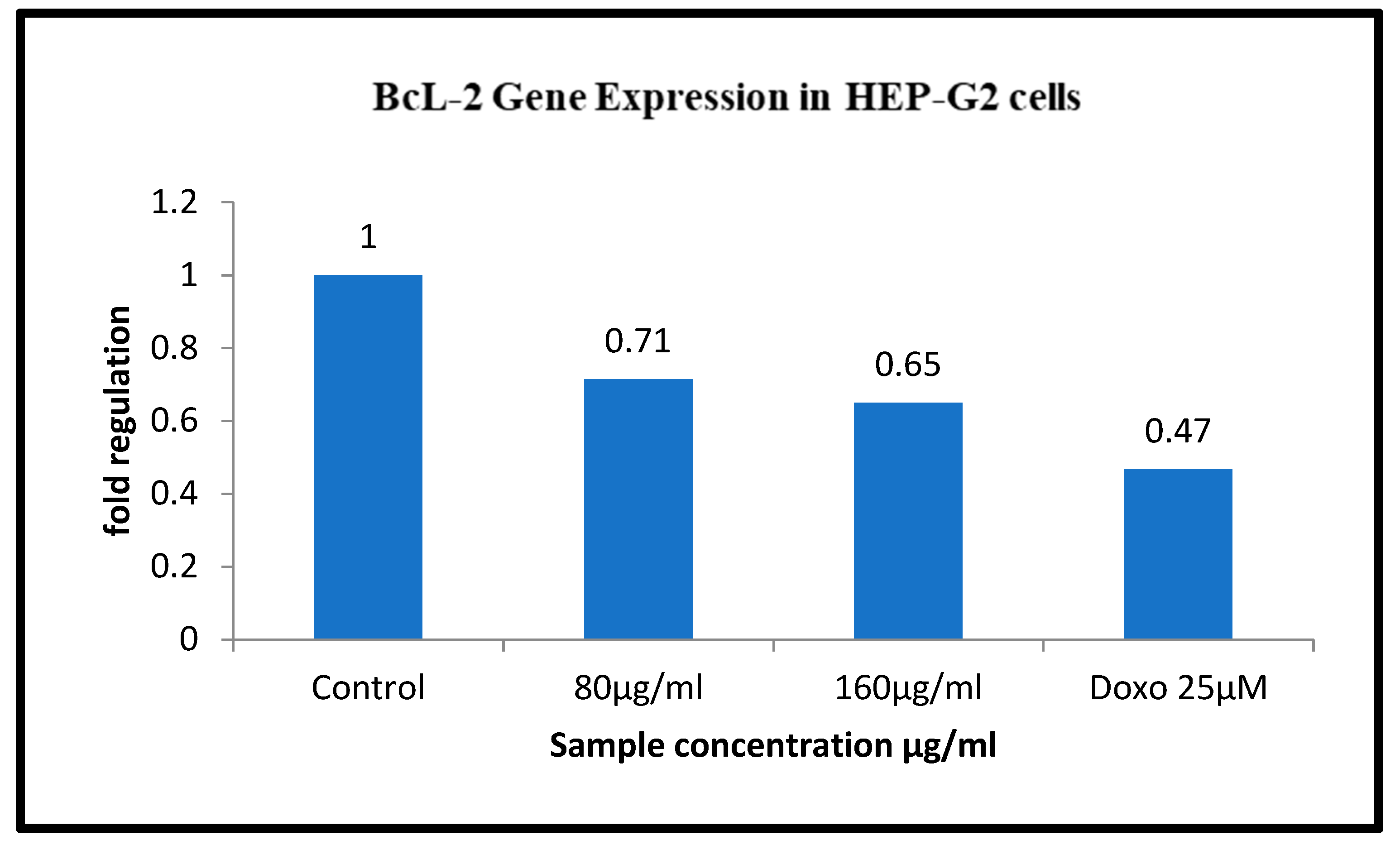
3.9. Gene Regulation of BAX and Bcl 2 in HepG2 Cell Line Treated with Sample Ashwagandha
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prasad, S.K.; Veeresh, P.M.; Ramesh, P.S.; Natraj, S.M.; Madhunapantula, S.V.; Devegowda, D. Phytochemical fractions from Annona muricata seeds and fruit pulp inhibited the growth of breast cancer cells through cell cycle arrest at G0/G1 phase. J. Cancer Res. Ther. 2020, 16, 1235–1249. [Google Scholar]
- Prasad, S.K.; Devananda, D. Evaluating the anticancer potentials of methanol extracted Annona muricata fruit pulp and seed(s) phytochemicals. Clin. Oncol. Res. 2020, 3, 2–8. [Google Scholar]
- Elsayed, E.A.; Sharaf-Eldin, M.A.; Wadaan, M. In vitro evaluation of cytotoxic activities of essential oil from Moringa oleifera seeds on HeLa, HepG2, MCF-7, CACO-2, and L929 cell lines. Asian Pac. J. Cancer Prev. 2015, 16, 4671–4675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elsayed, E.A.; Sharaf-Eldin, M.A.; El-Enshasy, H.A.; Wadaan, M. In vitro assessment of anticancer properties of Moringa peregrine essential seed oil on different cell lines. Pak. J. Zool. 2016, 48, 853–859. [Google Scholar]
- Noureini, S.K.; Wink, M. Antiproliferative effects of crocin in HepG2 cells by telomerase inhibition and hTERT down-regulation. Asian Pac. J. Cancer Prev. 2012, 13, 2305–2309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gavamukulya, Y.; Abou-Elella, F.; Wamunyokoli, F.; AEl-Shemy, H. Phytochemical screening, antioxidanactivity and in vitro anticancer potential of ethanolic and water leaves extracts of Annona muricata (Graviola). Asian Pac. J. Trop. Med. 2014, 1, S355–S363. [Google Scholar] [CrossRef] [Green Version]
- El-Wahab, A.E.; Ghareeb, D.A.; Sarhan, E.E.; Abu-Serie, M.M.; El Demellawy, M.A. In vitro biological assessment of Berberis vulgaris and its active constituent, berberine: Antioxidants, anti-acetylcholinesterase, anti-diabetic and anticancer effects. BMC Complement. Altern. Med. 2013, 13, 218–220. [Google Scholar] [CrossRef] [Green Version]
- Zuo, L.L.; Wang, Z.Y.; Fan, Z.L.; Tian, S.Q.; Liu, J.R. Evaluation of antioxidant and antiproliferative properties of three Actinidia (Actinidia kolomikta, Actinidia arguta, Actinidia chinensis) extracts in vitro. Int. J. Mol. Sci. 2012, 13, 5506–5518. [Google Scholar] [CrossRef] [Green Version]
- Hamilton-Miller, J.M. Anti-cariogenic properties of tea (Camellia sinensis). J. Med. Microbiol. 2001, 50, 299–302. [Google Scholar] [CrossRef]
- Wang, L.; Waltenberger, B.; Pferschy-Wenzig, E.M.; Blunder, M.; Liu, X.; Malainer, C.; Blazevic, T.; Schwaiger, S.; Rollinger, J.M.; Heiss, E.H.; et al. Natural product agonists of peroxisome proliferator-activated receptorgamma (PPARγ): A review. Biochem. Pharmacol. 2014, 92, 73–89. [Google Scholar] [CrossRef] [Green Version]
- Nassiri-Asl, M.; Hosseinzadeh, H. Review of the pharmacological effects of Vitis vinifera (Grape) and its bioactive compounds. Phytother. Res. 2009, 23, 1197–1204. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Schwaninger, H.R.; Baldo, A.M.; Labate, J.A.; Zhong, G.Y.; Simon, C.J. A phylogenetic analysis of the grape genus (Vitis L.) reveals broad reticulation and concurrent diversification during neogene and quaternary climate change. BMC Evolut. Biol. 2013, 13, 141. [Google Scholar] [CrossRef] [Green Version]
- Singh, N.; Bhalla, M.; de Jager, P.; Gilca, M. An overview on ashwagandha: A Rasayana (Rejuvenator) of Ayurveda. Afr. J. Tradit. Complement. Altern. Med. 2011, 8. [Google Scholar] [CrossRef]
- Kakaraparthi, P.S.; Rajput, D.K.; Komaraiah, K.; Kumar, N.; Kumar, R.R. Effect of Sowing Dates on Morphological Characteristics, Root Yield and Chemical Composition of the Root of Withania Somnifera Grown in the Semi-Arid Regions of Andhra Pradesh, India. J. Sci. Res. Rep. 2013, 121–132. [Google Scholar] [CrossRef]
- Rajakumar, D.V. Biochemical & Pharmacological Studies on the Antioxidant Properties of Dehydrozingerone and Its Analogs. Ph.D. Thesis, University of Mangalore, Mangalore, India, 1994. [Google Scholar]
- Crouch, S.P.M.; Kozlowski, R.; Slater, K.J.; Fletcher, J. The use of ATP bioluminescence as a measure of cell proliferation and cytotoxcity. J. Immunol. Methods 1993, 160, 81–88. [Google Scholar] [CrossRef]
- Gonzalez, R.J.; Tarloff, J.B. Evaluation of hepatic subcellular Fractions for alamar blue and MTT reductase activity. Toxicol. In Vitro 2001, 15, 257–259. [Google Scholar] [CrossRef]
- Hattori, N.; Sakakibara, T.; Kajiyama, N.; Igarashi, T.; Maeda, M.; Murakami, S. Enhanced microbial biomass assay using mutant luciferase resistant to benzalkonium chloride. Anal. Biochem. 2003, 319, 287–295. [Google Scholar] [CrossRef]
- Kangas, L.; Grönroos, M.; Nieminen, A.L. Bioluminescence of cellular ATP: A new method for evaluating cytotoxic agents in vitro. Med. Biol. 1984, 62, 338–343. [Google Scholar]
- Lundin, A.; Hasenson, M.; Persson, J.; Pousette, Å. [4] Estimation of biomass in growing cell lines by adenosine triphosphate assay. Methods Enzymol. 1986, 133, 27–42. [Google Scholar]
- Frei, M. Cell Viability and Proliferation—BioFiles. Centrifugation 2015, 6, 17–21. [Google Scholar]
- van de Loosdrecht, A.A.; Nennie, E.; Ossenkoppele, G.J.; Beelen, R.H.; Langenhuijsen, M.M. Cell mediated cytotoxicity against U 937 cells by human monocytes and macrophages in a modified colorimetric MTT assay: A methodological study. J. Immunol. Methods 1991, 141, 15–22. [Google Scholar] [CrossRef]
- van Engeland, M.; Ramaekers, F.C.; Schutte, B.; Reutelingsperger, C.P.M. A novel assay to measure loss of plasma membrane asymmetry during apoptosis of adherent cells in culture. Cytometry 1996, 24, 131–139. [Google Scholar] [CrossRef]
- Casciola-Rosen, L.; Rosen, A.; Petri, M.; Schlissel, M. Surface blebs on apoptotic cells are sites of enhanced procoagulant activity: Implications for coagulation events and antigenic spread in systemic lupus erythematosus. Proc. Nat. Acad. Sci. USA 1996, 93, 1624–1629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andree, H.A.; Reutelingsperger, C.P.; Hauptmann, R.; Hemker, H.C.; Hermens, W.T.; Willems, G.M. Binding of vascular anticoagulant alpha (VAC alpha) to planar phospholipid bilayers. J. Biol. Chem. 1990, 265, 4923–4928. [Google Scholar] [CrossRef]
- Homburg, C.H.; de Haas, M.; von dem Borne, A.E.; Verhoeven, A.J.; Reutelingsperger, C.P.; Roos, D. Human neutrophils lose their surface Fc gamma RIII and acquire Annexin V binding sites during apoptosis in vitro. Blood 1995, 85, 532–540. [Google Scholar] [CrossRef] [Green Version]
- Koopman, G.; Reutelingsperger, C.P.; Kuijten, G.A.; Keehnen, R.M.; Pals, S.T.; van Oers, M.H. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood 1994, 84, 1415–1420. [Google Scholar] [CrossRef] [Green Version]
- Martin, S.; Reutelingsperger, C.P.; McGahon, A.J.; Rader, J.A.; Van Schie, R.C.; Laface, D.M.; Green, D.R. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: Inhibition by overexpression of Bcl-2 and Abl. J. Exp. Med. 1995, 182, 1545–1556. [Google Scholar] [CrossRef] [Green Version]
- Raynal, P.; Pollard, H.B. Annexins: The problem of assessing the biological role for a gene family of multifunctional calcium- and phospholipid-binding proteins. Biochim Biophys Acta (BBA)-Rev. Biomembr. 1994, 1197, 63–93. [Google Scholar] [CrossRef]
- Vermes, I.; Haanen, C.; Steffens-Nakken, H.; Reutelingsperger, C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J. Immunol. Methods 1995, 184, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Kawakita, T.; Sasaki, H.; Hoshiba, T.; Asamoto, A.; Williamson, E. Amylase-producing ovarian carcinoma: A case report and a retrospective study. Gynecol. Oncol. Case Rep. 2012, 2, 112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casadei Gardini, A.; Mariotti, M.; Lucchesi, A.; Pini, S.; Valgiusti, M.; Bravaccini, S.; Del Monte, A.; Burgio, M.A.; Marisi, G.; Amadori, D.; et al. Paraneoplastic lipase and amylase production in a patient with small-cell lung cancer: Case report. BMC Cancer 2016, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ismail, A.; Ramli, N.S.; Mohamed, M.; Ahmad, W.A. Acute and sub-acute antihypertensive effects of Syzygium polyanthum leaf extracts with determination of gallic acid using HPLC analysis. Pharmacogn. J. 2018, 10, 663–671. [Google Scholar] [CrossRef]
- Yang, D.J.; Hwang, L.S.; Lin, J.T. Effects of different steeping methods and storage on caffeine, catechins and gallic acid in bag tea infusions. J. Chromatogr. A. 2007, 1156, 312–320. [Google Scholar] [CrossRef]
- Karamac, M.; Kosiñska, A.; Pegg, R.B. Content of gallic acid in selected plant extracts. Pol. J. Food Nutr. Sci. 2006, 15, 55. [Google Scholar]




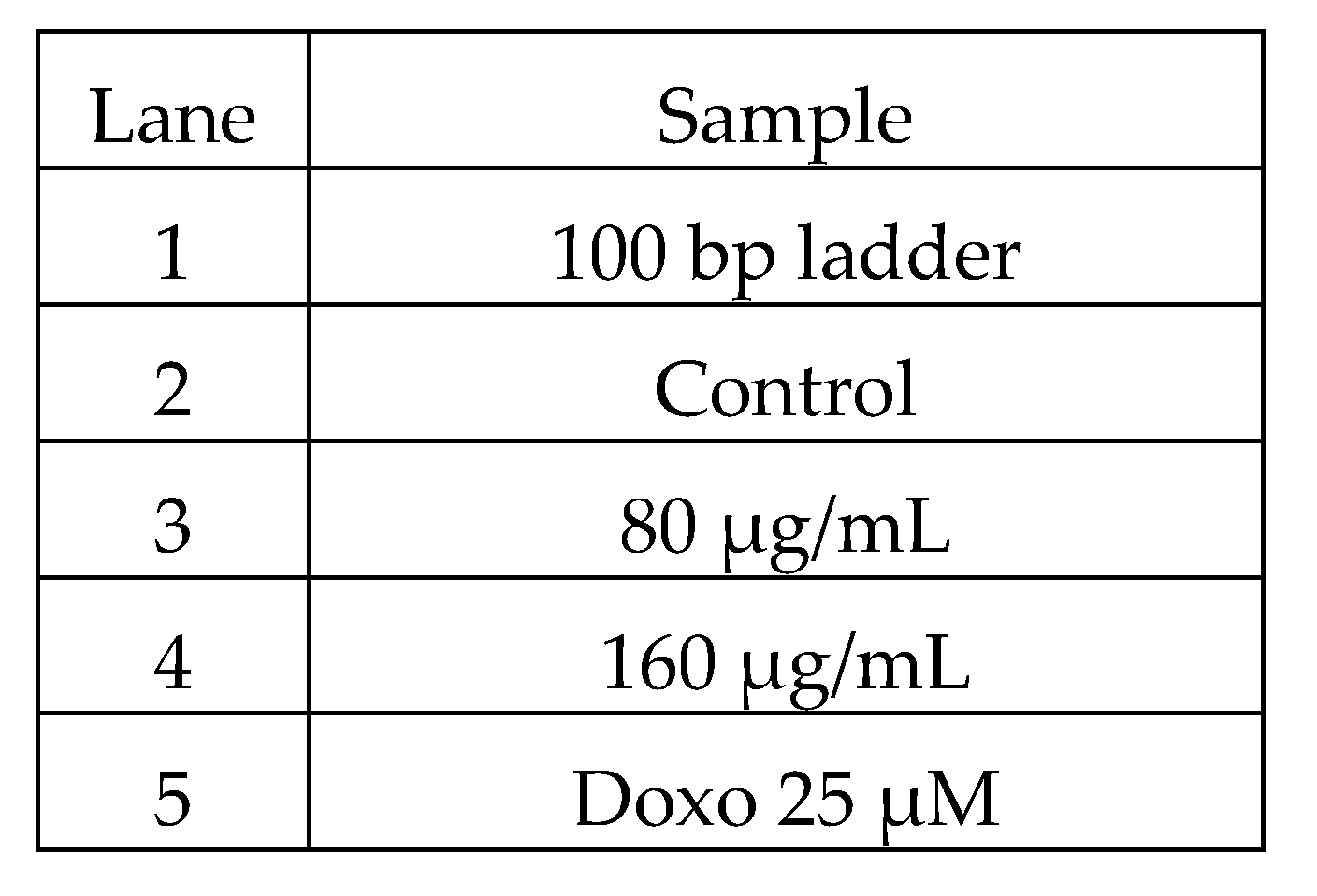


| Gene | Primer Pair | Sequence | Tm | Product Size (bp) |
|---|---|---|---|---|
| GAPDH | FP | GTCCAGTTAATTTCTGACCT | 47.7 | 162 bp |
| RP | GCTTTGTACATGGTATTCAC | 47.7 | ||
| BCL II | FP | CTGGTGGACAACATCGCTCTG | 61.4 | 131 bp |
| RP | GGTCTGCTCACCTCACTTGTG | 61.4 | ||
| BAX | FP | GGCACCTGAGCTGACCTTGG | 47.7 | 175 bp |
| RP | GAGGAAGTCCAGTGTCCAGC | 47.7 |
| Types of Tests | Green tea | Vitus vinefera | Ashwagandha |
|---|---|---|---|
| Alkaloids | + | + | + |
| Carbohydrates | - | - | - |
| Tannins | + | + | + |
| Terpenoids | + | + | + |
| Glycosides | - | - | - |
| Steroids | - | - | - |
| Saponins | - | - | - |
| Flavonoids | + | + | + |
| Cardiac glycosides | - | - | - |
| Mucilage test | - | - | - |
| Volatile oil | + | + | - |
| Sample | Conc. (μg/mL) | OD @ 510 nm | % Inhibition | IC50 (µg/mL) |
|---|---|---|---|---|
| Control | 0 | 0.912 | 0.00 | 2.866 |
| Quercetin | 0.3125 | 0.521 | 42.87 | |
| 0.625 | 0.430 | 52.85 | ||
| 1.25 | 0.391 | 57.13 | ||
| 2.5 | 0.324 | 64.47 | ||
| 5 | 0.224 | 75.44 | ||
| 10 | 0.145 | 84.09 | ||
| Vitus viniferaseeds | 3.125 | 0.845 | 7.35 | - |
| 6.25 | 0.746 | 18.20 | ||
| 12.5 | 0.684 | 25.00 | ||
| 25 | 0.568 | 37.72 | ||
| 50 | 0.532 | 41.67 | ||
| 100 | 0.488 | 46.49 | ||
| Camelia sinensis leaves | 3.125 | 0.806 | 11.62 | 14.16 |
| 6.25 | 0.698 | 23.46 | ||
| 12.5 | 0.497 | 45.50 | ||
| 25 | 0.361 | 60.42 | ||
| 50 | 0.231 | 74.67 | ||
| 100 | 0.104 | 88.60 | ||
| Withania somniferaleaves | 3.125 | 0.857 | 6.03 | 59.21 |
| 6.25 | 0.730 | 19.96 | ||
| 12.5 | 0.702 | 23.03 | ||
| 25 | 0.648 | 28.95 | ||
| 50 | 0.548 | 39.91 | ||
| 100 | 0.397 | 56.47 |
| HepG-2 | Standard | |||
|---|---|---|---|---|
| Compound Name | Conc. µM | OD @ 590 nm | % Inhibition | IC50 in µM |
| Control | 0 | 0.9731 | 0.00 | 18.8 |
| Doxorubicin | 3.125 | 0.692 | 28.88 | |
| 6.25 | 0.634 | 34.86 | ||
| 12.5 | 0.507 | 47.88 | ||
| 25 | 0.385 | 60.46 | ||
| 50 | 0.305 | 68.69 | ||
| 100 | 0.194 | 80.02 | ||
| HepG-2 | ||||
| Compound Name | Conc. µg/ml | OD @ 590 nm | % Inhibition | IC50 µg/mL |
| Control | 0 | 0.973 | 0.00 | IC50 was not calculated to lesser % inhibition |
| Vitis vinifera | 10 | 0.900 | 7.50 | |
| 20 | 0.865 | 11.03 | ||
| 40 | 0.753 | 22.58 | ||
| 80 | 0.720 | 26.01 | ||
| 160 | 0.667 | 31.38 | ||
| 320 | 0.577 | 40.62 | ||
| Camelia sinensis | 10 | 0.906 | 6.82 | IC50 was not calculated to lesser % inhibition |
| 20 | 0.848 | 12.76 | ||
| 40 | 0.776 | 20.19 | ||
| 80 | 0.684 | 29.68 | ||
| 160 | 0.642 | 34.00 | ||
| 320 | 0.610 | 37.29 | ||
| Ashwagandha leaves | 10 | 0.886 | 8.88 | 153.80 |
| 20 | 0.796 | 18.20 | ||
| 40 | 0.710 | 27.00 | ||
| 80 | 0.615 | 36.76 | ||
| 160 | 0.475 | 51.34 | ||
| 320 | 0.296 | 69.51 | ||
| HepG2 | ||||
|---|---|---|---|---|
| Sample Conc. µg/mL | Band Intensity of PCR Amplicons | Normalised | Relative Gene Expression | |
| GAPDH | BAX | |||
| Control | 12,631.08 | 17,305.61 | 1.370 | 1 |
| 80 µg/mL | 15,367.00 | 22,031.13 | 1.434 | 1.05 |
| 160 µg/mL | 14,598.85 | 24,165.42 | 1.655 | 1.21 |
| Doxo 25 µM | 16,118.44 | 29,342.88 | 1.820 | 1.33 |
| HepG2 | ||||
|---|---|---|---|---|
| Sample Conc. µg/mL | Band Intensity of PCR Amplicons | Normalised | Relative Gene Expression | |
| GAPDH | BCl2 | |||
| Control | 12,631.08 | 18,823.78 | 1.490 | 1 |
| 80 µg/mL | 15,367.00 | 16,342.77 | 1.063 | 0.71 |
| 160 µg/mL | 14,598.85 | 14,123.93 | 0.967 | 0.65 |
| Doxo 25 µM | 16118.44 | 11209.12 | 0.695 | 0.47 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venkatachalapathy, D.; Shivamallu, C.; Prasad, S.K.; Thangaraj Saradha, G.; Rudrapathy, P.; Amachawadi, R.G.; Patil, S.S.; Syed, A.; Elgorban, A.M.; Bahkali, A.H.; et al. Assessment of Chemopreventive Potential of the Plant Extracts against Liver Cancer Using HepG2 Cell Line. Molecules 2021, 26, 4593. https://doi.org/10.3390/molecules26154593
Venkatachalapathy D, Shivamallu C, Prasad SK, Thangaraj Saradha G, Rudrapathy P, Amachawadi RG, Patil SS, Syed A, Elgorban AM, Bahkali AH, et al. Assessment of Chemopreventive Potential of the Plant Extracts against Liver Cancer Using HepG2 Cell Line. Molecules. 2021; 26(15):4593. https://doi.org/10.3390/molecules26154593
Chicago/Turabian StyleVenkatachalapathy, Deepthi, Chandan Shivamallu, Shashanka K. Prasad, Gopenath Thangaraj Saradha, Parthiban Rudrapathy, Raghavendra G. Amachawadi, Sharanagouda S. Patil, Asad Syed, Abdallah M. Elgorban, Ali H. Bahkali, and et al. 2021. "Assessment of Chemopreventive Potential of the Plant Extracts against Liver Cancer Using HepG2 Cell Line" Molecules 26, no. 15: 4593. https://doi.org/10.3390/molecules26154593
APA StyleVenkatachalapathy, D., Shivamallu, C., Prasad, S. K., Thangaraj Saradha, G., Rudrapathy, P., Amachawadi, R. G., Patil, S. S., Syed, A., Elgorban, A. M., Bahkali, A. H., Kollur, S. P., & Basalingappa, K. M. (2021). Assessment of Chemopreventive Potential of the Plant Extracts against Liver Cancer Using HepG2 Cell Line. Molecules, 26(15), 4593. https://doi.org/10.3390/molecules26154593









