Abstract
Biofilms, the predominant growth mode of microorganisms, pose a significant risk to human health. The protective biofilm matrix, typically composed of exopolysaccharides, proteins, nucleic acids, and lipids, combined with biofilm-grown bacteria’s heterogenous physiology, leads to enhanced fitness and tolerance to traditional methods for treatment. There is a need to identify biofilm inhibitors using diverse approaches and targeting different stages of biofilm formation. This review discusses discovery strategies that successfully identified a wide range of inhibitors and the processes used to characterize their inhibition mechanism and further improvement. Additionally, we examine the structure–activity relationship (SAR) for some of these inhibitors to optimize inhibitor activity.
1. Introduction
Biofilms—microbial communities composed of microorganisms encased in an extracellular matrix—are the most abundant microbial growth mode on Earth [1]. Biofilms can have both beneficial and detrimental effects in environmental, industrial, and clinical settings. The ability to form biofilms is essential for the pathogenicity of several pathogenic bacteria. Biofilm-related infections are often associated with chronic diseases, and biofilms affect the virulence of acute infections and treatments’ efficacy [2,3,4,5]. When grown as biofilms, pathogenic microorganisms are recalcitrant to immune system clearance, inherently resistant to traditional antibiotics, rendering conventional antibiotics and therapeutics non-viable [6,7,8,9]. Biofilm formation represents another challenge in clinical settings, as pathogenic bacteria can form biofilms on implantable medical devices, resulting in device failure and chronic infections. Biofilms are involved in 65 to 80% of nosocomial infections [10,11,12,13]. There are no clinical antimicrobial agents currently existing that target biofilms; hence, the disruption of biofilm formation represents a valuable new target for antimicrobial discovery [14]. Furthermore, around the world, antibiotic resistance is increasing, and traditional treatment and prevention methods are becoming increasingly ineffective [15,16]. We need to identify and target different cellular processes for the development of new antimicrobial agents.
In this review, we discuss inhibitor discovery methods and strategies and approaches for characterizing novel inhibitors. We will first provide an overview of biofilm formation, then discuss successful strategies in discovering and developing biofilm inhibitors, the mechanism of action and structure–activity relationships (SAR) for well-characterized inhibitors, and the broad classification of antibiofilm compounds and shared characteristics. We primarily focused on compounds whose mode of biofilm inhibition occurred by altering biofilm regulation and biofilm-forming ability rather than antimicrobial activity. Finally, we also discuss the growing topic of therapeutic drug-conjugates, which improve cell/biofilm penetration and the delivery of antibiotics to eradicate biofilms through antimicrobial activity. While the inhibitors discussed are not a comprehensive list, they represent a wide range of diverse biofilm inhibitors with differing methods of discovery and inhibitory targets. As there is a growing interest in biofilms as antimicrobial targets, additional biofilm inhibitors and their known action mechanisms have been discussed in recent reviews, including synthetic biofilm inhibitors such as the phenazine and amino-immidozole inhibitor classes [17,18,19,20,21,22,23].
2. Biofilm Formation and Targets for Anti-Biofilm Interventions
In totality, biofilm formation is a series of events needed for the transition from the planktonic lifestyle to a surface-associated multicellular community. Biofilm formation begins with surface attachment using various cell-surface appendages such as pili, flagella, and surface proteins [24,25,26,27,28,29,30,31,32]. The initial stage of biofilm formation is crucial, as the inhibition of surface attachment prevents surface colonization and circumvents the traditional advantages of the biofilm lifestyle. Following surface attachment, some bacteria move along the surfaces using different surface motilities, while others do not exhibit surface motility [33,34,35]. Surface-attached founder cells undergo cell-division and retain their progeny by producing an extracellular matrix resulting in microcolony formation and eventually mature biofilms [24,25,26,27,28,29,30,31,32,33,34]. Composed of exopolysaccharides, matrix proteins, and extracellular DNA, the biofilm matrix aids in cell–cell and cell-surface adherence, structural integrity, and results in emergent biofilm properties such as resistance to physicochemical stresses, including resistance to antimicrobial agents [30,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51]. This resistance is due to multiple factors, including altered physiology of biofilm-grown cells, chemical inactivation of antimicrobials by the extracellular matrix, and decreased penetration of antibiotics within the biofilm [52,53]. As intact biofilms are difficult to eradicate, there is an interest in identifying compounds that prevent biofilm formation by targeting biofilm matrix production and its assembly.
Canonical regulatory circuitries controlling biofilm formation are another target of interest, as the inhibition of such circuitries can prevent biofilm formation. The regulation of biofilm formation is complex and involves several signaling molecules and regulatory circuitries. Nucleotide-based intracellular signaling molecule cyclic di-guanosine monophosphate (c-di-GMP), a broadly conserved bacterial signaling molecule, is the central regulator of biofilm formation [54,55,56,57]. C-di-GMP is produced by diguanylate cyclases (DGCs) and is degraded by phosphodiesterases (PDEs) [54,55,56,57]. C-di-GMP is sensed by receptor proteins or by c-di-GMP riboswitches to control cellular processes through transcriptional, post-transcriptional, and translational mechanisms that govern all stages of biofilm formation, including surface attachment, surface motility, and matrix formation [54,55,56,57,58]. Modulation of c-di-GMP production, c-di-GMP receptor function, and their cognate cellular targets are, therefore, one of the most crucial targets for developing anti-biofilm interventions. Other regulatory networks such as those involved in sensing cell density by autoinducers, i.e., quorum sensing (QS), or those involved in sensing and responding to environmental conditions, i.e., two-component regulatory systems (TCS), also govern the regulation of biofilm formation [59,60]. Compounds that inhibit these regulatory networks are also of interest and are widely explored [60,61,62,63] (Figure 1).
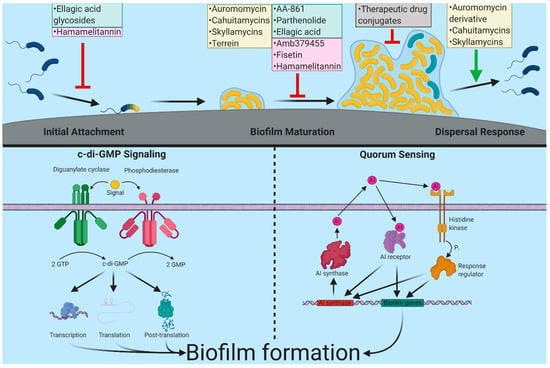
Figure 1.
Biofilm inhibitors and their targets discussed in this review. Biofilm formation starts when planktonic cells (colored blue) sense and attach to the surface. Following this, surface-attached cells (colored yellow) begin to divide, recruit nearby cells, and produce biofilm matrix components and form microcolonies and eventually mature biofilms. In response to cellular signals or external cues, biofilms can disperse, and dispersed cells can colonize new environments. Major inhibitors covered in this review, as well as their discovery method, are shown by color. High throughput discovery is represented by yellow, targeted/pathway discovery by green, in silico discovery by pink, and therapeutic drug conjugates in grey. A schematic of two major regulatory systems governing biofilm formation c-di-GMP signaling and quorum sensing (QS) are indicated. C-di-GMP is produced by diguanylate cyclases (DGCs) and degraded by phosphodiesterases (PDEs). Signals can impact the enzymatic activity of these enzymes allowing for control over the intracellular concentrations of c-di-GMP. These changes in c-di-GMP can be sensed by receptors that affect transcription, translation, and activity or stability of biofilm-associated genes/proteins. Bacteria use quorum sensing to determine cell density. These systems produce an autoinducer compound, which is secreted into the environment. QS signals are then sensed by periplasmic proteins, membrane-bound histidine kinases or cytosolic receptors and initiate signal transduction pathways regulating biofilm gene expression. Created with BioRender.com.
3. Approaches to Discover Anti-Biofilm Compounds
Biofilm inhibitors have been identified using various screening, high-throughput (HTS) and targeted/pathway screening, and in silico analysis approaches. Each of these strategies has its unique advantages and disadvantages, enhancing inhibitor discovery attempts. Upon discovery, potential inhibitors are tested for antibacterial activity, which is determined by measuring biomass or metabolic activity using methods evaluating cell density, cell counts, or ATP levels. Analysis of antibacterial activity is important in determining if a compound inhibits biofilm formation by modulating a biofilm-specific process or by cell viability (Figure 2).
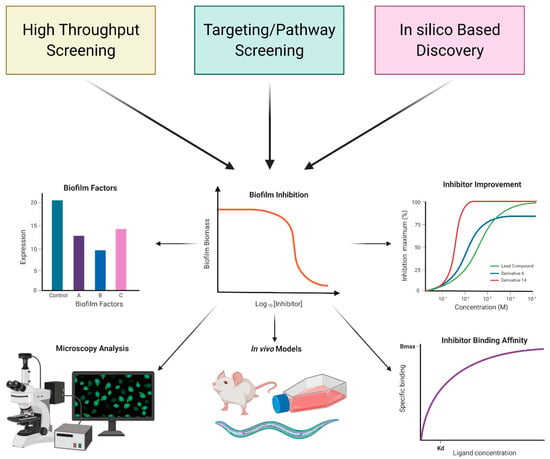
Figure 2.
Overview of approaches used identification of biofilm inhibitors. Screening-based and in silico discovery approaches are commonly used. After initial discovery, typically using biofilm biomass or biofilm gene expression as a readout, additional studies are performed to determine the impact of these inhibitors on biofilm matrix production, biofilm structure, biofilm driven infection, inhibitor binding affinity, and inhibitor improvement. Such studies provide an insight into the mechanism of action of the identified compounds. Created with BioRender.com.
One major group of biofilm inhibitors, inspired by scaffolds found in natural products with reported activity, are a diverse range of heterocyclic containing families, which include indoles [64,65,66,67,68,69,70,71], 2-aminoimidazoles [72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89], phenazines [90,91,92,93,94,95,96,97,98], and quinolines [99,100,101,102,103,104,105,106]. These lead molecules are the results of significant medicinal chemistry effort to improve potency and other drug-like properties. A prominent example of such studies is the work done by C. Melander and colleagues, where multiple biofilm inhibitors were developed based on a 2-aminoimidazole core inspired by the natural product bromoageliferin. These families of heterocyclic biofilm inhibitors have recently been extensively reviewed and will not be covered in this review [107,108,109,110].
In this review, we will focus on screening and in silico discovery efforts on selected pathogenic bacteria: Acinetobacter baumannii, Vibrio cholerae, Pseudomonas aeruginosa, Escherichia coli, and Staphylococcus aureus.
4. Inhibitors Identified via Cell-Based or In Vitro Screens
Unbiased screens, typically using biofilm biomass measurements as a readout, are often used to identify compounds impacting biofilm formation from natural products or synthetic chemical libraries. Targeted/pathway screens utilize specific information on biofilm formation, such as biofilm gene expression, enzymatic activity, along with a screening of natural products or synthetic chemical libraries to identify compounds that impact particular systems or pathways. In the following section, we discuss a set of biofilm inhibitors identified using these approaches (Figure 3). These compounds, along with those discussed in later sections, will have their published inhibitory and disruptive activity listed, including their target and discovery method for ease of comparison (Table 1).
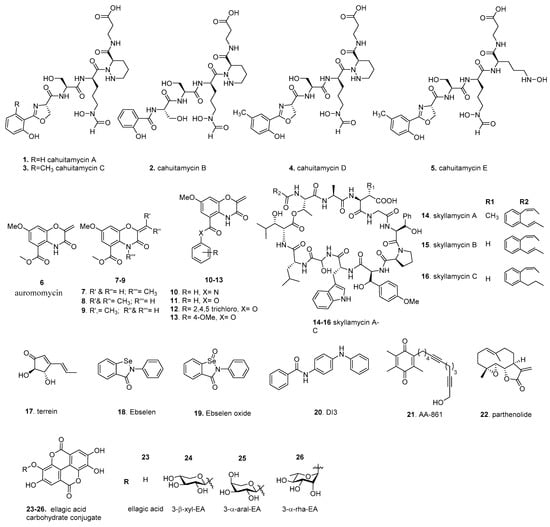
Figure 3.
Screening-based discovery of biofilm inhibitors.

Table 1.
Biofilm inhibitors discussed, their target pathogen, discovery method, and inhibitory activity. This includes biofilm inhibitory concentration (BIC50), enzymatic inhibitory concentration (IC50), biofilm dispersal concentration (BDC50), minimal biofilm eradication concentration (MBEC50), percent reduction in biofilm biomass (biofilm reduction), and percent inhibition of enzyme activity (enzyme inhibition).
4.1. Cahuitamycins
A widely utilized method for unbiased screening is biofilm staining, using crystal violet, to measure biofilm biomass [111,112]. In one such example, a library of 9831 marine microbial-derived extracts were screened to identify compounds that were able to inhibit A. baumannii biofilm formation. An extract from Streptomyces gandocaennsis was found to inhibit biofilm formation without impacting cell growth [113]. Subsequent studies identified the secondary metabolites cahuitamycins A–C (1–3) as the lead compounds (Figure 3). 3 had the most potent inhibitory potential of the three compounds with a half-maximal biofilm inhibitor concentration (BIC50) of 14.5 µM. Later, mutasynthetic studies, utilizing ribosomal engineering, led to the production of additional derivatives referred to as cahuitamycins D,E (4, 5), which improved the BIC50 to 8.54 and 10.5 µM, respectively [114]. Of the biofilm inhibiting compounds, Park et al. reported a minimal impact on cell viability. Further studies of the biosynthetic pathway of the cahuitamycins found that CahJ, an adenylation enzyme, was important in cahuitamycin diversification, and due to CahJ substrate promiscuity, could be used to generate further derivatives for evaluation as potent biofilm inhibitors [115]. These new compounds also gained the ability to disperse preformed biofilms at a relatively high concentration. The above biofilm inhibitory activity of cahuitamycins suggested that the terminal 2-hydroxybenzoyl-oxazoline group represents a key pharmacophore. As the cahuitamycins have siderophore-like properties, the authors tested the iron-complexed form of 1, which showed minimal biofilm inhibition. Interestingly, the loss of inhibitory activity over time occurred as a result of metal-complexed cahuitamycins forming. While the mechanism of action is currently unknown, Park et al. noted that the cahuitamycins impacted biofilm maturation and not initial attachment. These observations, including the gain of function for biofilm dispersal, suggests that this class of inhibitors primarily impacts biofilm maturation and integrity.
4.2. Auromomycin
In the past decade, high content screening has gained favor as a whole-cell approach. It provides direct measurements of the impact on biofilm formation, such as altered architecture or maturation dynamics. To identify biofilm inhibitors in V. cholerae, Peach et al. used fluorescently tagged V. cholerae rugose variant, which has enhanced biofilm-forming ability due to high c-di-GMP production, in a biofilm image-based screen. A unique marine microbial natural products library from 1248 unique prefractions was screened; the central chromophore of auromomycin (6) exhibited the most significant degree of biofilm inhibition among the lead compounds [116]. (Figure 3) We note that auromomycin has been studied previously as an antitumor natural product that prevented the growth of lymphoblastoma L5178Y cells and that auromomycin also showed antimicrobial activity against Gram-positive and Gram-negative cells [117,118]. The impact of 6 on biofilm formation was further investigated using confocal scanning laser microscopy (CSLM) to understand its effects on biofilm formation. In a dose-dependent manner, 6 altered the appearance of mature biofilm architecture and integrity and reduced the size of microcolonies with a BIC50 of 60.1 µM. Interestingly, Peach et al. found that 6 did not impact the cell growth of V. cholerae or the cell viability of HeLa cells at 250 µM. As 6 is structurally identical to an alkaline degradation product of the auromomycin chromophore, the antimicrobial and antitumor activity likely requires the intact chromophore [119]. It was shown that 6 is unable to disperse preformed biofilms [120], suggesting that 6 acts primarily against the early stages of biofilm formation. 6 is a structurally new class of biofilm inhibitor comprising a benzo[1,4]oxazines core with an exocyclic olefin and does not have cytotoxicity at BIC50 concentrations, making it a desirable inhibitor candidate.
A subsequent study focused on structural characterization and improvement of the auromomycin scaffold. Warner et al. reported the synthesis of 6, along with a series of structurally simplified analogs for SAR studies [121]. The library of 41 simplified analogs was examined for anti-biofilm activity against V. cholerae biofilms in relation to structural modifications. It was discovered that the removal of the exocyclic double bond or adding substituents (8,9) to the double bond was detrimental to activity. This is highlighted by the α, β unsaturated carbonyl that acts as a Michael acceptor with potential involvement in the mechanism of action. Similarly, the N-methyl analog 7 was also found to be inactive, suggesting that the hydrogen bond donor is required for its activity in the active site. The modification of substituents on the ester 11–13 resulted in an increase in biofilm inhibition, whereas its amide counterpart 10 was completely inactive. The lead compound 13 displayed strong biofilm dispersal activity and no bactericidal activity, with a biofilm dispersal concentration (BDC50) value of 13 µM, a BIC50 of 6 µM and no mammalian cell cytotoxicity against HeLa cells up to up to 200 µM. Compound 13 was capable of disrupting V. cholerae biofilms under both static and flow cell conditions, resulting in a seven-fold reduction in biofilm biomass. Additionally, co-dosing of compound 13 with 50 µM erythromycin or azithromycin showed enhanced detachment and subsequent clearance of preformed biofilms, suggesting a synergistic action of a dispersal agent such as 13 and antibiotics. SAR studies, such as that above, are powerful tools in the characterization and improvement of novel inhibitors.
4.3. Skyllamycins
Building upon the success of auromomycin’s discovery, Navarro et al. performed a modified version of the biofilm image-based screen against a fluorescently tagged P. aeruginosa strain [122]. For this screen, a biofilm image-based screen was coupled with an XTT ((2,3-Bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide) assay, which measures cellular metabolic activity via the reduction of XTT. Thus, this screen evaluates the impact of a compound on both biofilm formation and cell viability. The compounds were then sorted into quadrant-based bins based on biofilm inhibition and cellular activity to identify non-antibiotic activity inhibitors. Subsequently, two versions of this screen were used to model the modes of inhibitory action: the biofilm inhibition model (BIM) and the biofilm dispersal model (BDM). Such analysis provides insights into mechanisms of action. It allows the classification of compounds based on their ability to prevent biofilm formation via BIM and their ability to clear preformed biofilms via BDM. The authors focused on compounds corresponding to extracts with low biofilm coverage but with relatively normal cellular activity. They identified an extract containing skyllamycins A–C (14–16) as the most desirable compounds for biofilm inhibition and dispersal (Figure 3). 15 and 16 were found to have 50% effective concentrations (EC50), a term synonymous with BIC50, of 30 and 60 µM, respectively, where 14 had a relatively high EC50 of >250 µM. In all instances tested, 14–16 did not reduce cell activity, indicating that these compounds do not inhibit biofilms through antibiotic-mediated action. While 14 and 16 were unable to function as dispersal agents, 15 could clear preformed biofilms with an EC50 of 60 µM. The skyllamycins belong to a family of non-ribosomal cyclic depsipeptide natural products containing the very rare α-OH-glycine and the added functionality of three β-OH amino acids. Recent developments have led to the successful synthesis of skyllamycins opening the possibility of additional derivative studies for further inhibitory improvement [123]. As the skyllamycins are structurally distinct from quorum sensing mimics, these compounds likely act on a non-quorum sensing pathway, representing the first known class of cyclic depsipeptide biofilm inhibitors/dispersers. Co-dosing experiments of 15 and azithromycin, an antibiotic unable to clear pre-attached biofilms, demonstrated that, in combination, these compounds were able to eliminate surface-associated biofilms and lower cellular metabolic activity. A further study developed a solid phase peptide synthesis protocol and solution phase macrolactamization strategy to access the skyllamycins and four simplified analogs [124]. Interestingly, 14–16 and their deshydroxy analogs are found to have moderate biofilm inhibitory activity, suggesting that these variations are well-tolerated.
4.4. Terrein
Screening methodologies can be targeted towards particular aspects of biofilm formation [125]. This bias provides important contextual clues about the mechanism of inhibition. An example of this approach focused on an extracellular elastase activity, an important biofilm factor for late-stage biofilm maintenance and biomass accumulation in P. aeruginosa, as a readout of biofilm formation [126,127]. By using an elastin-Congo red conjugate, elastase activity can be measured as a function of increasing absorbance as the Congo red dye is released. From a microbial extract library of 12,300 compounds, terrein, (17) a compound isolated from the fungus Aspergillus terreus, was found to decrease elastase activity by 29.1% and 81.1% at concentrations of 30 and 100 µM, respectively, and could reduce biofilm biomass at those concentrations (Figure 3). Discovered in 1935, 17 has documented anti-cancer activity and minor antimicrobial activity; however, these effects were demonstrated at concentrations higher than those tested by Kim et al., suggesting that lower concentrations may allow for biofilm inhibition with minimal antimicrobial action [128,129,130]. 17 decreased elastase production, biofilm matrix production, and biofilm thickness. It was shown that the hydroxyl moieties on terrein are essential for such a decrease. Since 17 is structurally similar to the QS molecule acyl homeserine lactone (AHL), an AHL-based in vitro quorum sensing competition assay was utilized to determine whether terrein can block AHL receptors using homologs of the P. aeruginosa QS receptors LasR and RhlR. From this, it was found that 17 antagonistically inhibited both QS receptors’ ability to bind to their respective AHL compounds and subsequently inhibited QS-regulated genes. As QS and c-di-GMP signaling regulatory circuits regulate each other, exposure to terrein negatively impacted biofilm matrix production, decreased intracellular c-di-GMP, and attenuated virulence in Caenorhabditis elegans and the murine airway infection model [76].
4.5. Ebselen
C-di-GMP is a central regulator of biofilm formation; hence, inhibitors targeting the activity of c-di-GMP metabolizing enzymes and c-di-GMP receptors are of great interest. A recent study used a targeted c-di-GMP receptor and the differential radial capillary action of ligand assay (DRaCALA), which determines the fraction of the ligand bound to a specific protein, to identify competitive inhibitors [131]. This method was utilized to screen the NIH clinical collection 1 library for compounds that could reduce the fraction of bound 32P-c-di-GMP to PelD, a P. aeruginosa c-di-GMP receptor required for the synthesis of the exopolysaccharide Pel [132,133]. Ebselen (18), an organoselenium compound with drug-like properties and antioxidant, anti-inflammatory, and cytoprotective activity, was the only compound found to reduce c-di-GMP binding beyond 3 standard deviations of the positive cutoff. (Figure 3) Commercially purchased 18 and its oxide variant, the selenone analog (19), was able to reduce 32P-c-di-GMP binding to PelD and the P. aeruginosa DGC WspR by 80% and 90%, respectively, although this was not the case for PilZ domain c-di-GMP receptors or PDEs. Furthermore, 18 supplementation altered c-di-GMP dependent phenotypes, such as the motility and biofilm formation, while not impacting P. aeruginosa growth. Based on previous observations of ebselen modifying cysteine residues, Lieberman et al. proposed that 18 covalently modifies cysteine residues in the active site of DGCs. WspR has two cysteines at positions 49 and 240, one of which is adjacent to the RxxD I-site, which is important for c-di-GMP allosteric regulation of the catalytic domain [134].
4.6. DI-3
In another study targeting c-di-GMP metabolizing enzymes, Sambanthamoorthy et al. utilized a c-di-GMP inducible promoter, the upstream region of the V. cholerae gene VC1673, fused to a luciferase operon as a transcriptional reporter to identify compounds that decrease luminescence as a function of decreased c-di-GMP levels [135]. Over 66,000 unique compounds and natural product extracts were screened for a reduction in luminescence, which is also an indirect readout of decreased cellular-di-GMP levels. This study identified 184 compounds as being highly effective in reducing luminescence. Subsequently, Sambanthamoorthy et al. tested these compounds for their ability to inhibit the activity of a select group of purified DGCs in an in vitro analysis and impact on cell growth. The lead compound DI-3 (20) was found to inhibit the V. cholerae DGC VC2370 and the P. aeruginosa DGC WspR in a dose-dependent manner with in vitro IC50 values of 1.0 µM and 17.83 µM, respectively, without negatively impacting cell growth. (Figure 3) 20 is a linear molecule containing a biphenyl amine and one amide bond functional group and is in compliance with the Lipinski rule of five. All the lead compounds from this screen are linear and have a similar steric bulk, and 13 to 15 have the longest countable atomic linkages end to end. The presence of aromatic rings at both ends of the inhibitor suggests the folding of the molecule in half to achieve pi-pi stacking. This mimics pi-pi stacking of c-di-GMP to form higher-order multimers, and these multimers are required for binding to RXXD allosteric sites. Supplementation of 20 was able to reduce V. cholerae biofilm biomass, which was attributed to a decrease in c-di-GMP, as measured by mass spectrometry quantification. Using quantitative crystal violet staining on minimum-biofilm-eliminating concentration (MBEC) plates, the BIC50 for V. cholerae was found to be 26.2 µM.
4.7. AA-861 and Parthenolide
Bacteria with increased ability to produce biofilm matrix components form biofilms with distinct corrugated patterns resulting from matrix production and assembly [39,136,137,138]. This readily screenable phenotype of biofilm formation is used to identify biofilm inhibitors that can visibly impact these phenotypes. In one such example, a collection of bioactive compounds were screened for their impact on B. subtilis ability to form corrugated biofilms at the air–liquid interface, also known as pellicles [139]. The biofilm matrix of B. subtilis is composed primarily of exopolysaccharides (eps) and amyloid-like fibers formed from the protein TasA. This approach’s advantage is that wrinkles are a distinguishable critical feature and can be used to screen for molecules with anti-EPS and/or anti-amyloid activity. From the 480 compound BIOMOL–ICCB collection, two compounds were identified as having strong biofilm inhibitory activity and no impact on cell growth: AA-861 (21), a benzoquinone derivative with anti-inflammatory activity, and parthenolide (22), a germacrane sesquiterpene lactone obtained from the feverfew plant (Figure 3). By analyzing biofilm inhibition in the absence of one of the two major biofilm components, it was found that 21 and 22 inhibited biofilm formation via TasA. Further investigation of in vitro TasA polymerization, via a thioflavin T in vitro assay (fluorescence increases as thioflavin T binds to the β sheet-rich fold found in amyloids), showed that supplementation of either 21 or 22 at 50 µM to TasA led to a drastic reduction in fluorescence signal accumulation, indicating that these compounds inhibited TasA polymerization.
Furthermore, 21 and 22 had an additive effect in preventing TasA from forming amyloid-like fibers and could reduce biofilm biomass for B. cereus and E. coli. Though the mode of inhibition in those species is unclear, E. coli produces Curli, an amyloid fiber, and B. cereus has a homolog of TasA [48,139]. These findings suggest that these compounds may function as broad inhibitors of amyloid polymerization. Supplement of 100 µM of 22 was also able to disrupt preformed pellicles, indicating that it has dispersal qualities in addition to inhibition. The mechanism of action by which 21 and 22 reduce amyloidogenesis largely remains unclear. However, Romero et al. proposed that direct interaction of the inhibitors with different forms of the amyloid proteins could hamper the polymerization of the fiber.
Besides impacting amyloid-like fiber assembly, 22 affected the expression of quorum sensing systems and reduced the production of biofilm matrix components for P. aeruginosa [140]. Additionally, in silico analysis showed that 22 has a strong potential of binding to the ligand-binding domain of LasR, offering a potential explanation for biofilm inhibition in P. aeruginosa. 22 may be a useful inhibitor, as it can inhibit biofilm formation among a wide range of bacteria and through different mechanisms and targets.
4.8. Ellagic Acid Carbohydrate Conjugates
One of the earlier examples of biofilm inhibitors is ellagic acid (23), a plant-derived compound [141]. (Figure 3) 23 and other polyphenolic compounds with a gallic acid moiety were shown to inhibit biofilm formation when investigating plant extracts for anti-quorum sensing activity. The root extract of Rubus ulmifolius Schott, a wild shrub from the Mediterranean used in traditional medicine, specifically the fraction 220D-F2, was found to contain 23 and its derivatives [142]. This fraction displayed biofilm inhibitory activity against S. aureus and increased biofilms’ susceptibility to daptomycin and clindamycin. Using liquid chromatography coupled to UV detection and tandem mass spectrometry to identify the chemical contents of 220D-F2 revealed 23 (EA) and several ellagic acid derivatives (EADs) or sapogenin-related compounds [143]. A subsequent study reported that fraction 220D-F2 kills Streptococcus pneumonia planktonic cells and pneumococcal biofilms, and that 200 µg/mL of 220DF2 markedly decreased preformed pneumococcal biofilms. At the same time, a higher amount (i.e. 800 µg/mL of 220D-F2) was necessary to kill S. pneumonia biofilms formed on human pharyngeal cells. To identify the active component in 220D-F2, Fontaine et al. synthesized compounds 24 and 26 based on the molecular weight and ubiquity of rhamnose and xylose in plants [144]. It was hypothesized that antibiofilm activity associated with 220D-F2 is due to the 3-α-rha-EA 26 with the sugar moiety playing a pivotal role by making critical interactions with cellular components, thereby increasing anti-biofilm activity.
Recent studies reported the synthesis of ellagic acid carbohydrate derivatives 24–26 through a copper-mediated Ullmann coupling followed by a phenolic O-glycosylation [145]. The ellagic acid glycosides 24 and 25 displayed antibiofilm properties at concentrations of 512 µg/mL against Streptococcus agalactaie (Group B Streptococcus, GBS). The mechanism of action of ellagic acid glycosides remains mostly unknown, but scanning electron microscopy (SEM) has revealed that it limits initial adhesion in biofilm formation in GBS, and the biofilm to biomass ratios indicate that 24 and 25 impact biofilm formation but not planktonic cell growth.
5. In Silico Discovery
In silico-based studies are being increasingly utilized to identify biofilm inhibitors. By nature, this allows for the screening of extensive libraries without initial wet lab work. There are multiple approaches to in silico analysis, such as scaffold screens, analog development, and docking studies, which affords a degree of customization when employing computational screening techniques while offering opportunities to bias the screen towards particular targets [146] (Figure 4).
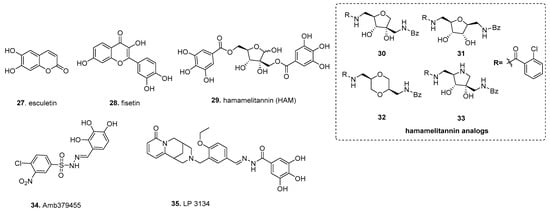
Figure 4.
In silico-based discovery of biofilm inhibitors.
5.1. Fisetin (from Ellagic Acid)
Building upon direct testing of gallic acid derivatives, in silico structure-based virtual screening using ellagic acid 23, the initial scaffold discovered more potent biofilm inhibitors with broad species activity. Using multigenerational screening with a MAACC fingerprint database of the 57,346 member Chinese Natural Product Database, esculetin (27) was identified in the first round of screening, and fisetin (28), a plant flavonol from the flavonoid group of polyphenols, was identified in the second round (Figure 4) [147]. Both compounds inhibited biofilm formation of S. aureus; however, 28 had a substantially lower inhibitory dose, inhibiting biofilm formation by 90% at 16 µg/mL, whereas esculetin reduced biofilm formation by 77% with 128 µg/mL. The concentrations tested were deemed as being well below antibacterial levels. Additionally, 27 only exhibited inhibitory activity against S. aureus by inhibiting biofilm maturation, whereas 28 inhibited biofilm formation for S. aureus and multiple strains of Streptococcus dysgalactiae while also impacting biofilm initiation and development. With this approach, one can utilize a well-established inhibitor as a scaffold, and through multiple generations of in silico screening can enrich for potent biofilm inhibitors.
5.2. Hamamelitannin
In silico-based studies can be used to identify analogs of substrates for bacterial systems, allowing for the discovery of competitive inhibitors. An example of this approach is hamamelitannin (29), which targets the S. aureus RAP/TRAP quorum-sensing (QS) system (Figure 4). This QS system uses the secreted autoinducer peptide (AIP) “RNAIII-activating protein” (RAP) to induce phosphorylation of the “target of RAP” (TRAP) to impact gene expression as a function of cell density [148]. Interestingly, non-self AIPs have shown the ability to inhibit quorum sensing through the RAP/TRAP system by antagonizing RAP binding to TRAP [148]. Based on this, Kiran et al. sought to discover RIP analogs to inhibit biofilm formation using the ribosomal protein L2, an ortholog of TRAP, to develop a model of RIP, which was then used to screen 300,000 compounds from the available chemicals database (ACD) [149]. From this, 29 was identified as a non-peptide analog of RIP that exhibited no growth impact on S. aureus, but could compete against RAP, reducing surface attachment and virulence in the rat device-associated biofilm model. A later study found that 29 increases S. aureus biofilm susceptibility to antibiotics through agr and the traP QS system [150]. This was attributed to a decrease in cell wall thickness and a decrease in eDNA release through inhibition of TraP, resulting in inhibition of antibiotic-associated virulence and in vivo virulence of C. elegans and the mouse mammary glands infection model. Detailed medicinal chemistry efforts around the HAM scaffold found that analog 30 modulates quorum sensing as a potential antibiotic against S. aureus without showing cytotoxicity [151]. The EC50 of 30 is 0.389 µM, an approximately 400-fold improvement compared to 29 (EC50 = 145.5 µm). The key structural difference between the 29 and 30 includes the central tetrahydrofuran core and benzamides replace the labile ester. Follow-up studies by the same group centered around designing different hamamelitannin analogs by using ligand-based virtual screening where they modified only the hamamelose central core to different heterocyclic cores [152]. The 2,5-anhydro-D-allitol 31 and dioxane-derived 32 showed similar activity to that of lead analog 30 in the combination treatment setup. Finally, the pyrrolidine-derived analog 33 with different N-alkylation was found to be completely inactive in inhibiting S. aureus biofilm formation [153].
5.3. Amb379455
Another option for screening is through in silico docking of compounds into the three-dimensional structure of the intended target, which allows screening extensive virtual libraries for compounds that likely can bind with specificity [154]. In one study, a diguanylate cyclase was used as a target protein. Fernicola et al. created a 14-point pharmacophore hypothesis of Caulobacter crescentus’s PleD, representing one of the first DGCs whose crystal structure is known. It is a common template used for DGC-based in silico docking, in complex with a GTP analog to define docking parameters to identify competitive antagonists for the active site [155,156]. A 3D representation of these docking parameters was presented in Figure 1 of Fernicola et al. This led to the identification of Amb379455 (34), a pyrogallol unit containing a sulfonohydrazide functional group, as a potential inhibitor from the 2.3 × 107 member ZINC database (Figure 4). The close structural analysis with other analogs suggests that phenols and nitro position is crucial for the activity. As this in silico screen utilized a known DGC, the Fernicola et al. investigated whether Amb379455 34 can specifically impact PleD. Not only was Amb379455 able to inhibit PleD with a 50% enzymatic inhibitory concentration (IC50) of 11.1 ± 1.1 µM, in a dose-dependant manner, it inhibited the P. aeruginosa DGCs WspR and YfiN, which are critical in c-di-GMP signaling and biofilm formation [157,158]. As YfiN lacks an inhibitory site (I-sites), a common feature among DGCs needed for allosteric inhibition of the DGC via the substrate c-di-GMP, the mechanism of inhibition for 34 is likely specific for the active site of these DGCs. Based on the pharmacophore hypothesis, 34 likely targets the active sites of both PleD and YfiN; the phenolic OH, which coordinates with magnesium, whereas sulphonamide, nitro functional groups interact with N335 and R366 residue, respectively. This notion is supported by the fact that 34 can competitively displace MANT-GTP from YfiN. This example illustrates that in silico discovery can achieve large gains in structural knowledge in tandem with inhibitor discovery, making this a valuable discovery tool.
5.4. LP 3134
Another study used the same target, C. crescentus’s PleD, in complex with c-di-GMP, to identify an active site’s inhibitor [159]. Sambanthamoorthy et al. used the active site and residues with GMP interactions to screen 15,000 compounds from a guanine/oroidin moiety-focused library which are illustrated in their Figure 1. Lead compounds were further refined with MOE software based on electrostatic energy, binding restrictions, and energy minimization to generate the final PleD-compound complexes. Four compounds were found to decrease biofilm formation, without impacting cell growth, in P. aeruginosa and A. baumannii using both crystal violet staining and confocal microscopy, but only LP-3134 (35), a heterocyclic compound containing N-benzylidenebenzohydrazide and pyridine-2 one fused with bicyclic amine moiety, was able to disperse intact biofilms for both P. aeruginosa and A. baumanii. (Figure 4) Based on the in silico complex of 35 and PleD, three of the four GMP-PleD hydrogen bonds are satisfied by the phenolic OH from N-benzylidenebenzohydrazide form. The fourth hydrogen occurs between the singular oxygen of the pyridine-2 fused ring and the amide backbone of R366. Additionally, 35 was found to inhibit surface attachment for P. aeruginosa and prevent biofilm formation on silicone catheters. Sambanthamoorthy et al. also reported that 35 had an IC50 of 44.9 µM against the P. aeruginosa DGC WpsR.
6. Therapeutic Drug Conjugates
Beyond the discovery of novel biofilm inhibitors, there has been increasing interest in the advances of therapeutic drug conjugates for cancer and drug delivery and utilizing that class of therapeutics to target biofilms [160,161]. Here, antimicrobials can be linked to antibodies, peptides, or various compounds to improve (i) target specificity, (ii) penetration of the biofilm matrix, or (iii) condition-specific activity [162,163,164,165,166,167,168]. This approach can reduce the efficacy of biofilms’ protective and resistant nature when challenged by traditional antibiotics and has found success in recent studies and clinical trials [169,170,171].
An early example of therapeutic drug conjugates to target biofilms utilized chitosan, the N-deacetylated derivative of chitin, as a delivery method for the aminoglycoside antibiotic streptomycin to target cell membranes and penetrate preformed biofilms [172,173]. (Figure 5) The conjugate 36 was synthesized by the reduction of Schiff base formed by the reaction of aldehyde from streptomycin and amine from chitosan. Dose-dependent analysis indicated that the C-S conjugate 36 (at 0.125, 0.25, and 0.5 mg/mL) was more efficient in disruption of L. monocytogenes biofilms than chitosan, streptomycin, or a mixture of both. The anti-biofilm efficacy of C- S conjugate 36 was effective against Gram-positive organisms (L. innocua, L. welshimeri, E. faecalis and S. aureus) but not Gram-negative organisms (P. aeruginosa, S. typhimurium).
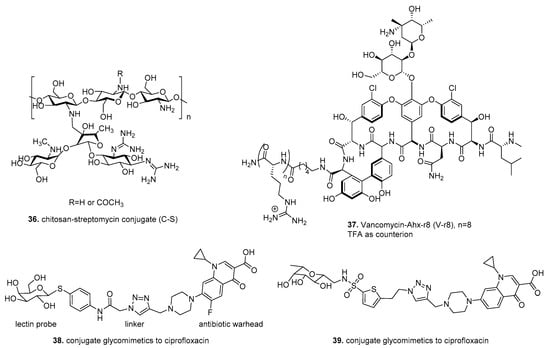
Figure 5.
Therapeutic drug conjugates.
Another study designed to improve antibiotic penetration into biofilms conjugated the antibiotic vancomycin to a molecular transporter (MoTr), which has been previously shown to penetrate cell membranes [169,174]. Antonoplis et al. utilized the guanidinium-rich MoTr D-octoarginine, which has 30-fold higher MIC than vancomycin for planktonic cells, and alone was ineffective against biofilm grown cells. The standard EDC condition used for coupling of unprotected NH2- Ahx−r8 to the C terminus of vancomycin to produce V−r8 (37) (Figure 5). Vancomycin-octoarginine (V-r8) was markedly more effective at eradicating methicillin-sensitive and methicillin-resistant S. aureus biofilm-grown cells compared to unconjugated vancomycin. However, V-r8 did not improve vancomycin’s ability to kill planktonic cells suggesting that V-r8 primarily aids in biofilm penetration, not cell penetration. Additional studies using S. aureus biofilms showed that 37 killed 91.6% of biofilm-associated cells, while unconjugated vancomycin resulted in only 35% killing. In a murine wound infection model, 37 eliminated 97% of biofilm-associated MRSA without acute dermal toxicity.
In contrast to the previous drug conjugates, which improved cell and biofilm penetration, Meiers et al. explored the conjugation of ciprofloxacin to glycomimetics to target LecA LecB, extracellular biofilm-related virulence factors of P. aeruginosa [175]. This strategy targets ciprofloxacin delivery to the infection site, reducing off-target effects and gaining antibiotic resistance. Sulfonamide-capped mannosides, and C-glycosides combining pharmacophores of its natural ligands, fucose, and mannose showed excellent binding affinity against LecB and inhibited in vitro biofilm formation [176].
Drug conjugates 38, 39 were synthesized using copper-catalyzed [3 + 2] cycloaddition of terminal azides from lectin probe and terminal alkyene from the antibiotic ciprofloxacin. (Figure 5) Utilizing P. aeruginosa biofilms formed on pegs, the two lectin-targeting conjugates (38 and 39) were compared with unconjugated ciprofloxacin at 100 μM. The lectin-targeted conjugates 38, 39 reached higher concentrations in the bacterial biofilm than the unmodified ciprofloxacin when analyzed with LC/MS. In vitro, ADMET data, which determine pharmacological properties of compounds, showed 38, 39 conjugates are metabolically stable and showed no cytotoxicity at 100 μM after 48 h incubation against a human embryonic kidney cell line (HEK 293) and adenocarcinoma human alveolar basal epithelial cells (A549).
Collectively, these studies revealed that therapeutic drug conjugates present desirable therapeutic options utilizing a potent antimicrobial warhead with a higher specific delivery method.
7. Conclusions
Biofilm formation is a beneficial growth mode that protects microorganisms from antimicrobial agents and immune responses, making biofilm-associated cells challenging to eradicate. Thus, there is a need for biofilm inhibitors that can render these bacteria susceptible to treatment strategies and removal. The discovery of novel inhibitors is an essential step in reaching this goal, and numerous approaches have proven successful. This review covered the two major approaches to inhibitor discovery, screening-based and in silico, and discussed structural information obtained through SAR strategies. We have provided examples of high-throughput screening strategies using "whole-cell" and targeted approaches that identified cahuitamycins A-E, auromomycin, skyllamycins A-C, and terrein.
Additionally, we discussed targeted/pathway screens, which discovered ebselen, DI-3, AA-861, parthenolide, and ellagic acid. Furthermore, we examined three distinct in silico discovery methods that identified fisetin, hamamelitannin, Amb379455, and LP 3134 as biofilm inhibitors. Finally, we explored the realm of drug conjugates that utilize antibiotics linked to compounds that aid in cell/biofilm penetration and target specificity such as chitosan-streptomycin, V-r8, and the glycomimetic-ciprofloxacin conjugates. Though not expansive, these examples each present a unique approach to compound discovery with particular advantages, methods for inhibitor characterization, and current SAR knowledge. As our understanding of mechanisms of biofilm formation improves, we anticipate seeing an expansion of biofilm inhibitor studies beyond the typical target of quorum sensing and into other critical biofilm regulatory systems such as c-di-GMP signaling. Similarly, mechanistic understanding of biofilm formation will aid in the refinement and development of in silico-based and therapeutic drug conjugate studies. Finally, the integration of in vitro model systems of biofilm-associated diseases, such as organoid models, in the development of screening platforms will lead to the identification of novel biofilm inhibitors.
Author Contributions
The M.A.T., R.D.S., J.B.M. and F.H.Y. contributed equally to this review. All authors have read and agreed to the published version of the manuscript.
Funding
We acknowledge NIH grant R01AI102584 for supporting this work.
Acknowledgments
Figures in this review were created with BioRender.com and Chemdraw.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ADMET | Absorption, distribution, metabolism, and excretion properties and toxicities which are useful in evaluating pharmecuetical properties of prospective therapeutics. |
| Antibacterial | A compound that kills bacteria or prevents bacterial growth. |
| BDC50 | The concentration at which 50% of the preformed biofilm is dispersed. |
| BIC50 | The concentration at which 50% of biofilm formation is inhibited. |
| Biofilm inhibitor | A compound that inhibits or negatively impacts biofilm formation. This includes the prevention of biofilm formation as well as dispersal and disruption of preformed biofilms. |
| DGC | Diguanylate cyclase |
| EC50 | The concentration at which treatment is 50% effective. |
| EDC condition | 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) is used as a carboxyl activating agent to form amide bonds in coupling reactions. |
| IC50 | The concentration at which enzymatic activity is 50% inhibited. |
| MBEC | Minimal biofilm eradication concentration. |
| MIC | Minimal inhibitory concentration. |
| MoTr | Cell associated or cell-penetrating molecular transporter. |
| PDE | Phosphodiesterase |
| QS | Quorum sensing |
References
- Flemming, H.C.; Wuertz, S. Bacteria and Archaea on Earth and Their Abundance in Biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Bjarnsholt, T. The Role of Bacterial Biofilms in Chronic Infections. APMIS Suppl. 2013, 121, 1–51. [Google Scholar] [CrossRef]
- Stewart, P.S. Mechanisms of Antibiotic Resistance in Bacterial Biofilms. Int. J. Med. Microbiol. 2002, 292, 107–113. [Google Scholar] [CrossRef]
- Tamayo, R.; Patimalla, B.; Camilli, A. Growth in a Biofilm Induces a Hyperinfectious Phenotype in Vibrio cholerae. Infect. Immun. 2010, 78, 3560–3569. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Hernandez, A.L.; Depas, W.H.; Park, J.H.; Teschler, J.K.; Hartmann, R.; Jeckel, H.; Drescher, K.; Beyhan, S.; Newman, D.K.; Yildiz, F.H. Upregulation of Virulence Genes Promotes Vibrio cholerae Biofilm Hyperinfectivity. Proc. Natl. Acad. Sci. USA 2020, 117, 11010–11017. [Google Scholar] [CrossRef]
- González, J.F.; Hahn, M.M.; Gunn, J.S. Chronic Biofilm-Based Infections: Skewing of the Immune Response. Pathog. Dis. 2018, 76, 1–7. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T.F. Molecular Mechanisms of Biofilm-Based Antibiotic Resistance and Tolerance in Pathogenic Bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Gunn, J.S.; Bakaletz, L.O.; Wozniak, D.J. What’s on the Outside Matters: The Role of the Extracellular Polymeric Substance of Gram-Negative Biofilms in Evading Host Immunity and as a Target for Therapeutic Intervention. J. Biol. Chem. 2016, 291, 12538–12546. [Google Scholar] [CrossRef] [PubMed]
- Crabbé, A.; Jensen, P.Ø.; Bjarnsholt, T.; Coenye, T. Antimicrobial Tolerance and Metabolic Adaptations in Microbial Biofilms. Trends Microbiol. 2019, 27, 850–863. [Google Scholar] [CrossRef]
- Lewis, K. Riddle of Biofilm Resistance. Antimicrob. Agents Chemother. 2001, 45, 999–1007. [Google Scholar] [CrossRef]
- Kirketerp-Møller, K.; Zulkowski, K.; James, G. Chronic Wound Colonization, Infection, and Biofilms. In Biofilm Infections; Springer: New York, NY, USA, 2011; pp. 11–24. [Google Scholar] [CrossRef]
- Hidron, A.I.; Edwards, J.R.; Patel, J.; Horan, T.C.; Sievert, D.M.; Pollock, D.A.; Fridkin, S.K.; National Healthcare Safety Network Team; Participating National Healthcare Safety Network Facilities. Antimicrobial-Resistant Pathogens Associated With Healthcare-Associated Infections: Annual Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect. Control. Hosp. Epidemiol. 2008, 29, 996–1011. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Bjarnsholt, T. Risk Factors for Chronic Biofilm-Related Infection Associated with Implanted Medical Devices. Clin. Microbiol. Infect. 2020, 26, 1034–1038. [Google Scholar] [CrossRef]
- Bjarnsholt, T.; Ciofu, O.; Molin, S.; Givskov, M.; Høiby, N. Applying Insights from Biofilm Biology to Drug Development-Can a New Approach Be Developed? Nat. Rev. Drug Discov. 2013, 12, 791–808. [Google Scholar] [CrossRef]
- Ventola, C.L. The Antibiotic Resistance Crisis: Causes and Threats. P T. 2015, 40, 277–283. [Google Scholar] [PubMed]
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Allan, R.N.; Howlin, R.P.; Stoodley, P.; Hall-Stoodley, L. Targeting Microbial Biofilms: Current and Prospective Therapeutic Strategies. Nat. Rev. Microbiol. 2017, 15, 740–755. [Google Scholar] [CrossRef]
- Ghosh, A.; Jayaraman, N.; Chatterji, D. Small-Molecule Inhibition of Bacterial Biofilm. ACS Omega 2020, 5, 3108–3115. [Google Scholar] [CrossRef]
- Qvortrup, K.; Hultqvist, L.D.; Nilsson, M.; Jakobsen, T.H.; Jansen, C.U.; Uhd, J.; Andersen, J.B.; Nielsen, T.E.; Givskov, M.; Tolker-Nielsen, T. Small Molecule Anti-Biofilm Agents Developed on the Basis of Mechanistic Understanding of Biofilm Formation. Front. Chem. 2019, 7, 742. [Google Scholar] [CrossRef]
- Cho, K.H.; Tryon, R.G.; Kim, J.H. Screening for Diguanylate Cyclase (DGC) Inhibitors Mitigating Bacterial Biofilm Formation. Front. Chem. 2020, 8, 264. [Google Scholar] [CrossRef]
- Verderosa, A.D.; Totsika, M.; Fairfull-Smith, K.E. Bacterial Biofilm Eradication Agents: A Current Review. Front. Chem. 2019, 7, 824. [Google Scholar] [CrossRef] [PubMed]
- Melander, R.; Basak, A.; Melander, C. Natural Products as Inspiration for the Development of Bacterial Antibiofilm Agents. Nat. Prod. Rep. 2020, 37, 1454–1477. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Xu, J.; Rapior, S.; Jeewon, R.; Lumyong, S.; Niego, A.G.T.; Abeywickrama, P.D.; Aluthmuhandiram, J.V.S.; Brahamanage, R.S.; Brooks, S.; et al. The Amazing Potential of Fungi: 50 Ways We Can Exploit Fungi Industrially. Fungal Divers. 2019, 97, 1–136. [Google Scholar] [CrossRef]
- Utada, A.S.; Bennett, R.R.; Fong, J.C.N.; Gibiansky, M.L.; Yildiz, F.H.; Golestanian, R.; Wong, G.C.L. Vibrio cholerae Use Pili and Flagella Synergistically to Effect Motility Switching and Conditional Surface Attachment. Nat. Commun. 2014, 5, 4913. [Google Scholar] [CrossRef] [PubMed]
- Gibiansky, M.L.; Conrad, J.C.; Jin, F.; Gordon, V.D.; Motto, D.A.; Mathewson, M.A.; Stopka, W.G.; Zelasko, D.C.; Shrout, J.D.; Wong, G.C.L. Bacteria Use Type IV Pili to Walk Upright and Detach from Surfaces. Science 2010, 330, 197. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.C.; Zamorano-Sánchez, D.; Pagliai, F.A.; Park, J.; Id, K.A.F.; Lee, C.K.; Kitts, G.; Rose, C.B.; Bilotta, E.M.; Wong, G.C.L.; et al. Reciprocal C-di-GMP Signaling: Incomplete Flagellum Biogenesis Triggers c-di-GMP Signaling Pathways That Promote Biofilm Formation. PLoS Genet. 2020, 16, e1008703. [Google Scholar] [CrossRef]
- Belas, R. Biofilms, Flagella, and Mechanosensing of Surfaces by Bacteria. Trends Microbiol. 2014, 22, 517–527. [Google Scholar] [CrossRef]
- Guttenplan, S.B.; Kearns, D.B. Regulation of Flagellar Motility during Biofilm Formation. FEMS Microbiol. Rev. 2013, 37, 849–871. [Google Scholar] [CrossRef]
- Laverty, G.; Gorman, S.P.; Gilmore, B.F. Biomolecular Mechanisms of Staphylococcal Biofilm Formation. Future Microbiol. 2013, 8, 509–524. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal Biofilms. Microbiol. Spectr. 2018, 6, 1–17. [Google Scholar] [CrossRef]
- Le, K.Y.; Park, M.D.; Otto, M. Immune Evasion Mechanisms of Staphylococcus epidermidis Biofilm Infection. Front. Microbiol. 2018, 9, 359. [Google Scholar] [CrossRef]
- Ageorges, V.; Monteiro, R.; Leroy, S.; Burgess, C.M.; Pizza, M.; Chaucheyras-Durand, F.; Desvaux, M. Molecular Determinants of Surface Colonisation in Diarrhoeagenic Escherichia coli (DEC): From Bacterial Adhesion to Biofilm Formation. FEMS Microbiol. Rev. 2020, 44, 314–350. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.; Conrad, J.C.; Gibiansky, M.L.; Wong, G.C.L. Bacteria Use Type-IV Pili to Slingshot on Surfaces. Proc. Natl. Acad. Sci. USA 2011, 108, 12617–12622. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Tseng, B.S.; Beckerman, B.; Jin, F.; Gibiansky, M.L.; Harrison, J.J.; Luijten, E.; Parsek, M.R.; Wong, G.C.L. Psl Trails Guide Exploration and Microcolony Formation in Pseudomonas aeruginosa Biofilms. Nature 2013, 497, 388–391. [Google Scholar] [CrossRef]
- Maier, B.; Wong, G.C.L. How Bacteria Use Type IV Pili Machinery on Surfaces. Trends Microbiol. 2015, 23, 775–788. [Google Scholar] [CrossRef] [PubMed]
- Jackson, K.D.; Starkey, M.; Kremer, S.; Parsek, M.R.; Wozniak, D.J. Identification of Psl, a Locus Encoding a Potential Exopolysaccharide That Is Essential for Pseudomonas aeruginosa PAO1 Biofilm Formation. J. Bacteriol. 2004, 186, 4466–4475. [Google Scholar] [CrossRef]
- Hobley, L.; Ostrowski, A.; Rao, F.V.; Bromley, K.M.; Porter, M.; Prescott, A.R.; MacPhee, C.E.; Van Aalten, D.M.F.; Stanley-Wall, N.R. BslA Is a Self-Assembling Bacterial Hydrophobin That Coats the Bacillus subtilis Biofilm. Proc. Natl. Acad. Sci. USA 2013, 110, 13600–13605. [Google Scholar] [CrossRef]
- Zogaj, X.; Nimtz, M.; Rohde, M.; Bokranz, W.; Römling, U. The Multicellular Morphotypes of Salmonella Typhimurium and Escherichia coli Produce Cellulose as the Second Component of the Extracellular Matrix. Mol. Microbiol. 2001, 39, 1452–1463. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, F.H.; Schoolnik, G.K. Vibrio cholerae O1 El Tor: Identification of a Gene Cluster Required for the Rugose Colony Type, Exopolysaccharide Production, Chlorine Resistance, and Biofilm Formation. Proc. Natl. Acad. Sci. USA 1999, 96, 4028–4033. [Google Scholar] [CrossRef]
- Hobley, L.; Harkins, C.; Macphee, C.E.; Stanley-Wall, N.R. Giving Structure to the Biofilm Matrix: An Overview of Individual Strategies and Emerging Common Themes. FEMS Microbiol. Rev. 2015, 39, 649–669. [Google Scholar] [CrossRef]
- Whitchurch, C.B.; Tolker-Nielsen, T.; Ragas, P.C.; Mattick, J.S. Extracellular DNA Required for Bacterial Biofilm Formation. Science 2002, 295, 1487. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.J.; Jarrod Smith, T.; Sondermann, H.; O’Toole, G.A. From Input to Output: The Lap/c-di-GMP Biofilm Regulatory Circuit. Annu. Rev. Microbiol. 2020, 74, 607–631. [Google Scholar] [CrossRef] [PubMed]
- Kitts, G.; Giglio, K.M.; Zamorano-Sánchez, D.; Park, J.H.; Townsley, L.; Cooley, R.B.; Wucher, B.R.; Klose, K.E.; Nadell, C.D.; Yildiz, F.H.; et al. A Conserved Regulatory Circuit Controls Large Adhesins in Vibrio cholerae. mBio 2019, 10, e02822-19. [Google Scholar] [CrossRef]
- Jones, C.J.; Wozniaka, D.J. Psl Produced by Mucoid Pseudomonas aeruginosa Contributes to the Establishment of Biofilms and Immune Evasion. mBio 2017, 8, e00864-17. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, D.P.; Matwichuk, M.L.; Townsend, D.O.; Reichhardt, C.; Lamba, D.; Wozniak, D.J.; Parsek, M.R. The Pseudomonas aeruginosa Lectin LecB Binds to the Exopolysaccharide Psl and Stabilizes the Biofilm Matrix. Nat. Commun. 2019, 10, 2183. [Google Scholar] [CrossRef] [PubMed]
- Reichhardt, C.; Wong, C.; da Silva, D.P.; Wozniak, D.J.; Parsek, M.R. CdrA Interactions within the Pseudomonas aeruginosa Biofilm Matrix Safeguard It from Proteolysis and Promote Cellular Packing. mBio 2018, 9, e01376-18. [Google Scholar] [CrossRef]
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel Is a Cationic Exopolysaccharide That Cross-Links Extracellular DNA in the Pseudomonas aeruginosa Biofilm Matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358. [Google Scholar] [CrossRef]
- Jain, N.; Chapman, M.R. Bacterial Functional Amyloids: Order from Disorder. Science 2019, 295, 954–960. [Google Scholar] [CrossRef] [PubMed]
- Berk, V.; Fong, J.C.N.; Dempsey, G.T.; Develioglu, O.N.; Zhuang, X.; Liphardt, J.; Yildiz, F.H.; Chu, S. Molecular Architecture and Assembly Principles of Vibrio cholerae Biofilms. Science 2012, 337, 236–239. [Google Scholar] [CrossRef] [PubMed]
- Colvin, K.M.; Gordon, V.D.; Murakami, K.; Borlee, B.R.; Wozniak, D.J.; Wong, G.C.L.; Parsek, M.R. The Pel Polysaccharide Can Serve a Structural and Protective Role in the Biofilm Matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011, 7, e1001264. [Google Scholar] [CrossRef]
- Earl, C.; Arnaouteli, S.; Bamford, N.C.; Porter, M.; Sukhodub, T.; MacPhee, C.E.; Stanley-Wall, N.R. The Majority of the Matrix Protein TapA Is Dispensable for Bacillus subtilis Colony Biofilm Architecture. Mol. Microbiol. 2020, 114, 920–933. [Google Scholar] [CrossRef]
- Stewart, P.S.; Franklin, M.J. Physiological Heterogeneity in Biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef]
- Walters, M.C.; Roe, F.; Bugnicourt, A.; Franklin, M.J.; Stewart, P.S. Contributions of Antibiotic Penetration, Oxygen Limitation, and Low Metabolic Activity to Tolerance of Pseudomonas aeruginosa Biofilms to Ciprofloxacin and Tobramycin. Antimicrob. Agents Chemother. 2003, 47, 317–323. [Google Scholar] [CrossRef]
- Jenal, U.; Reinders, A.; Lori, C. Cyclic Di-GMP: Second Messenger Extraordinaire. Nat. Rev. Microbiol. 2017, 15, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Conner, J.G.; Zamorano-Sánchez, D.; Park, J.H.; Sondermann, H.; Yildiz, F.H. The Ins and Outs of Cyclic Di-GMP Signaling in Vibrio cholerae. Curr. Opin. Microbiol. 2017, 36, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Hengge, R. Linking Bacterial Growth, Survival, and Multicellularity—Small Signaling Molecules as Triggers and Drivers. Curr. Opin. Microbiol. 2020, 55, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Valentini, M.; Filloux, A. Multiple Roles of C-di-GMP Signaling in Bacterial Pathogenesis. Annu. Rev. Microbiol. 2019, 8, 387–406. [Google Scholar] [CrossRef]
- Orr, M.W.; Galperin, M.Y.; Lee, V.T. Sustained Sensing as an Emerging Principle in Second Messenger Signaling Systems. Curr. Opin. Microbiol. 2016, 34, 119–126. [Google Scholar] [CrossRef]
- Papenfort, K.; Bassler, B.L. Quorum Sensing Signal-Response Systems in Gram-Negative Bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Jamal, S.B.; Hassan, S.S.; Carvalho, P.V.S.D.; Almeida, S.; Barh, D.; Ghosh, P.; Silva, A.; Castro, T.L.P.; Azevedo, V. Two-Component Signal Transduction Systems of Pathogenic Bacteria as Targets for Antimicrobial Therapy: An Overview. Front. Microbiol. 2017, 8, 1878. [Google Scholar] [CrossRef]
- Hirakawa, H.; Kurushima, J.; Hashimoto, Y.; Tomita, H. Progress Overview of Bacterial Two-Component Regulatory Systems as Potential Targets for Antimicrobial Chemotherapy. Antibiotics 2020, 9, 635. [Google Scholar] [CrossRef]
- Fong, J.; Yuan, M.; Jakobsen, T.H.; Mortensen, K.T.; Delos Santos, M.M.S.; Chua, S.L.; Yang, L.; Tan, C.H.; Nielsen, T.E.; Givskov, M. Disulfide Bond-Containing Ajoene Analogues As Novel Quorum Sensing Inhibitors of Pseudomonas aeruginosa. J. Med. Chem. 2017, 60, 215–227. [Google Scholar] [CrossRef] [PubMed]
- O’Loughlin, C.T.; Miller, L.C.; Siryaporn, A.; Drescher, K.; Semmelhack, M.F.; Bassler, B.L. A Quorum-Sensing Inhibitor Blocks Pseudomonas aeruginosa Virulence and Biofilm Formation. Proc. Natl. Acad. Sci. USA 2013, 110, 17981–17986. [Google Scholar] [CrossRef] [PubMed]
- Carle, J.S.; Christophersen, C. Bromo-Substituted Physostigmine Alkaloids from a Marine Bryozoa Flustra foliacea. J. Am. Chem. Soc. 1979, 101, 4012–4013. [Google Scholar] [CrossRef]
- Lee, J.; Bansal, T.; Jayaraman, A.; Bentley, W.E.; Wood, T.K. Enterohemorrhagic Escherichia coli Biofilms Are Inhibited by 7-Hydroxyindole and Stimulated by Isatin. Appl. Environ. Microbiol. 2007, 73, 4100–4109. [Google Scholar] [CrossRef]
- Bunders, C.; Cavanagh, J.; Melander, C. Flustramine Inspired Synthesis and Biological Evaluation of Pyrroloindoline Triazole Amides as Novel Inhibitors of Bacterial Biofilms. Org. Biomol. Chem. 2011, 9, 5476–5481. [Google Scholar] [CrossRef] [PubMed]
- Minvielle, M.J.; Bunders, C.A.; Melander, C. Indole-Triazole Conjugates are Selective Inhibitors and Inducers of Bacterial Biofilms. Med. Chem. Comm. 2013, 4, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Bunders, C.A.; Minvielle, M.J.; Worthington, R.J.; Ortiz, M.; Cavanagh, J.; Melander, C. Intercepting Bacterial Indole Signaling with Flustramine Derivatives. J. Am. Chem. Soc. 2011, 133, 20160–20163. [Google Scholar] [CrossRef]
- Minvielle, M.J.K.; Eguren, K.; Melander, C. Highly Active Modulators of Indole Signaling Alter Pathogenic Behaviors in Gram-Negative and Gram-Positive Bacteria. Chem. Eur. J. 2013, 19, 17595–17602. [Google Scholar] [CrossRef]
- Huggins, W.M.; Barker, W.T.; Baker, J.T.; Hahn, N.A.; Melander, R.J.; Melander, C. Meridianin D Analogues Display Antibiofilm Activity against MRSA and Increase Colistin Efficacy in Gram-Negative Bacteria. ACS Med. Chem. Lett. 2018, 9, 702–707. [Google Scholar] [CrossRef]
- Brackett, S.M.; Cox, K.E.; Barlock, S.L.; Huggins, W.M.; Ackart, D.F.; Bassaraba, R.J.; Melander, R.J.; Melander, C. Meridianin D Analogues Possess Antibiofilm Activity Against Mycobacterium smegmatis. RSC Med. Chem. 2020, 11, 92–97. [Google Scholar] [CrossRef]
- Huigens, R.W.; Richards, J.J.; Parise, G.; Ballard, T.E.; Zeng, W.; Deora, R.; Melander, C. Inhibition of Pseudomonas aeruginosa Biofilm Formation with Bromoageliferin Anlogues. J. Am. Chem. Soc. 2007, 129, 6966–6967. [Google Scholar] [CrossRef]
- Huigens, R.W.; Ma, L.; Gambino, C.; Moeller, P.D.R.; Basso, A.; Cavanagh, J.; Wozniak, D.J.; Melander, C. Control of Bacterial Biofilms with Marine Alkaloid Derivatives. Mol. BioSyst. 2008, 4, 614–621. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, E.A.; Brackett, C.M.; Mullikin, T.; Alcaraz, C.; Melander, C. The Discovery of N-1 Substituted 2-Aminobenzimidazoles as Zinc-Dependent, S. Aureus Biofilm Inhibitors. Med. Chem. Comm. 2012, 3, 1462–1465. [Google Scholar] [CrossRef][Green Version]
- Nguyen, T.V.; Peszko, M.T.; Melander, R.J.; Melander, C. Using 2-aminobenzimidazole Derivatives to Inhibit Mycobacterium Smegmatis Biofilm Formation. Med. Chem. Comm. 2019, 10, 456–459. [Google Scholar] [CrossRef] [PubMed]
- Huggins, W.M.; Nguyen, T.V.; Hahn, N.A.; Baker, J.T.; Kuo, L.G.; Kaur, D.; Melander, R.J.; Gunn, J.S.; Melander, C. 2-Aminobenzimidazoles as Antibiofilm Agents Against Salmonella enterica Serovar Typhimurium. Med. Chem. Comm. 2018, 9, 1547–1552. [Google Scholar] [CrossRef]
- Frei, R.; Breitbach, A.S.; Blackwell, H.E. 2-Aminobenzimidazole Derivatives Strongly Inhibit and Disperse Pseudomonas aeruginosa Biofilms. Angew. Chem. Int. Ed. 2012, 51, 5226–5229. [Google Scholar] [CrossRef]
- Richards, J.J.; Huigens, R.W.; Ballard, T.E.; Basso, A.; Cavanagh, J.; Melander, C. Inhibition and Dispersion of Proteobacterial Biofilms. Chem. Commun. 2008, 14, 1698–1700. [Google Scholar] [CrossRef]
- Richards, J.J.; Reed, C.S.; Melander, C. Effects of N-pyrrole Substitution on The Anti-biofilm Activities of Oroidin Derivatives Against Acinetobacter baumannii. Bioorg. Med. Chem. Lett. 2008, 18, 4325–4327. [Google Scholar] [CrossRef]
- Ballard, T.E.; Richards, J.J.; Wolfe, A.L.; Melander, C. Synthesis and Antibiofilm Activity of a Second-Generation Reverse-amide Oroidin Library: A Structure-Activity Relationship Study. Chem. Eur. J. 2008, 14, 10745–10761. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Worthington, R.J.; Melander, C.; Wu, H. A New Small Molecule Specifically Inhibits The Cariogenic Bacterium Streptococcus Mutans in Multispecies Biofilms. Antimicrob. Agents Chemother. 2011, 55, 2679–2687. [Google Scholar] [CrossRef] [PubMed]
- Milton, M.E.; Minrovic, B.M.; Harris, D.L.; Kang, B.; Jung, D.; Lewis, C.P.; Thompson, R.J.; Melander, R.J.; Zeng, D.; Melander, C.; et al. Re-sensitizing Multidrug Resistant Bacteria to Antibiotics by Targeting Bacterial Response Regulators: Characterization and Comparison of Interactions Between 2-Aminoimidazoles and The Response Regulators BfmR from Acinetobacter baumannii and QseB from Francisella spp. Front. Mol. Biosci. 2018, 5, 15. [Google Scholar] [CrossRef]
- Milton, M.E.; Allen, C.L.; Feldmann, E.A.; Bobay, B.G.; Jung, D.K.; Stephens, M.D.; Melander, R.J.; Theisen, K.E.; Zeng, D.; Thompson, R.J.; et al. Structure of the Francisella Response Regulator QseB Receiver Domain, and Characterization of QseB Inhibition by Antibiofilm 2-Aminoimidazole-based Compounds. Mol. Microbiol. 2017, 106, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Steenackers, H.P.L.; Ermolat’ev, D.S.; Savaliya, B.; De Weerdt, A.; De Costert, D.; Shah, A.; Van der Eycken, E.V.; De Vos, D.E.; Vanderleyden, J.; De Keersmaecker, S.C.J. Structure Activity Relationship of 4 (5)- aryl-2-amino-1H-imidazoles, N1-Substituted 2-Aminoimidazoles and Imidazo [1,2-a] pyrimidinium Salts as Inhibitors of Biofilm Formation by Salmonella typhimurium and Pseudomonas aeruginosa. J. Med. Chem. 2011, 54, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Ermolat’ev, D.S.; Bariwal, J.B.; Steenackers, H.P.L.; De Keersmaecker, S.C.J.; Van der Eycken, E.V. Concise and Diversity-Oriented Route toward Polysubstituted 2-Aminoimidazole Alkaloids and Their Analogues. Angew. Chem. Int. Ed. 2010, 49, 9465–9468. [Google Scholar] [CrossRef] [PubMed]
- Trang, T.T.T.; Dieltjens, L.; Hooyberghs, G.; Waldrant, K.; Ermolat’ev, D.S.; Van der Eycken, E.V.; Steenackers, H.P.L. Enhancing the Anti-biofilm Activity of 5-aryl-2-aminoimidazoles Through Nature Inspired Dimerisation. Bioorg. Med. Chem. 2018, 26, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, E.A.; Worthington, R.J.; Alcaraz, C.; Melander, C. 2-Aminopyrimidine as a Novel Scaffold for Biofilm Modulation. Org. Biomol. Chem. 2012, 10, 2552–2561. [Google Scholar] [CrossRef] [PubMed]
- Cox, K.E.; Melander, C. Anti-biofilm Activity of Quinazoline Derivatives against Mycobacterium smegmatis. Med. Chem. Comm. 2019, 10, 1177–1179. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.J.; Bobay, B.G.; Stowe, S.D.; Olson, A.L.; Peng, L.; Su, Z.; Actis, L.A.; Melander, C.; Cavanagh, J. Identification of BfmR, a Response Regulator Involved in Biofilm Development, as a Target for a 2-aminoimidazole-based Antibiofilm Agent. Biochemistry 2012, 51, 9776–9778. [Google Scholar] [CrossRef] [PubMed]
- Borrero, N.V.; Bai, F.; Perez, C.; Duong, B.Q.; Rocca, J.R.; Jin, S.; Huigens, R.W. Phenazine Antibiotic Inspired Discovery of Potent Bromophenazine Antibacterial Agents Against Staphylococcus aureus and Staphylococcus epidermidis. Org. Biomol. Chem. 2014, 12, 881–886. [Google Scholar] [CrossRef]
- Garrison, A.T.; Abouelhassan, Y.; Kallifidas, D.; Bai, F.; Ukhanova, M.; Mai, V.; Jin, S.; Luesch, H.; Huigens, R.W. Halogenated Phenazines that Potently Eradicate Biofilms, MRSA Persister Cells in Non-Biofilm Cultures, and Mycobacterium tuberculosis. Angew. Chem. Int. Ed. 2015, 54, 14819–14823. [Google Scholar] [CrossRef]
- Garrison, A.T.; Bai, F.; Abouelhassan, Y.; Paciaroni, N.G.; Jin, S.; Huigens, R.W. Bromophenazine Derivatives with Potent Inhibition, Dispersion and Eradication Activities against Staphylococcus aureus Biofilms. RSC Adv. 2014, 5, 1120–1124. [Google Scholar] [CrossRef]
- Garrison, A.T.; Abouelhassan, Y.; Norwood, V.M.; Kallifidas, D.; Bai, F.; Nguyen, M.T.; Rolfe, M.; Burch, G.M.; Jin, S.; Luesch, H.; et al. Structure-Activity Relationships of a Diverse Class of Halogenated Phenazines That Targets Persistent, Antibiotic-Tolerant Bacterial Biofilms and Mycobacterium tuberculosis. J. Med. Chem. 2016, 59, 3808–3825. [Google Scholar] [CrossRef]
- Garrison, A.T.; Abouelhassan, Y.; Kallifidas, D.; Tan, H.; Kim, Y.S.; Jin, S.; Luesch, H.; Huigens, R.W. An Efficient BuchwaldHartwig/Reductive Cyclization for the Scaffold Diversification of Halogenated Phenazines: Potent Antibacterial Targeting, Biofilm Eradication and Prodrug Exploration. J. Med. Chem. 2018, 61, 3962–3983. [Google Scholar] [CrossRef] [PubMed]
- Hifnawy, M.S.; Hassan, H.M.; Mohammed, R.; Fouda, M.M.; Sayed, A.M.; Hamed, A.A.; AbouZid, S.F.; Rateb, M.E.; Alhadrami, H.A.; Abdelmohsen, U.R. Induction of Antibacterial Metabolites by Co-Cultivation of Two Red-Sea-Sponge-Associated Actinomycetes Micromonospora sp. UR56 and Actinokinespora sp. EG49. Mar. Drugs 2020, 18, 243. [Google Scholar] [CrossRef]
- Garrison, A.T.; Abouelhassan, Y.; Yang, H.; Yousaf, H.H.; Nguyen, T.J.; Huigens Iii, R.W. Microwave-Enhanced Friedländer Synthesis for the Rapid Assembly of Halogenated Quinolines with Antibacterial and Biofilm Eradication Activities against Drug Resistant and Tolerant Bacteria. Med. Chem. Comm. 2017, 8, 720–724. [Google Scholar] [CrossRef]
- Basak, A.; Abouelhassan, Y.; Huigens, R.W. Halogenated Quinolines Discovered Through Reductive Amination with Potent eradication Activities Against MRSA, MRSE and VRE Biofilms. Org. Biomol. Chem. 2015, 13, 10290–10294. [Google Scholar] [CrossRef] [PubMed]
- Basak, A.; Abouelhassan, Y.; Norwood, V.M., IV; Bai, F.; Nguyen, M.T.; Jin, S.; Huigens, R.W. Synthetically Tuning the 2-Position of Halogenated Quinolines: Optimizing Antibacterial and Biofilm Eradication Activities via Alkylation and Reductive Amination Pathways. Chem. Eur. J. 2016, 22, 9181–9189. [Google Scholar] [CrossRef]
- Basak, A.; Abouelhassan, Y.; Kim, Y.S.; Norwood, V.M.; Jin, S.; Huigens, R.W. Halogenated Quinolines Bearing Polar Functionality at the 2-Position: Identification of New Antibacterial Agents with Enhanced Activity Against Staphylococcus epidermidis. Eur. J. Med. Chem. 2018, 155, 705–713. [Google Scholar] [CrossRef]
- Kamal, A.; Rahim, A.; Riyaz, S.; Poornachandra, Y.; Balakrishna, M.; Kumar, C.G.; Hussaini, S.M.A.; Sridhar, B.; Machiraju, P.K. Regioselective Synthesis, Antimicrobial Evaluation and Theoretical Studies of 2-styryl Quinolines. Org. Biomol. Chem. 2015, 13, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- Leon, B.; Fong, J.C.N.; Peach, K.C.; Wong, W.R.; Yildiz, F.H.; Linington, R.G. Development of Quinoline-Based Disruptors of Biofilm Formation Against Vibrio cholera. Org. Lett. 2013, 15, 1234–1237. [Google Scholar] [CrossRef]
- Abouelhassan, Y.; Garrison, A.T.; Burch, G.M.; Wong, W.; Norwood, V.M.; Huigens, R.W. Discovery of Quinoline Small Molecules with Potent Dispersal Activity Against Methicillin-Resistant Staphylococcus aureus and Staphylococcus epidermidis Biofilms Using a Scaffold Hopping Strategy. Bioorg. Med. Chem. Lett. 2014, 24, 5076–5080. [Google Scholar] [CrossRef]
- Leon, B.; Haeckl, F.P.; Linington, R.G. Optimized Quinoline Amino Alcohols as Disruptors and Dispersal Agents of Vibrio cholerae Biofilms. Org. Biomol. Chem. 2015, 13, 8495–8499. [Google Scholar] [CrossRef]
- Zaheer, Z.; Khan, F.A.; Sangshetti, J.N.; Patil, R.H.; Lohar, K.S. Novel Amalgamation of Phthalazine-quinolines as Biofilm Inhibitors: One-pot Synthesis, Biological Evaluation and in Silico ADME Prediction with Favorable Metabolic Fate. Bioorg. Med. Chem. Lett. 2016, 26, 1696–1703. [Google Scholar] [CrossRef] [PubMed]
- Aleksic, I.; Segan, S.; Andric, F.; Zlatovic, M.; Moric, I.; Opsenica, D.M.; Senerovic, L. Long-Chain 4-Aminoquinolines as Quorum Sensing Inhibitors in Serratia marcescens and Pseudomonas aeruginosa. ACS Chem. Biol. 2017, 12, 1425–1434. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.A.K.; Kaduskar, R.N.; Patil, R.; Patil, R.H.; Ansari, S.A.; Alkahtani, H.M.; Almehizia, A.A.; Shinde, D.B.; Sangshetti, J.N. Synthesis, Biological Evaluations and Computational Studies of N-(3-(-2-(7-Chloroquinolin-2-yl)vinyl) benzylidene)anilines as Fungal Biofilm Inhibitors. Bioorg. Med. Chem. Lett. 2019, 29, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.N.; Zimmer, K.R.; Macedo, A.J.; Trentin, D.S. Plant Natural Products Targeting Bacterial Virulence Factors. Chem. Rev. 2016, 116, 9162–9236. [Google Scholar] [CrossRef] [PubMed]
- Huigens, R.W.; Abouelhassan, Y.; Yang, H. Phenazine Antibiotic-Inspired Discovery of Bacterial Biofilm-Eradicating Agents. Chem. Bio. Chem. 2019, 20, 2885–2902. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, L.; Zhang, M.; Liu, H.; Lu, P.; Lin, K. Quorum Sensing Inhibitors: A Patent Review (2014−2018). Expert Opin. Ther. Pat. 2018, 28, 849–865. [Google Scholar] [CrossRef]
- Parrino, B.; Schillaci, D.; Carnevale, I.; Giovannetti, E.; Diana, P.; Cirrincione, G.; Cascioferro, S. Synthetic Small Molecules as AntiBiofilm Agents in the Struggle against Antibiotic Resistance. Eur. J. Med. Chem. 2019, 161, 154–178. [Google Scholar] [CrossRef]
- Gilbert-Girard, S.; Savijoki, K.; Yli-Kauhaluoma, J.; Fallarero, A. Optimization of a High-Throughput 384-Well Plate-Based Screening Platform with Staphylococcus aureus ATCC 25923 and Pseudomonas aeruginosa ATCC 15442 Biofilms. Int. J. Mol. Sci. 2020, 21, 3034. [Google Scholar] [CrossRef]
- Paytubi, S.; de La Cruz, M.; Tormo, J.R.; Martín, J.; González, I.; González-Menendez, V.; Genilloud, O.; Reyes, F.; Vicente, F.; Madrid, C.; et al. A High-Throughput Screening Platform of Microbial Natural Products for the Discovery of Molecules with Antibiofilm Properties against Salmonella. Front. Microbiol. 2017, 8, 326. [Google Scholar] [CrossRef]
- Park, S.R.; Tripathi, A.; Wu, J.; Schultz, P.J.; Yim, I.; McQuade, T.J.; Yu, F.; Arevang, C.J.; Mensah, A.Y.; Tamayo-Castillo, G.; et al. Discovery of Cahuitamycins as Biofilm Inhibitors Derived from a Convergent Biosynthetic Pathway. Nat. Commun. 2016, 7, 10710. [Google Scholar] [CrossRef] [PubMed]
- Ochi, K.; Okamoto, S.; Tozawa, Y.; Inaoka, T.; Hosaka, T.; Xu, J.; Kurosawa, K. Ribosome Engineering and Secondary Metabolite Production. Adv. Appl. Microbiol. 2004, 56, 155–184. [Google Scholar] [CrossRef]
- Tripathi, A.; Park, S.R.; Sikkema, A.P.; Cho, H.J.; Wu, J.; Lee, B.; Xi, C.; Smith, J.L.; Sherman, D.H. A Defined and Flexible Pocket Explains Aryl Substrate Promiscuity of the Cahuitamycin Starter Unit-Activating Enzyme CahJ. Chem. Bio. Chem. 2018, 19, 1595–1600. [Google Scholar] [CrossRef] [PubMed]
- Peach, K.C.; Cheng, A.T.; Oliver, A.G.; Yildiz, F.H.; Linington, R.G. Discovery and Biological Characterization of the Auromomycin Chromophore as an Inhibitor of Biofilm Formation in Vibrio cholerae. Chem. Bio. Chem. 2013, 14, 2209–2215. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Miura, K.; Kumada, Y.; Takeuchi, T.; Tanaka, N. Biological Activities of Non-Protein Chromophores of Antitumor Protein Antibiotics: Auromomycin and Neocarzinostatin. Biochem. Biophys. Res. Commun. 1980, 94, 255–261. [Google Scholar] [CrossRef]
- Shibuya, M.; Sakurai, H.; Maeda, T.; Nishiwaki, E.; Saito, M. Synthesis of the Degradation Product of Auromomycin Chromophore and DNA-Cleaving Activities of Its Derivatives. Tetrahedron Lett. 1986, 27, 1351–1354. [Google Scholar] [CrossRef]
- Kumada, Y.; Miwa, T.; Naoi, N.; Watanabe, K.; Naganawa, H.; Takita, T.; Umezawa, H.; Nnakamura, H.; Iitaka, Y. A Degradation Product of the Chromophore of Auromomycin. J. Antibiot. (Tokyo) 1983, 36, 200–202. [Google Scholar] [CrossRef]
- Peach, K.C.; Bray, W.M.; Shikuma, N.J.; Gassner, N.C.; Lokey, R.S.; Yildiz, F.H.; Linington, R.G. An Image-Based 384-Well High-Throughput Screening Method for the Discovery of Biofilm Inhibitors in Vibrio cholerae. Mol. Biosyst. 2011, 7, 1176. [Google Scholar] [CrossRef] [PubMed]
- Warner, C.J.A.; Cheng, A.T.; Yildiz, F.H.; Linington, R.G. Development of Benzo[1,4]Oxazines as Biofilm Inhibitors and Dispersal Agents against Vibrio cholerae. Chem. Commun. 2015, 51, 1305–1308. [Google Scholar] [CrossRef]
- Navarro, G.; Cheng, A.T.; Peach, K.C.; Bray, W.M.; Bernan, V.S.; Yildiz, F.H.; Linington, R.G. Image-Based 384-Well High-Throughput Screening Method for the Discovery of Skyllamycins A to C as Biofilm Inhibitors and Inducers of Biofilm Detachment in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2014, 58, 1092–1099. [Google Scholar] [CrossRef]
- Giltrap, A.M.; Haeckl, F.P.J.; Kurita, K.L.; Linington, R.G.; Payne, R.J. Total Synthesis of Skyllamycins A-C. J. Org. Chem. 2017, 23, 15046–15049. [Google Scholar] [CrossRef] [PubMed]
- Giltrap, A.M.; Haeckl, F.P.J.; Kurita, K.L.; Linington, R.G.; Payne, R.J. Synthetic Studies Toward the Skyllamycins: Total Synthesis and Generation of Simplified Analogues. J. Org. Chem. 2018, 83, 7250–7270. [Google Scholar] [CrossRef]
- Zhao, T.; Zhang, J.; Tang, M.; Ma, L.Z.; Lei, X. Development of an Effective Fluorescence Probe for Discovery of Aminopeptidase Inhibitors to Suppress Biofilm Formation. J. Antibiot. 2019, 72, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; He, X.; Xie, W.; Xiong, J.; Sheng, H.; Guo, S.; Huang, C.; Zhang, D.; Zhang, K. Elastase LasB of Pseudomonas aeruginosa Promotes Biofilm Formation Partly through Rhamnolipid-Mediated Regulation. Can. J. Microbiol. 2014, 60, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Park, J.S.; Choi, H.Y.; Yoon, S.S.; Kim, W.G. Terrein Is an Inhibitor of Quorum Sensing and C-di-GMP in Pseudomonas aeruginosa: A Connection between Quorum Sensing and c-di-GMP. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Raistrick, H.; Smith, G. Studies in the Biochemistry of Micro-Organisms. Biochem. J. 1935, 29, 606–611. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Kim, D.S.; Kim, W.G.; Ryoo, I.J.; Lee, D.H.; Huh, C.H.; Youn, S.W.; Yoo, I.D.; Park, K.C. Terrein: A New Melanogenesis Inhibitor and Its Mechanism. Cell. Mol. Life Sci. 2004, 61, 2878–2885. [Google Scholar] [CrossRef]
- Goutam, J.; Sharma, G.; Tiwari, V.K.; Mishra, A.; Kharwar, R.N.; Ramaraj, V.; Koch, B. Isolation and Characterization of “Terrein” an Antimicrobial and Antitumor Compound from Endophytic Fungus Aspergillus Terreus (JAS-2) Associated from Achyranthus Aspera Varanasi, India. Front. Microbiol. 2017, 8, 1334. [Google Scholar] [CrossRef]
- Roelofs, K.G.; Wang, J.; Sintim, H.O.; Lee, V.T. Differential Radial Capillary Action of Ligand Assay for High-Throughput Detection of Protein-Metabolite Interactions. Proc. Natl. Acad. Sci. USA 2011, 108, 15528–15533. [Google Scholar] [CrossRef]
- Lieberman, O.J.; Orr, M.W.; Wang, Y.; Lee, V.T. High-Throughput Screening Using the Differential Radial Capillary Action of Ligand Assay Identifies Ebselen as an Inhibitor of Diguanylate Cyclases. ACS Chem. Biol. 2014, 9, 183–192. [Google Scholar] [CrossRef]
- Lee, V.T.; Matewish, J.M.; Kessler, J.L.; Hyodo, M.; Hayakawa, Y.; Lory, S. A Cyclic-di-GMP Receptor Required for Bacterial Exopolysaccharide Production. Mol. Microbiol. 2007, 65, 1474–1484. [Google Scholar] [CrossRef]
- Schewe, T. Molecular Actions of Ebselen-an Antiinflammatory Antioxidant. Gen. Pharm. 1995, 26, 1153–1169. [Google Scholar] [CrossRef]
- Sambanthamoorthy, K.; Sloup, R.E.; Parashar, V.; Smith, J.M.; Kim, E.E.; Semmelhack, M.F.; Neiditch, M.B.; Waters, C.M. Identification of Small Molecules That Antagonize Diguanylate Cyclase Enzymes To Inhibit Biofilm Formation. Antimicrob. Agents Chemother. 2012, 56, 5202–5211. [Google Scholar] [CrossRef] [PubMed]
- Römling, U.; Sierralta, W.D.; Eriksson, K.; Normark, S. Multicellular and Aggregative Behaviour of Salmonella typhimurium Strains Is Controlled by Mutations in the AgfD Promoter. Mol. Microbiol. 1998, 28, 249–264. [Google Scholar] [CrossRef]
- Kirisits, M.J.; Prost, L.; Starkey, M.; Parsek, M.R. Characterization of Colony Morphology Variants Isolated from Pseudomonas aeruginosa Biofilms. Appl. Environ. Microbiol. 2005, 71, 4809–4821. [Google Scholar] [CrossRef]
- Romero, D.; Sanabria-Valentin, E.; Vlamakis, H.; Kolter, R. Biofilm Inhibitors That Target Amyloid Proteins. Chem. Biol. 2013, 20, 102–110. [Google Scholar] [CrossRef]
- Caro-Astorga, J.; Pérez-García, A.; de Vicente, A.; Romero, D. A Genomic Region Involved in the Formation of Adhesin Fibers in Bacillus cereus Biofilms. Front. Microbiol. 2014, 5, 745. [Google Scholar] [CrossRef] [PubMed]
- Kalia, M.; Yadav, V.K.; Singh, P.K.; Sharma, D.; Narvi, S.S.; Agarwal, V. Exploring the Impact of Parthenolide as Anti-Quorum Sensing and Anti-Biofilm Agent against Pseudomonas aeruginosa. Life Sci. 2018, 199, 96–103. [Google Scholar] [CrossRef]
- Huber, B.; Eberl, L.; Feucht, W.; Polster, J. Influence of Polyphenols on Bacterial Biofilm Formation and Quorum-Sensing. Z Naturforsch. C J. Biosci. 2003, 58, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Quave, C.L.; Estévez-Carmona, M.; Compadre, C.M.; Hobby, G.; Hendrickson, H.; Beenken, K.E.; Smeltzer, M.S. Ellagic Acid Derivatives from Rubus ulmifolius Inhibit Staphylococcus aureus Biofilm Formation and Improve Response to Antibiotics. PLoS ONE 2012, 7, e28737. [Google Scholar] [CrossRef] [PubMed]
- Talekar, S.J.; Chochua, S.; Nelson, K.; Klugman, K.P.; Quave, C.L.; Vidal, J.E. 220D-F2 from Rubus Ulmifolius Kills Streptococcus pneumoniae Planktonic Cells and Pneumococcal Biofilms. PLoS ONE 2014, 9, e97314. [Google Scholar] [CrossRef]
- Fontaine, B.M.; Nelson, K.; Lyles, J.T.; Jariwala, P.B.; García-Rodriguez, J.M.; Quave, C.L.; Weinert, E.E. Identification of Ellagic Acid Rhamnoside as a Bioactive Component of a Complex Botanical Extract with Anti-Biofilm Activity. Front. Microbiol. 2017, 8, 496. [Google Scholar] [CrossRef]
- Chambers, S.A.; Gaddy, J.A.; Townsend, S.D. Synthetic Ellagic Acid Glycosides Inhibit Early Stage Adhesion of Streptococcus agalactiae Biofilms as Observed by Scanning Electron Microscopy. Chem. A Eur. J. 2020, 26, 9923–9928. [Google Scholar] [CrossRef]
- Maia, E.H.B.; Assis, L.C.; de Oliveira, T.A.; da Silva, A.M.; Taranto, A.G. Structure-Based Virtual Screening: From Classical to Artificial Intelligence. Front. Chem. 2020, 8, 343. [Google Scholar] [CrossRef] [PubMed]
- Dürig, A.; Kouskoumvekaki, I.; Vejborg, R.M.; Klemm, P. Chemoinformatics-Assisted Development of New Anti-Biofilm Compounds. Appl. Microbiol. Biotechnol. 2010, 87, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Balaban, I.; Novick, R.P. Autocrine Regulation of Toxin Synthesis by Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 1995, 92, 1619–1623. [Google Scholar] [CrossRef] [PubMed]
- Kiran, M.D.; Adikesavan, N.V.; Cirioni, O.; Giacometti, A.; Silvestri, C.; Scalise, G.; Ghiselli, R.; Saba, V.; Orlando, F.; Shoham, M.; et al. Discovery of a Quorum-Sensing Inhibitor of Drug-Resistant Staphylococcal Infections by Structure-Based Virtual Screening. Mol. Pharmacol. 2008, 73, 1578–1586. [Google Scholar] [CrossRef]
- Brackman, G.; Breyne, K.; De Rycke, R.; Vermote, A.; Van Nieuwerburgh, F.; Meyer, E.; Van Calenbergh, S.; Coenye, T. The Quorum Sensing Inhibitor Hamamelitannin Increases Antibiotic Susceptibility of Staphylococcus aureus Biofilms by Affecting Peptidoglycan Biosynthesis and EDNA Release. Sci. Rep. 2016, 6, 20321. [Google Scholar] [CrossRef]
- Vermote, A.; Brackman, G.; Risseeuw, M.D.P.; Vanhoutte, B.; Cos, P.; Van Hecke, K.; Breyne, K.; Meyer, E.; Coenye, T.; Van Calenbergh, S. Hamamelitannin Analogues That Modulate Quorum Sensing as Potentiators of Antibiotics against Staphylococcus aureus. Angew.Chem. Int. Ed. 2016, 55, 6551–6555. [Google Scholar] [CrossRef]
- Vermote, A.; Brackman, G.; Risseeuw, M.D.P.; Coenye, T.; Van Calenbergh, S. Novel Hamamelitannin Analogues for the Treatment of Biofilm Related MRSA Infections–A Scaffold Hopping Approach. Eur. J. Med. Chem. 2017, 127, 757–770. [Google Scholar] [CrossRef]
- Bouton, J.; Van Hecke, K.; Rasooly, R.; Van Calenbergh, S. Synthesis of Pyrrolidine-Based Hamamelitannin Analogues as Quorum Sensing Inhibitors in Staphylococcus aureus. Beilstein J. Org. Chem. 2018, 14, 2822–2828. [Google Scholar] [CrossRef] [PubMed]
- Soukarieh, F.; Oton, E.V.; Dubern, J.F.; Gomes, J.; Halliday, N.; De Pilar Crespo, M.; Ramírez-Prada, J.; Insuasty, B.; Abonia, R.; Quiroga, J.; et al. In Silico and in Vitro-Guided Identification of Inhibitors of Alkylquinolone-Dependent Quorum Sensing in Pseudomonas aeruginosa. Molecules 2018, 23, 257. [Google Scholar] [CrossRef] [PubMed]
- Rinaldo, S.; Giardina, G.; Mantoni, F.; Paiardini, A.; Paone, A.; Cutruzzolà, F. Discovering Selective Diguanylate Cyclase Inhibitors: From PleD to Discrimination of the Active Site of Cyclic-di-GMP Phosphodiesterases. Methods Mol. Biol. 2017, 1657, 431–453. [Google Scholar] [CrossRef] [PubMed]
- Fernicola, S.; Paiardini, A.; Giardina, G.; Rampioni, G.; Leoni, L.; Cutruzzolà, F.; Rinaldo, S. In Silico Discovery and In Vitro Validation of Catechol-Containing Sulfonohydrazide Compounds as Potent Inhibitors of the Diguanylate Cyclase PleD. J. Bacteriol. 2016, 198, 147–156. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, J.R.; Kuwada, N.J.; Huangyutitham, V.; Wiggins, P.A.; Harwood, C.S. Surface Sensing and Lateral Subcellular Localization of WspA, the Receptor in a Chemosensory-like System Leading to c-di-GMP Production. Mol. Microbiol. 2012, 86, 720–729. [Google Scholar] [CrossRef]
- Ueda, A.; Wood, T.K. Connecting Quorum Sensing, c-di-GMP, Pel Polysaccharide, and Biofilm Formation in Pseudomonas aeruginosa through Tyrosine Phosphatase TpbA (PA3885). PLoS Pathog. 2009, 5, e1000483. [Google Scholar] [CrossRef]
- Sambanthamoorthy, K.; Luo, C.; Pattabiraman, N.; Feng, X.; Koestler, B.; Waters, C.M.; Palys, T.J. Identification of Small Molecules Inhibiting Diguanylate Cyclases to Control Bacterial Biofilm Development. Biofouling 2014, 30, 17–28. [Google Scholar] [CrossRef]
- Hafeez, U.; Parakh, S.; Gan, H.K.; Scott, A.M. Antibody–Drug Conjugates for Cancer Therapy. Molecules 2020, 25, 4764. [Google Scholar] [CrossRef]
- Drago, J.Z.; Modi, S.; Chandarlapaty, S. Unlocking the Potential of Antibody–Drug Conjugates for Cancer Therapy. Nat. Rev. Clin. Oncol. 2021, 18, 327–344. [Google Scholar] [CrossRef]
- Liu, Y.; Busscher, H.J.; Zhao, B.; Li, Y.; Zhang, Z.; Van Der Mei, H.C.; Ren, Y.; Shi, L. Surface-Adaptive, Antimicrobially Loaded, Micellar Nanocarriers with Enhanced Penetration and Killing Efficiency in Staphylococcal Biofilms. ACS Nano 2016, 10, 4779–4789. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, M.F.; Brezden, A.; Mohammad, H.; Chmielewski, J.; Seleem, M.N. Targeting Biofilms and Persisters of ESKAPE Pathogens with P14KanS, a Kanamycin Peptide Conjugate. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 848–859. [Google Scholar] [CrossRef] [PubMed]
- Sedghizadeh, P.P.; Sun, S.; Junka, A.F.; Richard, E.; Sadrerafi, K.; Mahabady, S.; Bakhshalian, N.; Tjokro, N.; Bartoszewicz, M.; Oleksy, M.; et al. Design, Synthesis, and Antimicrobial Evaluation of a Novel Bone-Targeting Bisphosphonate-Ciprofloxacin Conjugate for the Treatment of Osteomyelitis Biofilms. J. Med. Chem. 2017, 60, 2326–2343. [Google Scholar] [CrossRef]
- Konai, M.M.; Haldar, J. Fatty Acid Comprising Lysine Conjugates: Anti-MRSA Agents That Display in Vivo Efficacy by Disrupting Biofilms with No Resistance Development. Bioconjug. Chem. 2017, 28, 1194–1204. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Thakur, J.; Pal, S.; Gupta, R.; Mishra, D.; Kumar, S.; Yadav, K.; Saini, A.; Yavvari, P.S.; Vedantham, M.; et al. Cholic Acid-Peptide Conjugates as Potent Antimicrobials against Interkingdom Polymicrobial Biofilms. Antimicrob. Agents Chemother. 2019, 63, e00520-19. [Google Scholar] [CrossRef]
- Klahn, P.; Brönstrup, M. Bifunctional Antimicrobial Conjugates and Hybrid Antimicrobials. Nat. Prod. Rep. 2017, 34, 832–885. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Sommer, R.; Hinsberger, S.; Lu, C.; Hartmann, R.W.; Empting, M.; Titz, A. Novel Strategies for the Treatment of Pseudomonas Aeruginosa Infections. J. Med. Chem. 2016, 59, 5929–5969. [Google Scholar] [CrossRef]
- Stanzl, E.G.; Trantow, B.M.; Vargas, J.R.; Wender, P.A. Fifteen Years of Cell-Penetrating, Guanidinium-Rich Molecular Transporters: Basic Science, Research Tools, and Clinical Applications. Acc. Chem. Res. 2013, 46, 2944–2954. [Google Scholar] [CrossRef]
- Yang, H.; Liu, K.; Jin, S.; Huigens, R.W. Design, Synthesis and Biological Evaluation of a Halogenated Phenazine-Erythromycin Conjugate Prodrug for Antibacterial Applications. Org. Biomol. Chem. 2021, 19, 1483–1487. [Google Scholar] [CrossRef]
- Xiao, T.; Liu, K.; Huigens, R.W. Progress towards a Stable Cephalosporin-Halogenated Phenazine Conjugate for Antibacterial Prodrug Applications. Bioorg. Med. Chem. Lett. 2020, 30, 127515. [Google Scholar] [CrossRef]
- Yilmaz Atay, H. Antibacterial Activity of Chitosan-Based Systems. In Functional Chitosan: Drug Delivery and Biomedical Applications; Springer: Singapore, 2019; pp. 457–489. [Google Scholar] [CrossRef]
- Zhang, A.; Mu, H.; Zhang, W.; Cui, G.; Zhu, J.; Duan, J. Chitosan Coupling Makes Microbial Biofilms Susceptible to Antibiotics. Sci. Rep. 2013, 3, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Antonoplis, A.; Zang, X.; Huttner, M.A.; Chong, K.K.L.; Lee, Y.B.; Co, J.Y.; Amieva, M.R.; Kline, K.A.; Wender, P.A.; Cegelski, L. A Dual-Function Antibiotic-Transporter Conjugate Exhibits Superior Activity in Sterilizing MRSA Biofilms and Killing Persister Cells. J. Am. Chem. Soc. 2018, 140, 16140–16151. [Google Scholar] [CrossRef] [PubMed]
- Meiers, J.; Zahorska, E.; Röhrig, T.; Hauck, D.; Wagner, S.; Titz, A. Directing Drugs to Bugs: Antibiotic-Carbohydrate Conjugates Targeting Biofilm-Associated Lectins of Pseudomonas Aeruginosa. J. Med. Chem. 2020, 63, 11707–11724. [Google Scholar] [CrossRef] [PubMed]
- Sommer, R.; Wagner, S.; Rox, K.; Varrot, A.; Hauck, D.; Wamhoff, E.C.; Schreiber, J.; Ryckmans, T.; Brunner, T.; Rademacher, C.; et al. Glycomimetic, Orally Bioavailable LecB Inhibitors Block Biofilm Formation of Pseudomonas Aeruginosa. J. Am. Chem. Soc. 2018, 140, 2537–2545. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).