Metabolic Engineering of Microbial Cell Factories for Biosynthesis of Flavonoids: A Review
Abstract
:1. Introduction
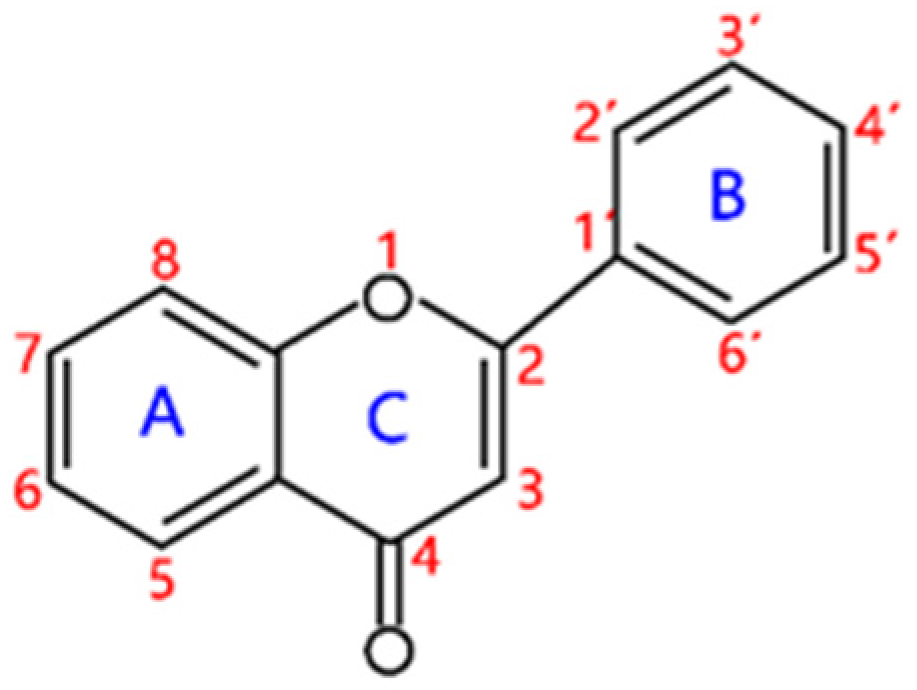
2. Structural Features and Classification
2.1. Flavones and Flavanones
2.2. Flavonols and Flavanonols
2.3. Flavanols
2.4. Anthocyanidins
2.5. Chalcone
2.6. Aurones
2.7. Isoflavonoids
2.8. Neoflavonoids
2.9. Biflavonoids
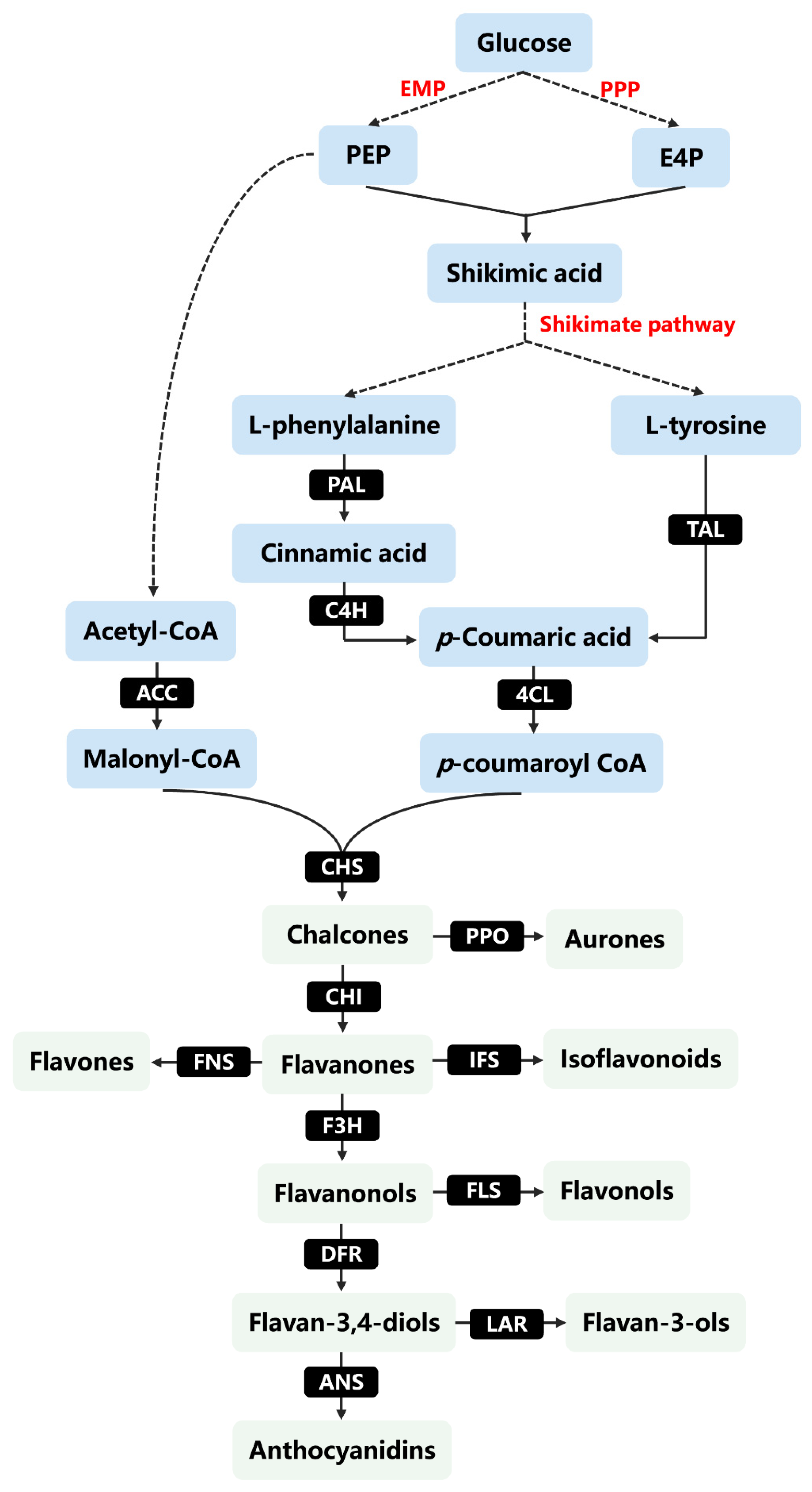
3. Biosynthesis of Flavonoids in Plants
4. Biosynthesis of Flavonoids by Metabolic Engineering
4.1. Microbial Mono-Culture for Flavonoid Synthesis
| Product | Substrate | Host Strain | Titer (mg/L) | Reference |
|---|---|---|---|---|
| Luteolin | Glucose | S. albus | 0.09 | [66] |
| Apigenin | Glucose | S. albus | 0.089 | [66] |
| Apigenin | Glucose and naringenin | S. albus | 0.384 | [66] |
| Scutellarin | Glucose | S. cerevisiae | 108 | [67] |
| Apigenin-7-O-glucuronide | Glucose | S. cerevisiae | 185 | [67] |
| Baicalein | L-phenylalanine | E. coli | 23.6 | [13] |
| Scutellarein | L-tyrosine | E. coli | 106.5 | [13] |
| Isoorientin | Luteolin | E. coli | 3829 | [68] |
| Isovitexin | Apigenin | E. coli | 3772 | [68] |
| Liquiritigenin | Glucose | S. cerevisiae | 5.31 | [16] |
| Eriodictyol | Glucose | S. albus | 0.002 | [66] |
| Eriodictyol | Naringenin | E. coli | 62.7 | [69] |
| Eriodictyol | Caffeic acid | C. glutamicum | 37 | [63] |
| Eriodictyol | Glucose | S. cerevisiae | 152 | [60] |
| Eriodictyol | Glucose | Y. lipolytica | 54.2 | [70] |
| Eriodictyol | Glucose | Y. lipolytica | 134.2 | [2] |
| Eriodictyol | Glycerol | E. coli | 88 | [19] |
| Homoeriodictyol | Glycerol | E. coli | 17 | [19] |
| Pinocembrin | Glycerol | E. coli | 214 | [19] |
| Naringenin | p-Coumaric acid | C. glutamicum | 35 | [63] |
| Naringenin | Tyrosine | S. cerevisiae | 90 | [71] |
| Naringenin | Tyrosine | E. coli | 191.9 | [22] |
| Naringenin | Glucose | Y. lipolytica | 71.2 | [70] |
| Naringenin | Glucose | Y. lipolytica | 252.4 | [2] |
| Naringenin | p-Coumaric acid | S. cerevisiae | 648.63 | [72] |
| Naringenin | Glycerol | E. coli | 484 | [19] |
| Naringenin | Glucose | E. coli | 588 | [61] |
| Naringenin | Glucose | E. coli | 523.7 | [62] |
| Fisetin | Glucose | S. cerevisiae | 2.3 | [16] |
| Resokaempferol | Glucose | S. cerevisiae | 14.54 | [16] |
| Kaempferol | Glucose | S. cerevisiae | 26.57 | [16] |
| Quercetin | Glucose | S. cerevisiae | 20.38 | [16] |
| Quercetin | Glucose | S. albus | 0.599 | [20] |
| Taxifolin | Glucose | Y. lipolytica | 48.1 | [70] |
| Taxifolin | Glucose | Y. lipolytica | 110.5 | [2] |
| Silybin and isosilybin | Eugenol and taxifolin | E. coli | 2580 | [73] |
| Taxifolin | Glucose | S. cerevisiae | 336.8 | [74] |
| Catechin | Afzelechin | E. coli | 34.7 | [69] |
| Peonidin 3-O-glucoside | Catechin | E. coli | 56.3 | [75] |
| Pelargonidin 3-O-glucoside | Glucose | S. cerevisiae | 0.85 | [60] |
| Cyanidin 3-O-glucoside | Glucose | S. cerevisiae | 1.55 | [60] |
| Delphinidin 3-O-glucoside | Glucose | S. cerevisiae | 1.86 | [60] |
| Cyanidin 3-O-glucoside | Catechin | C. glutamicum | 40 | [76] |
| Cyanidin 3-O-glucoside | Catechin | E. coli | 439 | [77] |
| Anthocyanins | Tea | L. lactis | 1.5 | [65] |
| Orobol | Genistein | P. pastoris | 23 | [64] |
| 4ʹ-O-methyl-genistein | Genistein | E. coli | 48.61 | [78] |
| 4ʹ-O-methyl-daidzein | Daidzein | E. coli | 102.88 | [78] |
| Icaritin | Glucose | S. cerevisiae | 7.2 | [57] |
4.2. Microbial Co-Culture for Flavonoid Synthesis
| Product | Substrate | Co-Culture System | Titer (mg/L) | Reference |
|---|---|---|---|---|
| Genistein | Tyrosine | E. coli–S. cerevisiae coculture | 6 | [82] |
| Genistein | Tyrosine | E. coli–S. cerevisiae coculture | 100 | [83] |
| (+)-Afzelechin | p-Coumaric acid | E. coli–E. coli coculture | 40.7 | [21] |
| (+)-Afzelechin | Glucose, glycerol and xylose | Three-strain E. coli polyculture | 26.1 | [90] |
| Naringenin | Glucose | E. coli–E. coli coculture | 41.5 | [84] |
| Naringenin | p-Coumaric acid | S. cerevisiae–S. cerevisiae coculture | 18.5 | [86] |
| Naringenin | Xylose | E. coli–S. cerevisiae coculture | 21.16 | [85] |
| Pelargonidin 3-O-glucoside | Glucose, glycerol and xylose | Four-strain E. coli polyculture | 9.5 | [90] |
| Apigetrin | p-Coumaric acid | E. coli–E. coli coculture | 16.6 | [87] |
| Pyranocyanidin-3-O-glucoside-phenol | Glucose, (+)- catechin and tyrosine | E. coli–E. coli coculture | 19.5 | [91] |
| Pyranocyanidin-3-O-glucoside-catechol | Glucose and (+)- catechin | E. coli–E. coli coculture | 13 | [91] |
| Sakuranetin | Glucose | E. coli–E. coli coculture | 79 | [92] |
| Vitexin | Luteolin and apigenin | E. coli–E. coli coculture | 5050 | [93] |
| Orientin | Luteolin and apigenin | E. coli–E. coli coculture | 7090 | [93] |
| Icaritin | Glucose | E. coli–S. cerevisiae coculture | 19.7 | [57] |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Y.; Chen, S.; Yu, O. Metabolic engineering of flavonoids in plants and microorganisms. Appl. Microbiol. Biotechnol. 2011, 91, 949–956. [Google Scholar] [CrossRef]
- Lv, Y.; Marsafari, M.; Koffas, M.; Zhou, J.; Xu, P. Optimizing Oleaginous Yeast Cell Factories for Flavonoids and Hydroxylated Flavonoids Biosynthesis. ACS Synth. Biol. 2019, 8, 2514–2523. [Google Scholar] [CrossRef]
- Kim, B.G.; Yang, S.M.; Kim, S.Y.; Cha, M.N.; Ahn, J.H. Biosynthesis and production of glycosylated flavonoids in Escherichia coli: Current state and perspectives. Appl. Microbiol. Biotechnol. 2015, 99, 2979–2988. [Google Scholar] [CrossRef]
- Nabavi, S.M.; Samec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Rastrelli, L.; Daglia, M.; Xiao, J.; et al. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 2020, 38, 107316. [Google Scholar] [CrossRef]
- Brazier-Hicks, M.; Evans, K.M.; Gershater, M.C.; Puschmann, H.; Steel, P.G.; Edwards, R. The C-glycosylation of flavonoids in cereals. J. Biol. Chem. 2009, 284, 17926–17934. [Google Scholar] [CrossRef] [Green Version]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, R.P.; Parajuli, P.; Koffas, M.A.G.; Sohng, J.K. Microbial production of natural and non-natural flavonoids: Pathway engineering, directed evolution and systems/synthetic biology. Biotechnol. Adv. 2016, 34, 634–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birchfield, A.S.; McIntosh, C.A. Metabolic engineering and synthetic biology of plant natural products—A minireview. Curr. Plant Biol. 2020, 24. [Google Scholar] [CrossRef]
- Maleki, S.J.; Crespo, J.F.; Cabanillas, B. Anti-inflammatory effects of flavonoids. Food Chem. 2019, 299, 125124. [Google Scholar] [CrossRef] [PubMed]
- Kopustinskiene, D.M.; Jakstas, V.; Savickas, A.; Bernatoniene, J. Flavonoids as Anticancer Agents. Nutrients 2020, 12, 457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weseler, A.R.; Bast, A. Masquelier’s grape seed extract: From basic flavonoid research to a well-characterized food supplement with health benefits. Nutr. J. 2017, 16, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, F.L.A.; Ramzi, A.B.; Baharum, S.N.; Noor, N.M.; Goh, H.H.; Leow, T.C.; Oslan, S.N.; Sabri, S. Recent advancement of engineering microbial hosts for the biotechnological production of flavonoids. Mol. Biol. Rep. 2019, 46, 6647–6659. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tian, C.; Xia, Y.; Mutanda, I.; Wang, K.; Wang, Y. Production of plant-specific flavones baicalein and scutellarein in an engineered E. coli from available phenylalanine and tyrosine. Metab. Eng. 2019, 52, 124–133. [Google Scholar] [CrossRef]
- Xu, P.; Marsafari, M.; Zha, J.; Koffas, M. Microbial Coculture for Flavonoid Synthesis. Trends Biotechnol. 2020, 38, 686–688. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, A.; Pandey, R.P.; Dhakal, D.; Parajuli, P.; Sohng, J.K. Biosynthesis of flavone C-glucosides in engineered Escherichia coli. Appl. Microbiol. Biotechnol. 2018, 102, 1251–1267. [Google Scholar] [CrossRef]
- Rodriguez, A.; Strucko, T.; Stahlhut, S.G.; Kristensen, M.; Svenssen, D.K.; Forster, J.; Nielsen, J.; Borodina, I. Metabolic engineering of yeast for fermentative production of flavonoids. Bioresour. Technol. 2017, 245, 1645–1654. [Google Scholar] [CrossRef] [Green Version]
- Levisson, M.; Patinios, C.; Hein, S.; de Groot, P.A.; Daran, J.M.; Hall, R.D.; Martens, S.; Beekwilder, J. Engineering de novo anthocyanin production in Saccharomyces cerevisiae. Microb. Cell Fact. 2018, 17, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, E.I.; Kaneko, M.; Ohnishi, Y.; Horinouchi, S. Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster. Appl. Environ. Microbiol. 2003, 69, 2699–2706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunstan, M.S.; Robinson, C.J.; Jervis, A.J.; Yan, C.; Carbonell, P.; Hollywood, K.A.; Currin, A.; Swainston, N.; Feuvre, R.L.; Micklefield, J.; et al. Engineering Escherichia coli towards de novo production of gatekeeper (2S)-flavanones: Naringenin, pinocembrin, eriodictyol and homoeriodictyol. ACS Synth. Biol. 2020, 5, ysaa012. [Google Scholar] [CrossRef] [PubMed]
- Marin, L.; Gutierrez-Del-Rio, I.; Entrialgo-Cadierno, R.; Villar, C.J.; Lombo, F. De novo biosynthesis of myricetin, kaempferol and quercetin in Streptomyces albus and Streptomyces coelicolor. PLoS ONE 2018, 13, e0207278. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.A.; Vernacchio, V.R.; Sinkoe, A.L.; Collins, S.M.; Ibrahim, M.H.A.; Lachance, D.M.; Hahn, J.; Koffas, M.A.G. Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids. Metab. Eng. 2016, 35, 55–63. [Google Scholar] [CrossRef]
- Zhou, S.; Lyu, Y.; Li, H.; Koffas, M.A.G.; Zhou, J. Fine-tuning the (2S)-naringenin synthetic pathway using an iterative high-throughput balancing strategy. Biotechnol. Bioeng. 2019, 116, 1392–1404. [Google Scholar] [CrossRef]
- Pandey, R.P.; Malla, S.; Simkhada, D.; Kim, B.G.; Sohng, J.K. Production of 3-O-xylosyl quercetin in Escherichia coli. Appl. Microbiol. Biotechnol. 2013, 97, 1889–1901. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.K.; Zill, E.H.; Dangles, O. A comprehensive review on flavanones, the major citrus polyphenols. J. Food Compos. Anal. 2014, 33, 85–104. [Google Scholar] [CrossRef]
- Peterson, J.J.; Beecher, G.R.; Bhagwat, S.A.; Dwyer, J.T.; Gebhardt, S.E.; Haytowitz, D.B.; Holden, J.M. Flavanones in grapefruit, lemons, and limes: A compilation and review of the data from the analytical literature. J. Food Compos. Anal. 2006, 19, S74–S80. [Google Scholar] [CrossRef]
- Pei, J.; Dong, P.; Wu, T.; Zhao, L.; Cao, F.; Tang, F. Characterization flavanone 3β-hydroxylase expressed from Populus euphratica in Escherichia coli and its application in dihydroflavonol production. Appl. Biochem. Microbiol. 2017, 53, 318–324. [Google Scholar] [CrossRef]
- Kang, J.H.; McRoberts, J.; Shi, F.; Moreno, J.E.; Jones, A.D.; Howe, G.A. The flavonoid biosynthetic enzyme chalcone isomerase modulates terpenoid production in glandular trichomes of tomato. Plant Physiol. 2014, 164, 1161–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Yuan, W.; Li, M.; Miao, M.; Mu, W. Purification and characterization of an intracellular alpha-l-rhamnosidase from a newly isolated strain, Alternaria alternata SK37.001. Food Chem. 2018, 269, 63–69. [Google Scholar] [CrossRef]
- Lee, J.W.; Kim, N.H.; Kim, J.Y.; Park, J.H.; Shin, S.Y.; Kwon, Y.S.; Lee, H.J.; Kim, S.S.; Chun, W. Aromadendrin Inhibits Lipopolysaccharide-Induced Nuclear Translocation of NF-kappaB and Phosphorylation of JNK in RAW 264.7 Macrophage Cells. Biomol. Ther. 2013, 21, 216–221. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Yang, L.; Wang, W.; Yang, F.; Zhao, C.; Zu, Y. Extraction of dihydroquercetin from Larix gmelinii with ultrasound-assisted and microwave-assisted alternant digestion. Int. J. Mol. Sci. 2012, 13, 8789–8804. [Google Scholar] [CrossRef] [Green Version]
- Kiehlmann, E.; Li, E.P.M. ISOMERIZATION OF DIHYDROQUERCETIN. J. Nat. Prod. 1995, 58, 450–455. [Google Scholar] [CrossRef]
- Monfoulet, L.E.; Ruskovska, T.; Ajdzanovic, V.; Havlik, J.; Vauzour, D.; Bayram, B.; Krga, I.; Fabiola, C.J.K.; Kistanova, E.; Abadjieva, D.; et al. Molecular Determinants of the Cardiometabolic Improvements of Dietary Flavanols Identified by an Integrative Analysis of Nutrigenomic Data From a Systematic Review of Animal Studies. Mol. Nutr. Food Res. 2021, e2100227. [Google Scholar] [CrossRef] [PubMed]
- Ruskovska, T.; Massaro, M.; Carluccio, M.A.; Arola-Arnal, A.; Muguerza, B.; Vanden Berghe, W.; Declerck, K.; Bravo, F.I.; Calabriso, N.; Combet, E.; et al. Systematic bioinformatic analysis of nutrigenomic data of flavanols in cell models of cardiometabolic disease. Food Funct. 2020, 11, 5040–5064. [Google Scholar] [CrossRef]
- Lee, K.W.; Kim, Y.J.; Lee, H.J.; Lee, C.Y. Cocoa has more phenolic phytochemicals and a higher antioxidant capacity than teas and red wine. J. Agric. Food Chem. 2003, 51, 7292–7295. [Google Scholar] [CrossRef]
- Karabay, A.; Saija, J.D.; Field, D.T.; Akyurek, E.G. The acute effects of cocoa flavanols on temporal and spatial attention. Psychopharmacology 2018, 235, 1497–1511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, M.A.; Ramos, S. Impact of cocoa flavanols on human health. Food Chem. Toxicol. 2021, 151, 112121. [Google Scholar] [CrossRef] [PubMed]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwashina, T. Flavonoid Properties of five Families newly Incorporated into the Order Caryophyllales (Review). Bull. Natl. Mus. Nat. Sci. Ser. B Bot. 2013, 39, 25–51. [Google Scholar]
- Castañeda-Ovando, A.; Pacheco-Hernández, M.d.L.; Páez-Hernández, M.E.; Rodríguez, J.A.; Galán-Vidal, C.A. Chemical studies of anthocyanins: A review. Food Chem. 2009, 113, 859–871. [Google Scholar] [CrossRef]
- Orlikova, B.; Tasdemir, D.; Golais, F.; Dicato, M.; Diederich, M. Dietary chalcones with chemopreventive and chemotherapeutic potential. Genes Nutr. 2011, 6, 125–147. [Google Scholar] [CrossRef] [Green Version]
- Michalkova, R.; Mirossay, L.; Gazdova, M.; Kello, M.; Mojzis, J. Molecular Mechanisms of Antiproliferative Effects of Natural Chalcones. Cancers 2021, 13, 2730. [Google Scholar] [CrossRef]
- Alves Borges Leal, A.L.; Teixeira da Silva, P.; Nunes da Rocha, M.; Marinho, E.M.; Marinho, E.S.; Marinho, M.M.; Bandeira, P.N.; Sampaio Nogueira, C.E.; Barreto, H.M.; Rodrigues Teixeira, A.M.; et al. Potentiating activity of Norfloxacin by synthetic chalcones against NorA overproducing Staphylococcus aureus. Microb. Pathog. 2021, 155, 104894. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.X.; Yuan, Y.L.; Cui, S.S.; Li, M.; Tan, X.; Qiu, Z.C.; Li, R.M. Dissection of the potential pharmacological function of neohesperidin dihydrochalcone—A food additive—by in vivo substances profiling and network pharmacology. Food Funct. 2021, 12, 4325–4336. [Google Scholar] [CrossRef]
- Boucherle, B.; Peuchmaur, M.; Boumendjel, A.; Haudecoeur, R. Occurrences, biosynthesis and properties of aurones as high-end evolutionary products. Phytochemistry 2017, 142, 92–111. [Google Scholar] [CrossRef] [PubMed]
- Popova, A.V.; Bondarenko, S.P.; Frasinyuk, M.S. Aurones: Synthesis and Properties. Chem. Heterocycl. Compd. 2019, 55, 285–299. [Google Scholar] [CrossRef]
- Sui, G.; Li, T.; Zhang, B.; Wang, R.; Hao, H.; Zhou, W. Recent advances on synthesis and biological activities of aurones. Bioorg. Med. Chem. 2021, 29, 115895. [Google Scholar] [CrossRef] [PubMed]
- Blicharski, T.; Oniszczuk, A. Extraction Methods for the Isolation of Isoflavonoids from Plant Material. Open Chem. 2017, 15, 34–45. [Google Scholar] [CrossRef]
- Wang, J.F.; Liu, S.S.; Song, Z.Q.; Xu, T.C.; Liu, C.S.; Hou, Y.G.; Huang, R.; Wu, S.H. Naturally Occurring Flavonoids and Isoflavonoids and Their Microbial Transformation: A Review. Molecules 2020, 25, 5112. [Google Scholar] [CrossRef]
- Kang, H.R.; Lee, D.; Benndorf, R.; Jung, W.H.; Beemelmanns, C.; Kang, K.S.; Kim, K.H. Termisoflavones A-C, Isoflavonoid Glycosides from Termite-Associated Streptomyces sp. RB1. J. Nat. Prod. 2016, 79, 3072–3078. [Google Scholar] [CrossRef]
- McCue, P.; Shetty, K. Health benefits of soy isoflavonoids and strategies for enhancement: A review. Crit. Rev. Food Sci. Nutr. 2004, 44, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, G.B.; Oliveira, G.L.S.; da Silva, J.C.C.L.; Oliveira, M.C.P.; da Silva, A.P.S.C.L.; de Lima David, J.P. Neoflavonoids as Prospective Compounds Against Parasitic Neglected Tropical Infections and Human Immunodeficiency Virus. Curr. Bioact. Compd. 2017, 13. [Google Scholar] [CrossRef]
- Garazd, M.M.; Garazd, Y.L.; Khilya, V.P. Neoflavones. 1. Natural distribution and spectral and biological properties. Chem. Nat. Compd. 2003, 39, 54–121. [Google Scholar] [CrossRef]
- Menezes, J.; Diederich, M.F. Bioactivity of natural biflavonoids in metabolism-related disease and cancer therapies. Pharmacol. Res. 2021, 167, 105525. [Google Scholar] [CrossRef]
- Menezes, J.; Campos, V.R. Natural biflavonoids as potential therapeutic agents against microbial diseases. Sci. Total Environ. 2021, 769, 145168. [Google Scholar] [CrossRef]
- Gontijo, V.S.; Dos Santos, M.H.; Viegas, C., Jr. Biological and Chemical Aspects of Natural Biflavonoids from Plants: A Brief Review. Mini Rev. Med. Chem. 2017, 17, 834–862. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, J.; Li, H.; Chiang, V.L.; Fu, Y. Cooperative Regulation of Flavonoid and Lignin Biosynthesis in Plants. Crit. Rev. Plant Sci. 2021, 40, 109–126. [Google Scholar] [CrossRef]
- Wang, P.; Li, C.; Li, X.; Huang, W.; Wang, Y.; Wang, J.; Zhang, Y.; Yang, X.; Yan, X.; Wang, Y.; et al. Complete biosynthesis of the potential medicine icaritin by engineered Saccharomyces cerevisiae and Escherichia coli. Sci. Bull. 2021. [Google Scholar] [CrossRef]
- Sheng, H.; Sun, X.; Yan, Y.; Yuan, Q.; Wang, J.; Shen, X. Metabolic Engineering of Microorganisms for the Production of Flavonoids. Front. Bioeng. Biotechnol. 2020, 8, 589069. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Park, S.Y.; Park, Y.S.; Eun, H.; Lee, S.Y. Metabolic Engineering of Escherichia coli for Natural Product Biosynthesis. Trends Biotechnol. 2020, 38, 745–765. [Google Scholar] [CrossRef] [PubMed]
- Eichenberger, M.; Hansson, A.; Fischer, D.; Durr, L.; Naesby, M. De novo biosynthesis of anthocyanins in Saccharomyces cerevisiae. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Hao, T.; Zhou, J. Fermentation and Metabolic Pathway Optimization to De Novo Synthesize (2S)-Naringenin in Escherichia coli. J. Microbiol. Biotechnol. 2020, 30, 1574–1582. [Google Scholar] [CrossRef]
- Zhou, S.; Yuan, S.F.; Nair, P.H.; Alper, H.S.; Deng, Y.; Zhou, J. Development of a growth coupled and multi-layered dynamic regulation network balancing malonyl-CoA node to enhance (2S)-naringenin biosynthesis in Escherichia coli. Metab. Eng. 2021, 67, 41–52. [Google Scholar] [CrossRef]
- Kallscheuer, N.; Vogt, M.; Stenzel, A.; Gatgens, J.; Bott, M.; Marienhagen, J. Construction of a Corynebacterium glutamicum platform strain for the production of stilbenes and (2S)-flavanones. Metab. Eng. 2016, 38, 47–55. [Google Scholar] [CrossRef]
- Wang, T.Y.; Tsai, Y.H.; Yu, I.Z.; Chang, T.S. Improving 3′-Hydroxygenistein Production in Recombinant Pichia pastoris Using Periodic Hydrogen Peroxide-Shocking Strategy. J. Microbiol. Biotechnol. 2016, 26, 498–502. [Google Scholar] [CrossRef]
- Solopova, A.; van Tilburg, A.Y.; Foito, A.; Allwood, J.W.; Stewart, D.; Kulakauskas, S.; Kuipers, O.P. Engineering Lactococcus lactis for the production of unusual anthocyanins using tea as substrate. Metab. Eng. 2019, 54, 160–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marin, L.; Gutierrez-Del-Rio, I.; Yague, P.; Manteca, A.; Villar, C.J.; Lombo, F. De Novo Biosynthesis of Apigenin, Luteolin, and Eriodictyol in the Actinomycete Streptomyces albus and Production Improvement by Feeding and Spore Conditioning. Front. Microbiol. 2017, 8, 921. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Cheng, J.; Zhang, G.; Ding, W.; Duan, L.; Yang, J.; Kui, L.; Cheng, X.; Ruan, J.; Fan, W.; et al. Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat. Commun. 2018, 9, 448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, J.; Sun, Q.; Gu, N.; Zhao, L.; Fang, X.; Tang, F.; Cao, F. Production of isoorientin and isovitexin from luteolin and apigenin using coupled catalysis of glycosyltransferase and sucrose synthase. Appl. Biochem. Biotechnol. 2020, 190, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.A.; Collins, S.M.; Vernacchio, V.R.; Lachance, D.M.; Koffas, M.A. Optimization of naringenin and p-coumaric acid hydroxylation using the native E. coli hydroxylase complex, HpaBC. Biotechnol. Prog. 2016, 32, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Edwards, H.; Zhou, J.; Xu, P. Combining 26s rDNA and the Cre-loxP System for Iterative Gene Integration and Efficient Marker Curation in Yarrowia lipolytica. ACS Synth. Biol. 2019, 8, 568–576. [Google Scholar] [CrossRef]
- Lyu, X.; Ng, K.R.; Lee, J.L.; Mark, R.; Chen, W.N. Enhancement of Naringenin Biosynthesis from Tyrosine by Metabolic Engineering of Saccharomyces cerevisiae. J. Agric. Food Chem. 2017, 65, 6638–6646. [Google Scholar] [CrossRef]
- Gao, S.; Lyu, Y.; Zeng, W.; Du, G.; Zhou, J.; Chen, J. Efficient Biosynthesis of (2S)-Naringenin from p-Coumaric Acid in Saccharomyces cerevisiae. J. Agric. Food Chem. 2020, 68, 1015–1021. [Google Scholar] [CrossRef]
- Lv, Y.; Xu, S.; Lyu, Y.; Zhou, S.; Du, G.; Chen, J.; Zhou, J. Engineering enzymatic cascades for the efficient biotransformation of eugenol and taxifolin to silybin and isosilybin. Green Chem. 2019, 21, 1660–1667. [Google Scholar] [CrossRef]
- Yang, J.; Liang, J.; Shao, L.; Liu, L.; Gao, K.; Zhang, J.L.; Sun, Z.; Xu, W.; Lin, P.; Yu, R.; et al. Green production of silybin and isosilybin by merging metabolic engineering approaches and enzymatic catalysis. Metab. Eng. 2020, 59, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Cress, B.F.; Leitz, Q.D.; Kim, D.C.; Amore, T.D.; Suzuki, J.Y.; Linhardt, R.J.; Koffas, M.A. CRISPRi-mediated metabolic engineering of E. coli for O-methylated anthocyanin production. Microb. Cell Fact. 2017, 16, 10. [Google Scholar] [CrossRef] [Green Version]
- Zha, J.; Zang, Y.; Mattozzi, M.; Plassmeier, J.; Gupta, M.; Wu, X.; Clarkson, S.; Koffas, M.A.G. Metabolic engineering of Corynebacterium glutamicum for anthocyanin production. Microb. Cell Fact. 2018, 17, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shrestha, B.; Pandey, R.P.; Darsandhari, S.; Parajuli, P.; Sohng, J.K. Combinatorial approach for improved cyanidin 3-O-glucoside production in Escherichia coli. Microb. Cell Fact. 2019, 18, 7. [Google Scholar] [CrossRef]
- Koirala, N.; Pandey, R.P.; Thuan, N.H.; Ghimire, G.P.; Jung, H.J.; Oh, T.J.; Sohng, J.K. Metabolic engineering of Escherichia coli for the production of isoflavonoid-4′-O-methoxides and their biological activities. Biotechnol. Appl. Biochem. 2019, 66, 484–493. [Google Scholar] [CrossRef]
- McCarty, N.S.; Ledesma-Amaro, R. Synthetic Biology Tools to Engineer Microbial Communities for Biotechnology. Trends Biotechnol. 2019, 37, 181–197. [Google Scholar] [CrossRef] [Green Version]
- Sgobba, E.; Wendisch, V.F. Synthetic microbial consortia for small molecule production. Curr. Opin. Biotechnol. 2020, 62, 72–79. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, S.; Wang, Z.; Koffas, M.A. Recent advances in modular co-culture engineering for synthesis of natural products. Curr. Opin. Biotechnol. 2020, 62, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Katsuyama, Y.; Miyahisa, I.; Funa, N.; Horinouchi, S. One-pot synthesis of genistein from tyrosine by coincubation of genetically engineered Escherichia coli and Saccharomyces cerevisiae cells. Appl. Microbiol. Biotechnol. 2007, 73, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, S. Combinatorial biosynthesis of plant medicinal polyketides by microorganisms. Curr. Opin. Chem. Biol. 2009, 13, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, V.; Li, Z.; Wang, X.; Zhang, H. Heterologous biosynthesis of natural product naringenin by co-culture engineering. Synth. Syst. Biotechnol. 2017, 2, 236–242. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, H.; Li, X.; Liu, D.; Dong, X.T.; Li, F.F.; Wang, E.X.; Li, B.Z.; Yuan, Y.J. Production of naringenin from D-xylose with co-culture of E. coli and S. cerevisiae. Eng. Life Sci. 2017, 17, 1021–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Song, Y.; Zhu, J.; Xu, X.; Pang, B.; Jin, H.; Jiang, C.; Liu, Y.; Shi, J. Potential application of CHS and 4CL genes from grape endophytic fungus in production of naringenin and resveratrol and the improvement of polyphenol profiles and flavour of wine. Food Chem. 2021, 347, 128972. [Google Scholar] [CrossRef]
- Thuan, N.H.; Chaudhary, A.K.; Van Cuong, D.; Cuong, N.X. Engineering co-culture system for production of apigetrin in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2018, 45, 175–185. [Google Scholar] [CrossRef]
- Yan, Y.J.; Chemler, J.; Huang, L.X.; Martens, S.; Koffas, M.A.G. Metabolic engineering of anthocyanin biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 2005, 71, 3617–3623. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.J.; Li, Z.; Koffas, M.A.G. High-yield anthocyanin biosynthesis in engineered Escherichia coli. Biotechnol. Bioeng. 2008, 100, 126–140. [Google Scholar] [CrossRef]
- Jones, J.A.; Vernacchio, V.R.; Collins, S.M.; Shirke, A.N.; Xiu, Y.; Englaender, J.A.; Cress, B.F.; McCutcheon, C.C.; Linhardt, R.J.; Gross, R.A.; et al. Complete Biosynthesis of Anthocyanins Using E. coli Polycultures. MBio 2017, 8, 9. [Google Scholar] [CrossRef] [Green Version]
- Akdemir, H.; Silva, A.; Zha, J.; Zagorevski, D.V.; Koffas, M.A.G. Production of pyranoanthocyanins using Escherichia coli co-cultures. Metab. Eng. 2019, 55, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Z.; Policarpio, L.; Koffas, M.A.G.; Zhang, H. De novo biosynthesis of complex natural product sakuranetin using modular co-culture engineering. Appl. Microbiol. Biotechnol. 2020, 104, 4849–4861. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Wang, H.; Zhao, L.; Pei, J. Orientin and vitexin production by a one-pot enzymatic cascade of a glycosyltransferase and sucrose synthase. Bioorg. Chem. 2021, 112, 104926. [Google Scholar] [CrossRef] [PubMed]
| Groups | Structures | Examples | |
|---|---|---|---|
| Flavones | 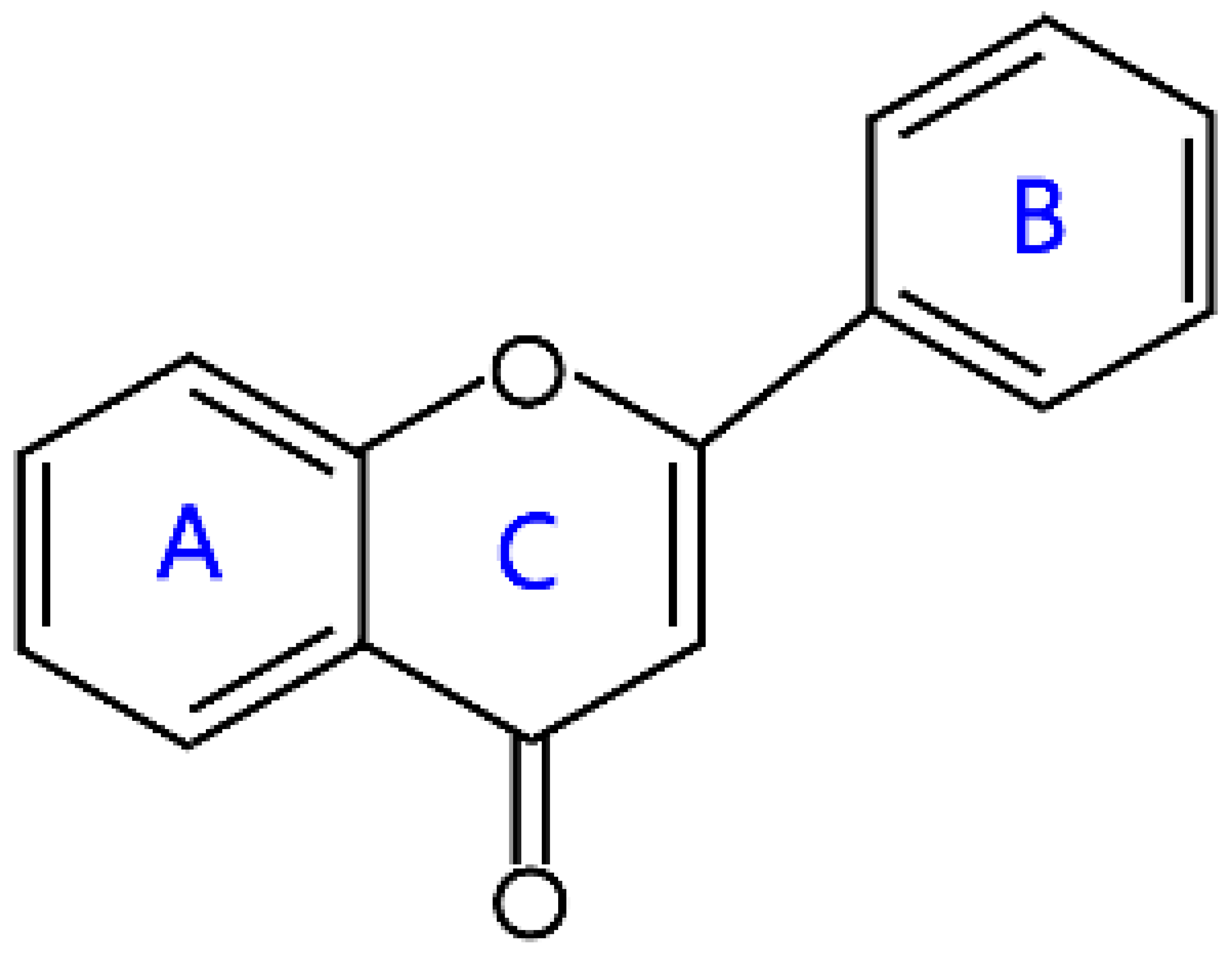 | Apigenin, luteolin, baicalein, and tangeritin | |
| Flavanones | 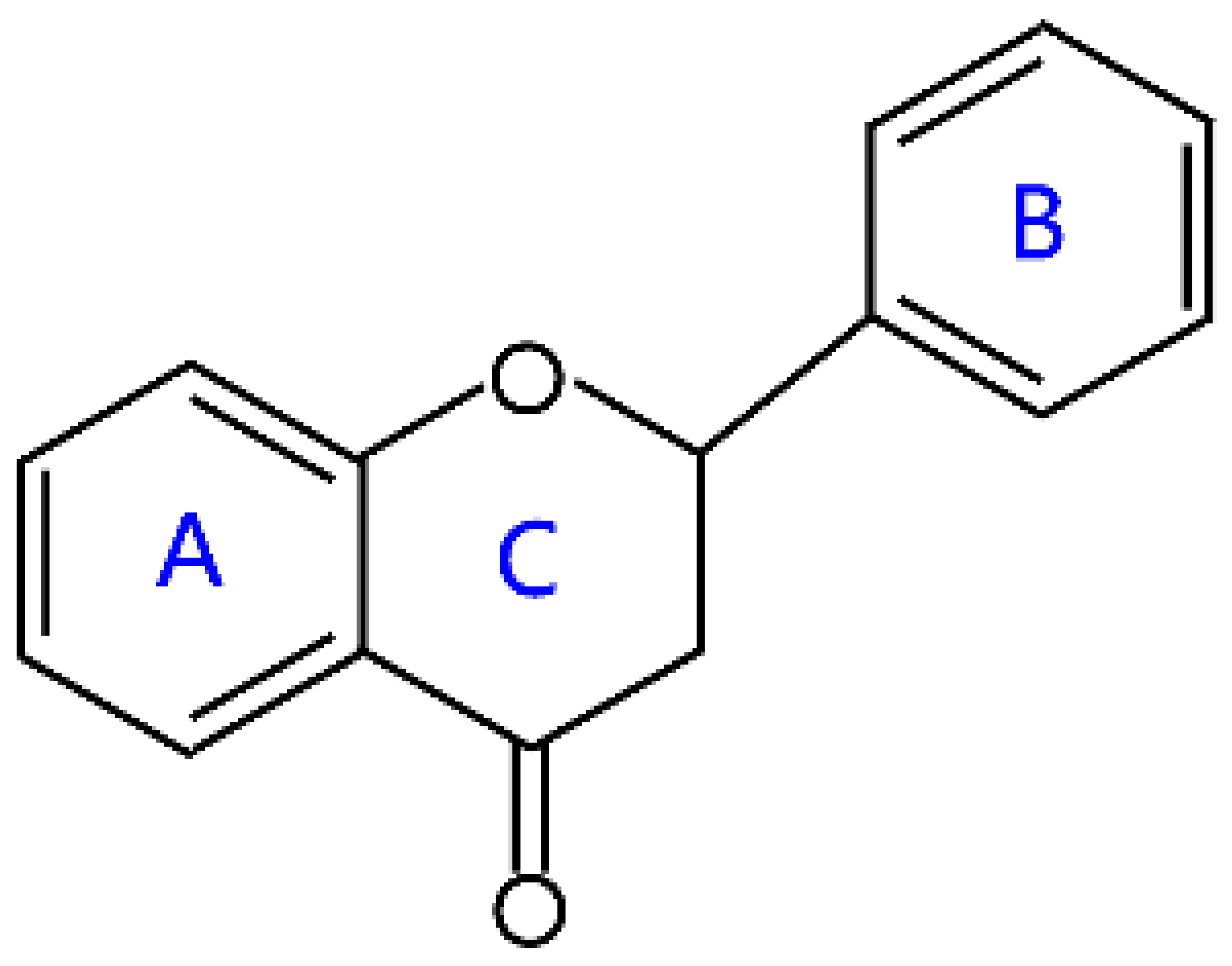 | Liquiritigenin, hesperitin, naringenin, and eriodictyol | |
| Flavonols | 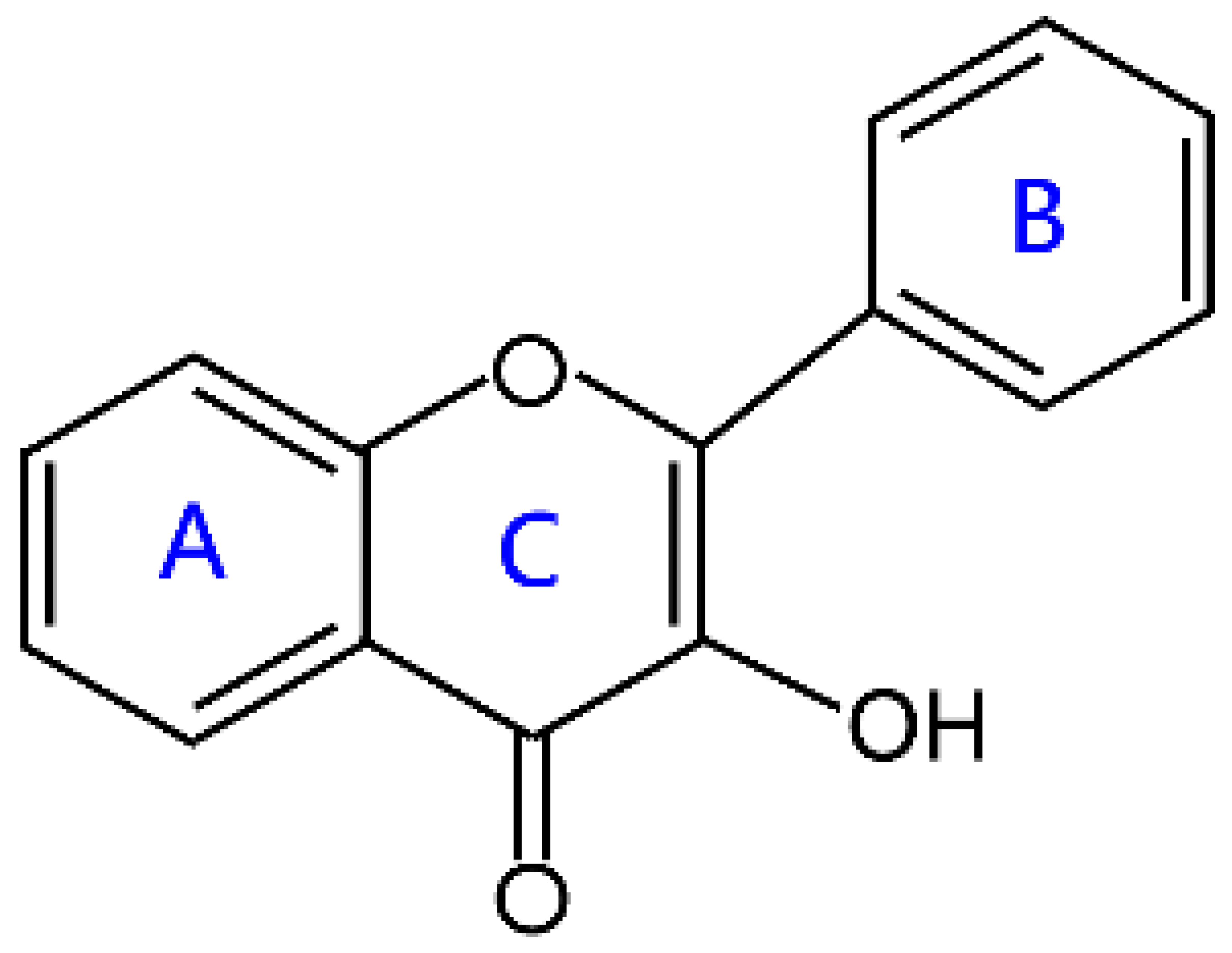 | Kaempferol, quercetin, myricetin, and rutin | |
| Flavanonols | 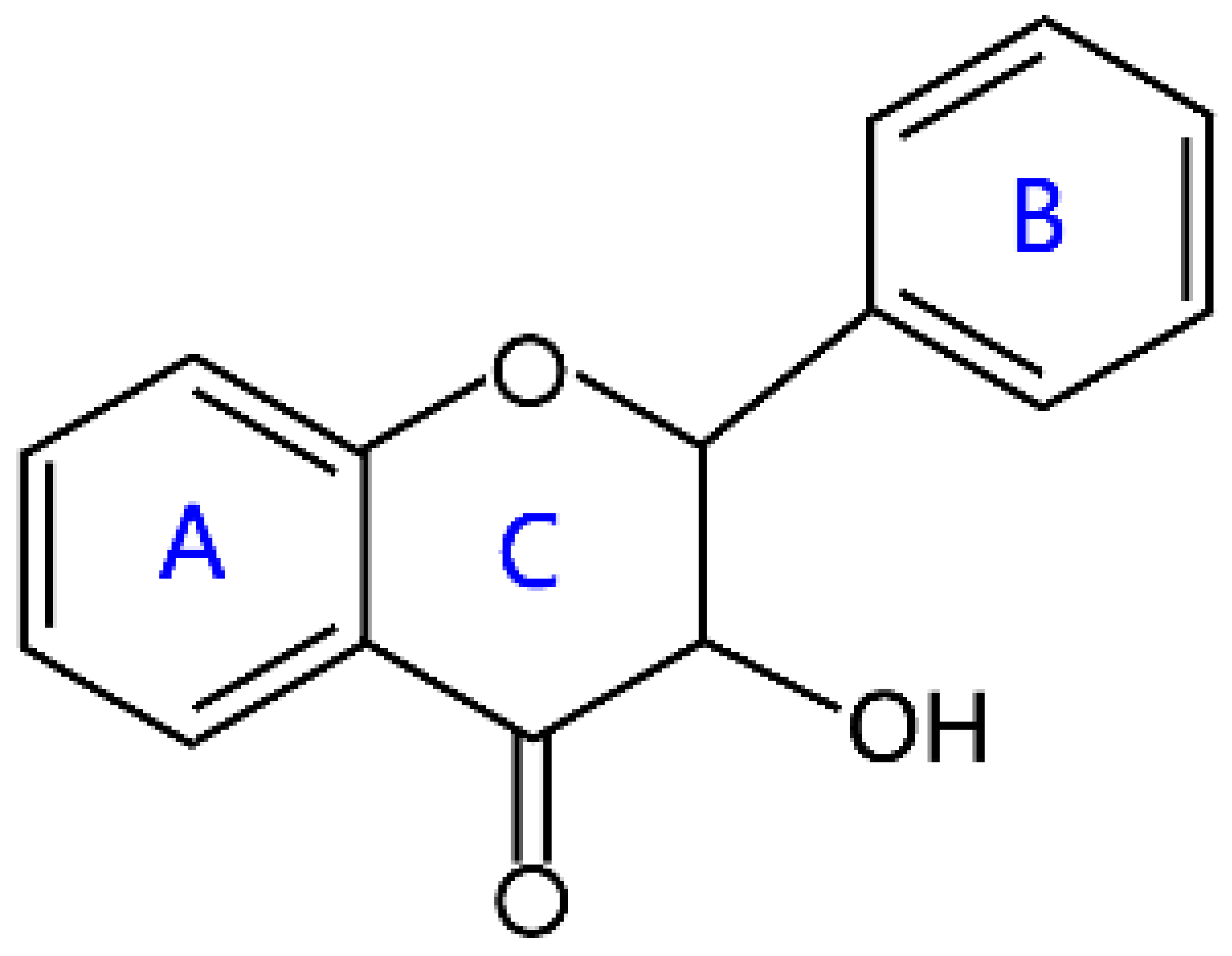 | Aromadendrin, distylin, dihydrokaempferol, and dihydroquercetin | |
| Flavanols | Flavan-3-ols | 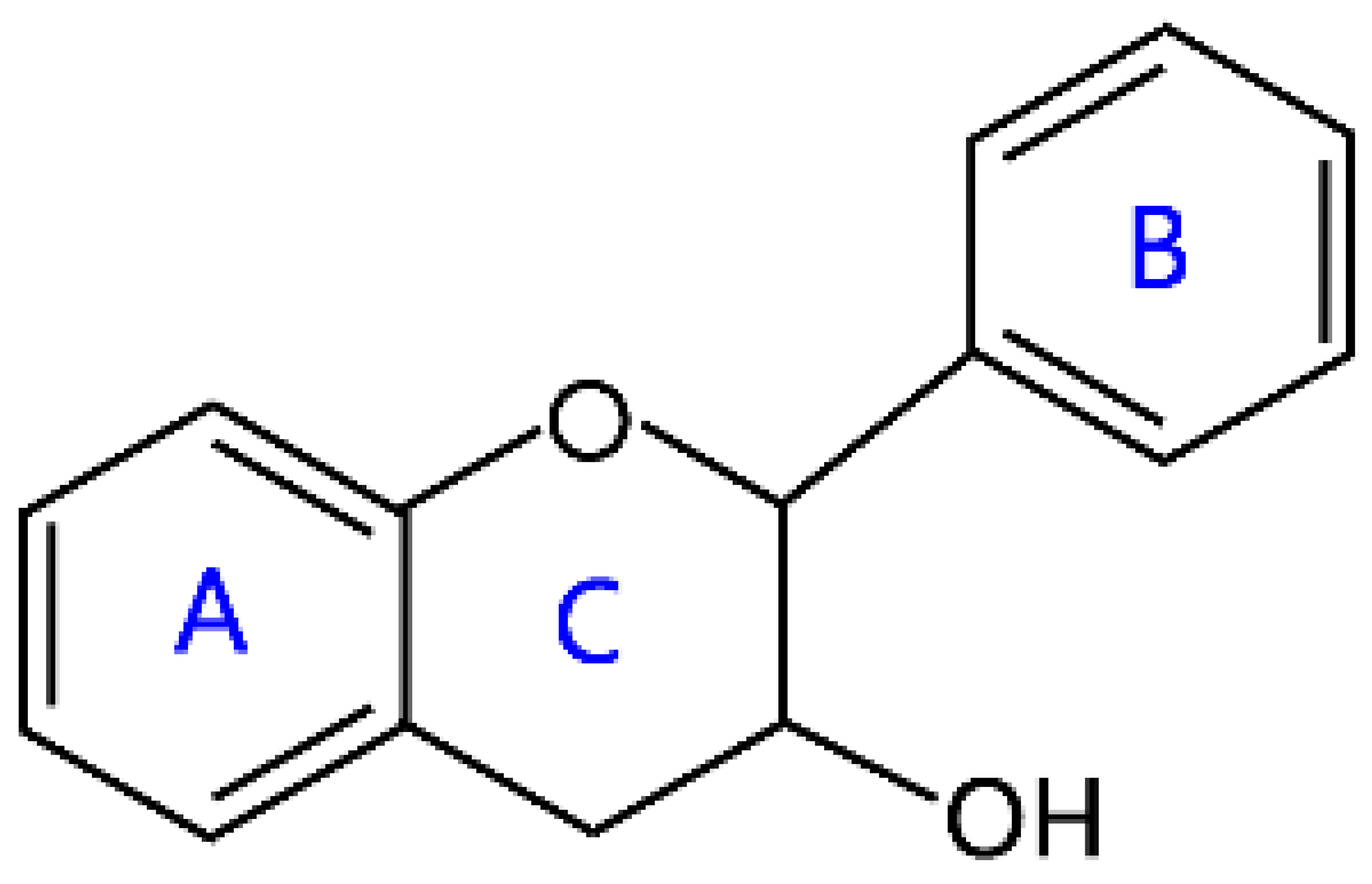 | Catechin, elephantorrhizol, guibourtinidol, and epirobinetinidol |
| Flavan-3,4-diols | 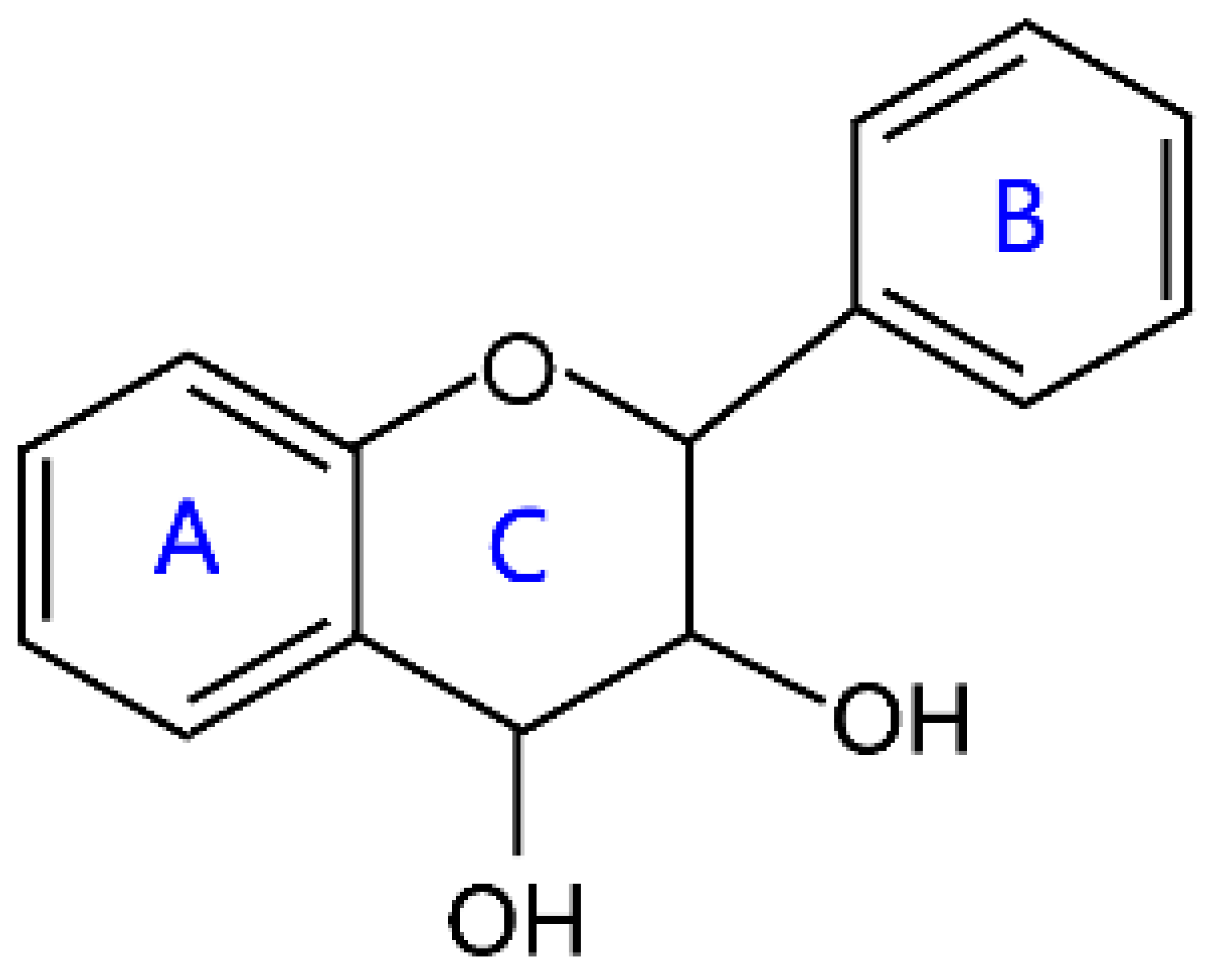 | Leucofisetinidin, leucocyanidin, melacacidin, and leucopeonidin | |
| Anthocyanidins |  | Cyanidin, delphinidin, malvidin, peonidin, pelargonidin, and petunidin | |
| Chalcones | 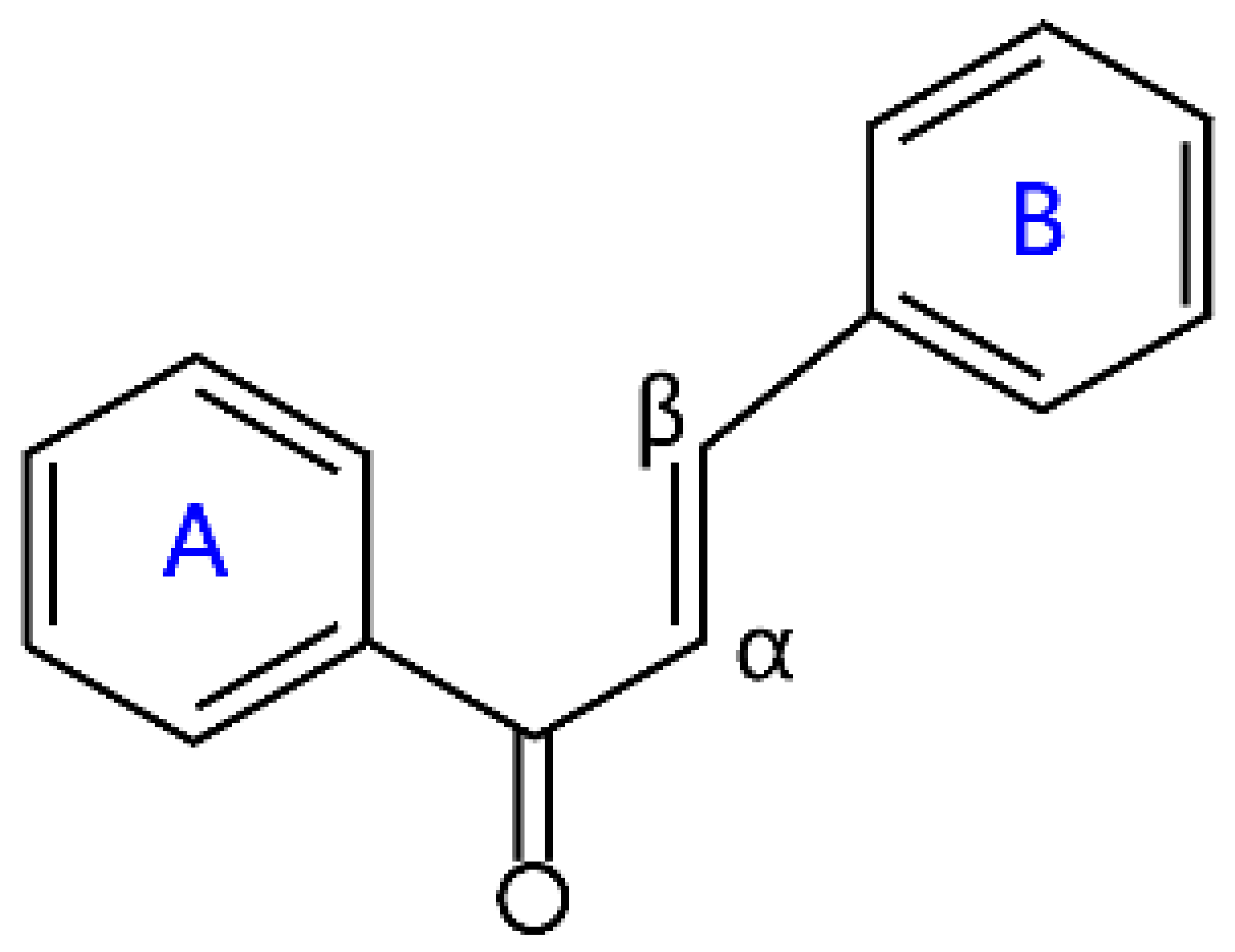 | Naringenin chalcone, pinocembrin chalcone, echinatin, and licochalcone A | |
| Dihydrochalcones | 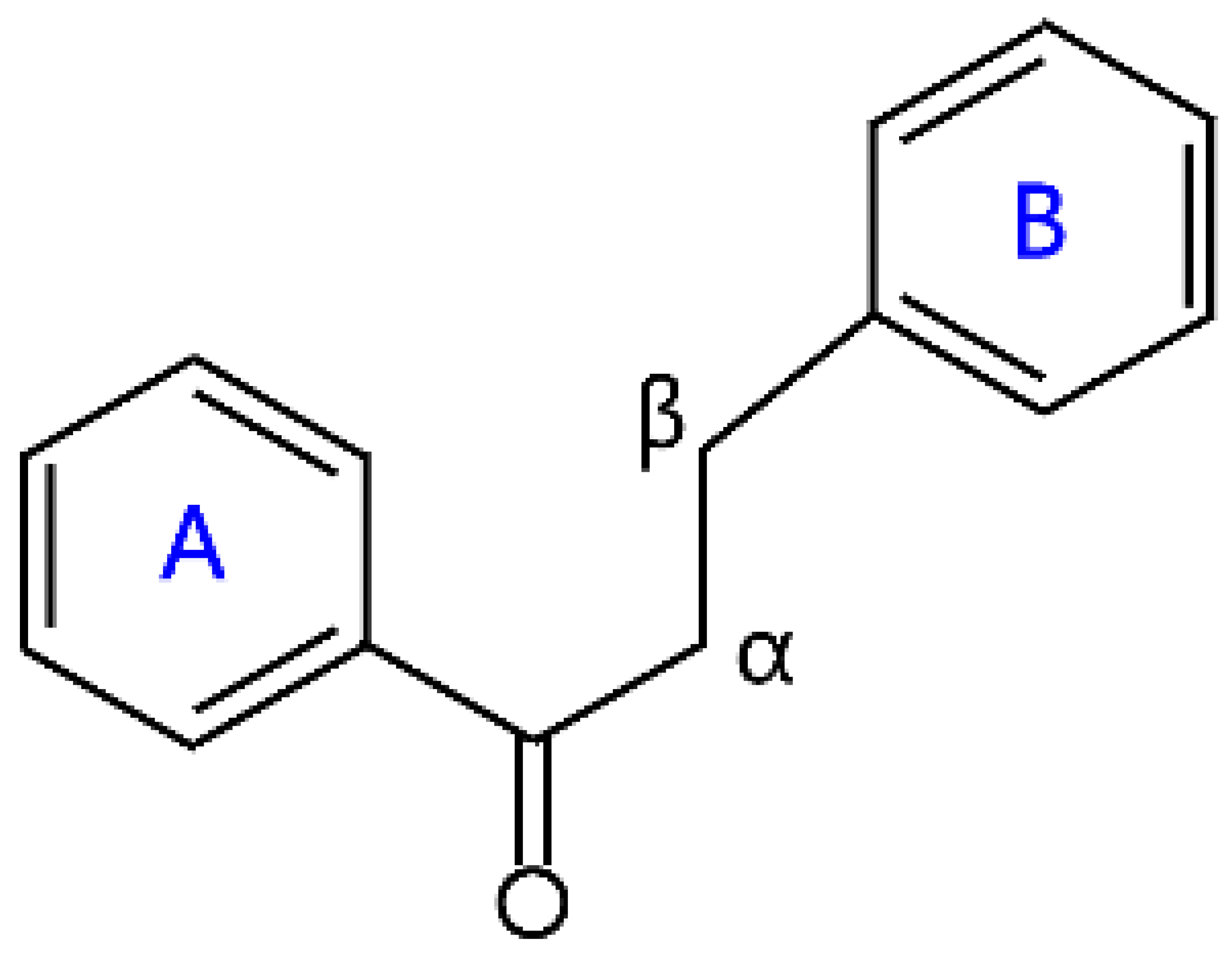 | Neohesperidin dihydrochalcone, phloridzin, phloretin, and naringin dihydrochalcone | |
| Aurones | 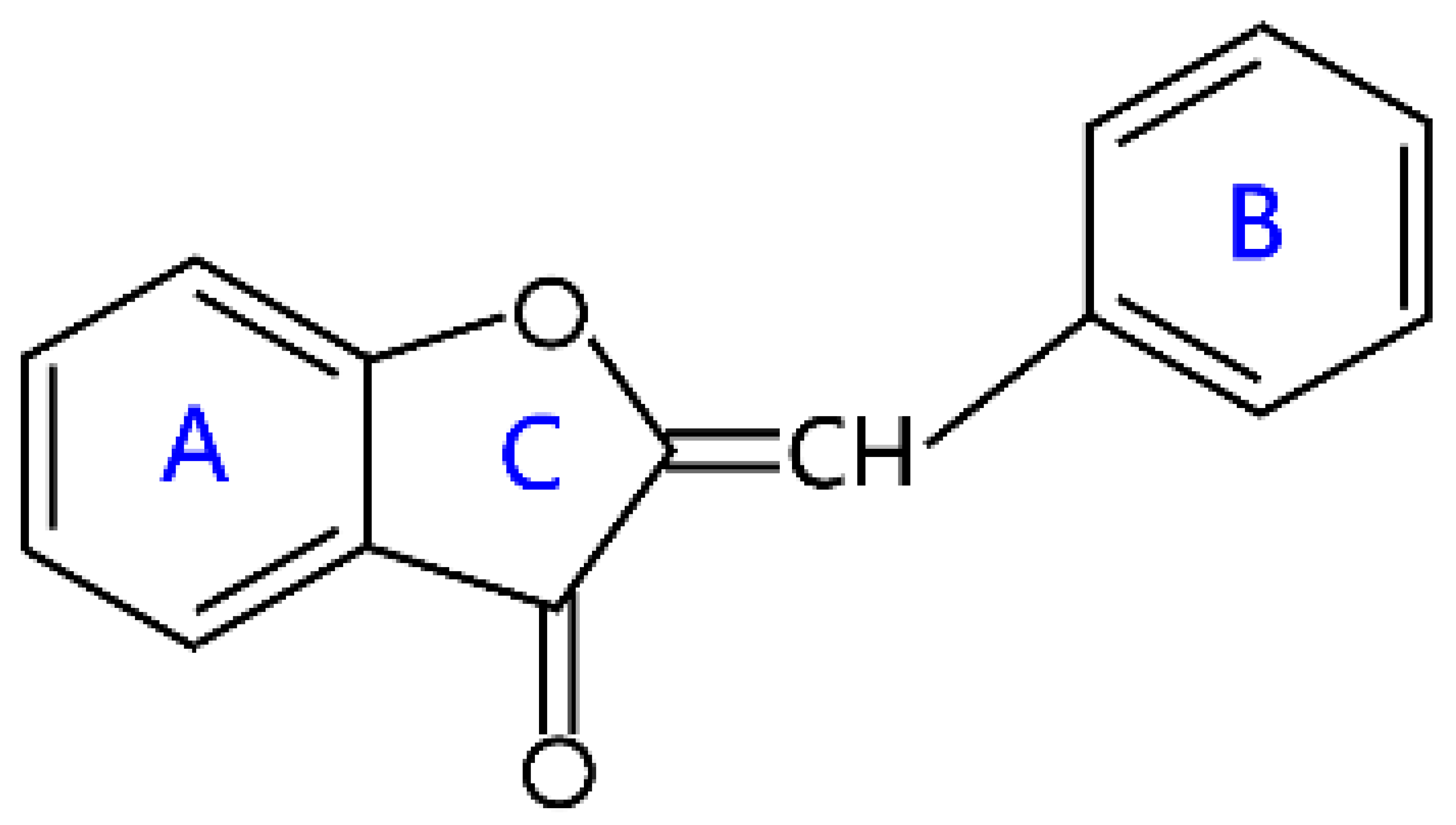 | Aureusidin, maritimetin, hispidol, and hamiltrone | |
| Isoflavonoids | Isoflavones | 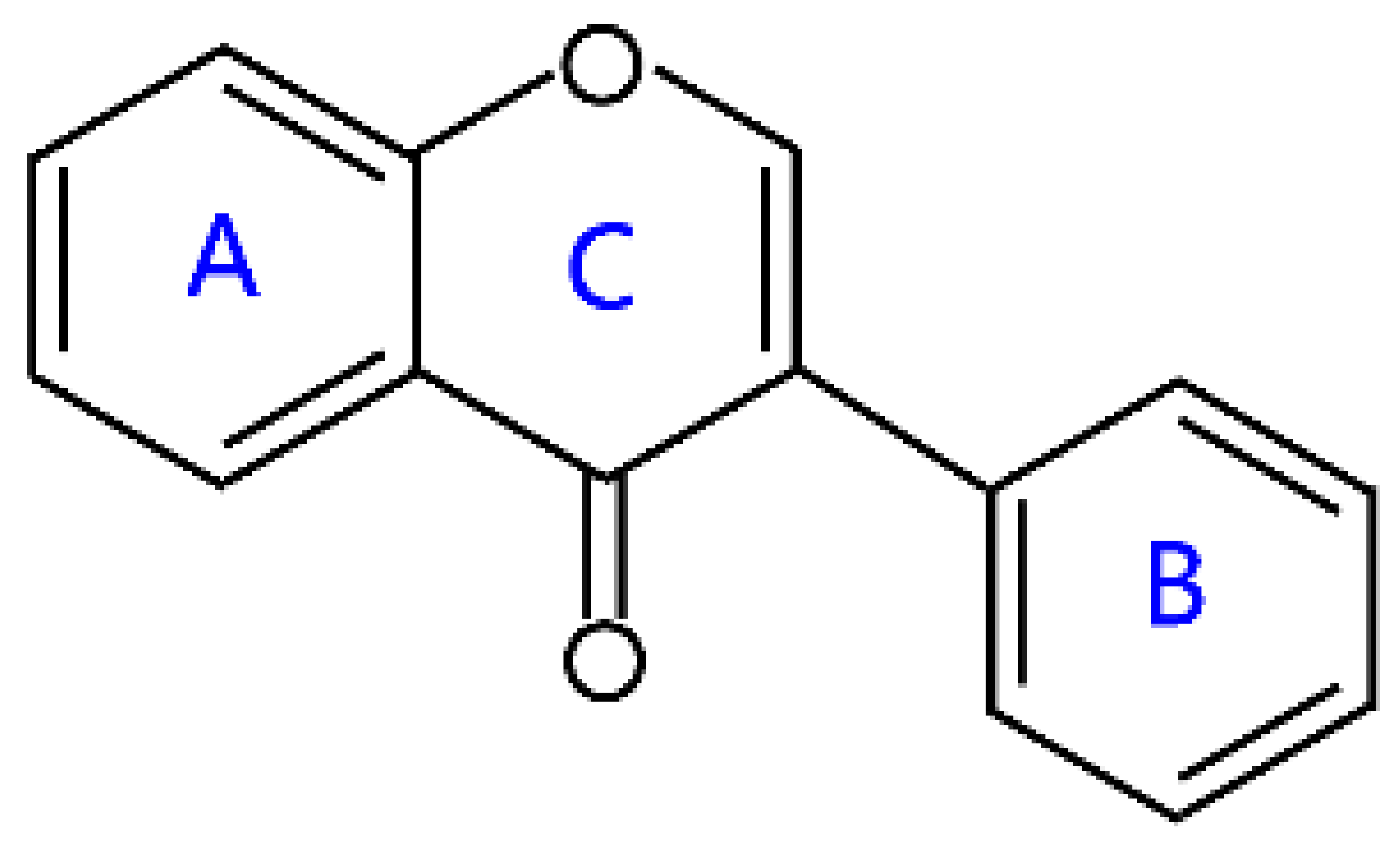 | Daidzein, genistein, biochanin A, glycitein, and formononetin |
| Isoflavanones |  | Pterostilbene, trifolium sophorin, and pterostilbene | |
| Isoflavans | 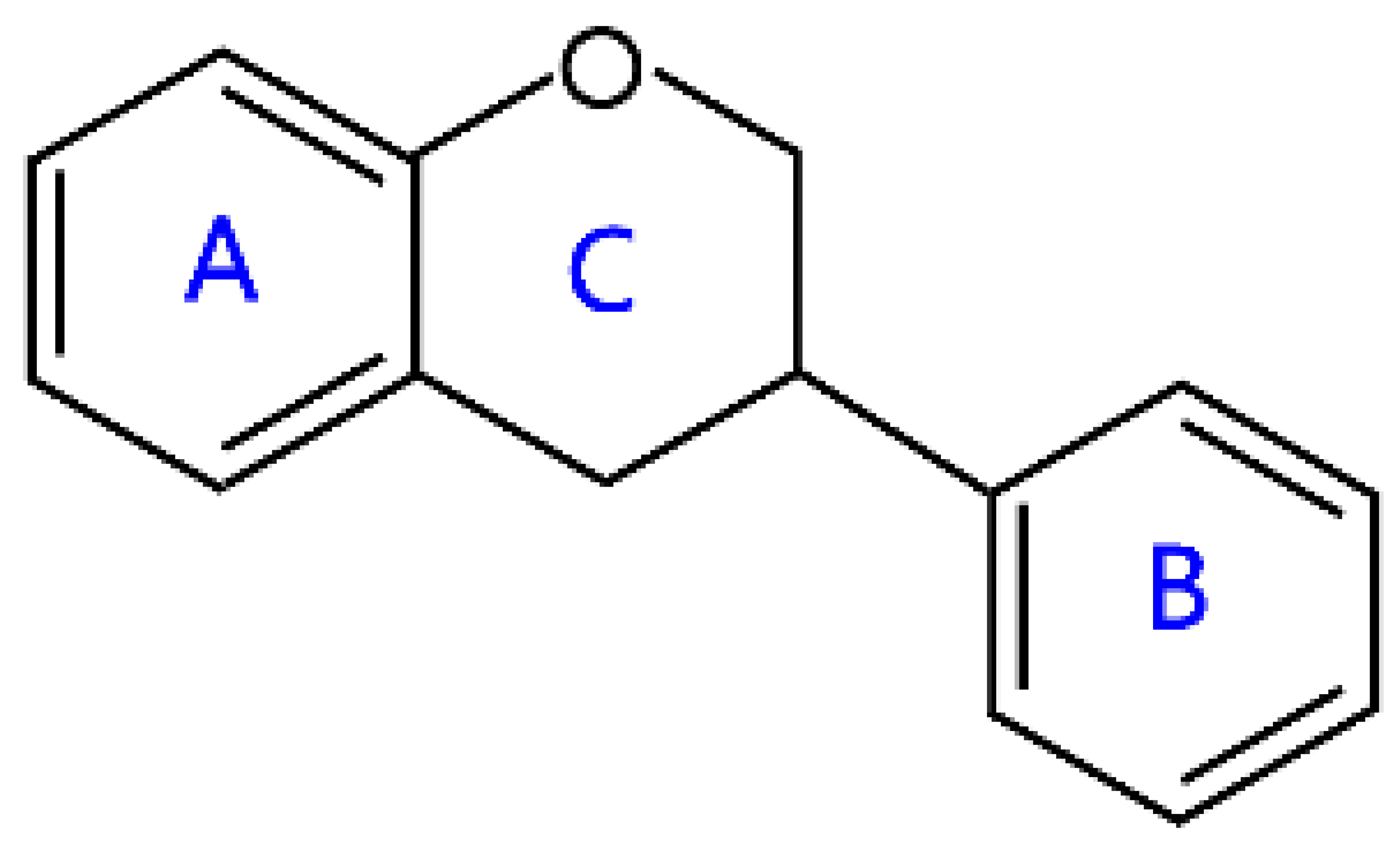 | Equol | |
| Neoflavonoids |  | Dalbergin and calophyllolide | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lou, H.; Hu, L.; Lu, H.; Wei, T.; Chen, Q. Metabolic Engineering of Microbial Cell Factories for Biosynthesis of Flavonoids: A Review. Molecules 2021, 26, 4522. https://doi.org/10.3390/molecules26154522
Lou H, Hu L, Lu H, Wei T, Chen Q. Metabolic Engineering of Microbial Cell Factories for Biosynthesis of Flavonoids: A Review. Molecules. 2021; 26(15):4522. https://doi.org/10.3390/molecules26154522
Chicago/Turabian StyleLou, Hanghang, Lifei Hu, Hongyun Lu, Tianyu Wei, and Qihe Chen. 2021. "Metabolic Engineering of Microbial Cell Factories for Biosynthesis of Flavonoids: A Review" Molecules 26, no. 15: 4522. https://doi.org/10.3390/molecules26154522
APA StyleLou, H., Hu, L., Lu, H., Wei, T., & Chen, Q. (2021). Metabolic Engineering of Microbial Cell Factories for Biosynthesis of Flavonoids: A Review. Molecules, 26(15), 4522. https://doi.org/10.3390/molecules26154522







