Point-Substitution of Phenylalanine Residues of 26RFa Neuropeptide: A Structure-Activity Relationship Study
Abstract
:1. Introduction
2. Results and Discussion
2.1. Impact of Modifications of Each Phenylalanine Residue
2.2. Impact of Concomitant Modifications of Gly20, Gly21 and Phe22 Residues
3. Materials and Methods
3.1. Reagents
3.2. Peptide Synthesis and Purification
3.3. Cell Culture
3.4. Calcium Mobilization Assays
3.5. Statistical Analysis
3.6. Nomenclature of Targets and Ligands
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
References
- Chartrel, N.; Dujardin, C.; Anouar, Y.; Leprince, J.; Decker, A.; Clerens, S.; Do-Régo, J.-C.; Vandesande, F.; Llorens-Cortes, C.; Costentin, J.; et al. Identification of 26RFa, a hypothalamic neuropeptide of the RFamide peptide family with orexigenic activity. Proc. Natl. Acad. Sci. USA 2003, 100, 15247–15252. [Google Scholar] [CrossRef] [Green Version]
- Fukusumi, S.; Yoshida, H.; Fujii, R.; Maruyama, M.; Komatsu, H.; Habata, Y.; Shintani, Y.; Hinuma, S.; Fujino, M. A new peptidic ligand and its receptor regulating adrenal function in rats. J. Biol. Chem. 2003, 278, 46387–46395. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Luo, L.; Gustafson, E.L.; Yadav, D.; Laverty, M.; Murgolo, N.; Vassileva, G.; Zeng, M.; Laz, T.M.; Behan, J.; et al. Identification and characterization of a novel RF-amide peptide ligand for orphan G-protein-coupled receptor SP9155. J. Biol. Chem. 2003, 278, 27652–27657. [Google Scholar] [CrossRef] [Green Version]
- Chartrel, N.; Dujardin, C.; Leprince, J.; Desrues, L.; Tonon, M.-C.; Cellier, E.; Cosette, P.; Jouenne, T.; Simonnet, G.; Vaudry, H. Isolation, characterization and distribution of a novel neuropeptide, Rana RFamide (R-RFa), in the brain of the European green frog Rana esculenta. J. Comp. Neurol. 2002, 448, 111–127. [Google Scholar] [CrossRef]
- Leprince, J.; Bagnol, D.; Bureau, R.; Fukusumi, S.; Granata, R.; Hinuma, S.; Larhammar, D.; Primeaux, S.; Sopkova-de Oliveira Santos, J.; Tsutsui, K.; et al. The Arg-Phe-amide peptide 26RFa/glutamine RF-amide peptide and its receptor. IUPHAR Review 24. Br. J. Pharmacol. 2017, 174, 3573–3607. [Google Scholar] [CrossRef] [PubMed]
- Ukena, K.; Tachibana, T.; Iwakoshi-Ukena, E.; Saito, Y.; Minakata, H.; Kawaguchi, R.; Osugi, T.; Tobari, Y.; Leprince, J.; Vaudry, H.; et al. Identification, localization, and function of a novel avian hypothalamic neuropeptide, 26RFa, and its cognate receptor, G protein-coupled receptor-103. Endocrinology 2010, 151, 2255–2264. [Google Scholar] [CrossRef] [PubMed]
- Tobari, Y.; Iijima, N.; Tsunekawa, K.; Osugi, T.; Haraguchi, S.; Ubuka, T.; Ukena, K.; Okanoya, K.; Tsutsui, K.; Ozawa, H. Identification, localisation and functional implication of 26RFa orthologue peptide in the brain of zebra finch (Taeniopygia guttata). J. Neuroendocrinol. 2011, 23, 791–803. [Google Scholar] [CrossRef]
- Takayasu, S.; Sakurai, T.; Iwasaki, S.; Teranishi, H.; Yamanaka, A.; Williams, S.C.; Iguchi, H.; Kawasawa, Y.I.; Ikeda, Y.; Sakakibara, I.; et al. A neuropeptide ligand of the G protein-coupled receptor GPR103 regulates feeding, behavioral arousal, and blood pressure in mice. Proc. Natl. Acad. Sci. USA 2006, 103, 7438–7443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruzzone, F.; Lectez, B.; Tollemer, H.; Leprince, J.; Dujardin, C.; Rachidi, W.; Chatenet, D.; Baroncini, M.; Beauvillain, J.-C.; Vallarino, M.; et al. Anatomical distribution and biochemical characterization of the novel RFamide peptide 26RFa in the human hypothalamus and spinal cord. J. Neurochem. 2006, 99, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Kampe, J.; Wiedmer, P.; Pfluger, P.T.; Castaneda, T.R.; Burget, L.; Mondala, H.; Kerr, J.; Liaw, C.; Oldfield, B.J.; Tschöp, M.H.; et al. Effect of central administration of QRFP(26) peptide on energy balance and characterization of a second QRFP receptor in rat. Brain Res. 2006, 1119, 133–149. [Google Scholar] [CrossRef]
- Lectez, B.; Jeandel, L.; El-Yamani, F.-Z.; Arthaud, S.; Alexandre, D.; Mardargent, A.; Jégou, S.; Mounien, L.; Bizet, P.; Magoul, R.; et al. The orexigenic activity of the hypothalamic neuropeptide 26RFa is mediated by the neuropeptide Y and proopiomelanocortin neurons of the arcuate nucleus. Endocrinology 2009, 150, 2342–2350. [Google Scholar] [CrossRef] [PubMed]
- Do Rego, J.-C.; Leprince, J.; Chartrel, N.; Vaudry, H.; Costentin, J. Behavioral effects of 26RFamide and related peptides. Peptides 2006, 27, 2715–2721. [Google Scholar] [CrossRef] [PubMed]
- Moriya, R.; Sano, H.; Umeda, T.; Ito, M.; Takahashi, Y.; Matsuda, M.; Ishihara, A.; Kanatani, A.; Iwaasa, H. RFamide peptide QRFP43 causes obesity with hyperphagia and reduced thermogenesis in mice. Endocrinology 2006, 147, 2916–2922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Primeaux, S.D.; Blackmon, C.; Barnes, M.J.; Braymer, H.D.; Bray, G.A. Central administration of the RFamide peptides, QRFP-26 and QRFP-43, increases high fat food intake in rats. Peptides 2008, 29, 1994–2000. [Google Scholar] [CrossRef] [Green Version]
- Primeaux, S.D.; Barnes, M.J.; Braymer, H.D. Hypothalamic QRFP: Regulation of food intake and fat selection. Horm. Metab. Res. 2013, 45, 967–974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zagorácz, O.; Kovács, A.; László, K.; Ollmann, T.; Péczely, L.; Lénárd, L. Effects of direct QRFP-26 administration into the medial hypothalamic area on food intake in rats. Brain Res. Bull. 2015, 118, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Navarro, V.M.; Fernández-Fernández, R.; Nogueiras, R.; Vigo, E.; Tovar, S.; Chartrel, N.; Le Marec, O.; Leprince, J.; Aguilar, E.; Pinilla, L.; et al. Novel role of 26RFa, a hypothalamic RFamide orexigenic peptide, as putative regulator of the gonadotropic axis. J. Physiol. 2006, 573, 237–249. [Google Scholar] [CrossRef]
- Patel, S.R.; Murphy, K.G.; Thompson, E.L.; Patterson, M.; Curtis, A.E.; Ghatei, M.A.; Bloom, S.R. Pyroglutamylated RFamide peptide 43 stimulates the hypothalamic-pituitary-gonadal axis via gonadotropin-releasing hormone in rats. Endocrinology 2008, 149, 4747–4754. [Google Scholar] [CrossRef]
- Palotai, M.; Telegdy, G. Anxiolytic effect of the GPR103 receptor agonist peptide P550 (homolog of neuropeptide 26RFa) in mice. Involvement of neurotransmitters. Peptides 2016, 82, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Zagorácz, O.; Ollmann, T.; Péczely, L.; László, K.; Kovács, A.; Berta, B.; Kállai, V.; Kertes, E.; Lénárd, L. QRFP administration into the medial hypothalamic nuclei improves memory in rats. Brain Res. 2020, 1727, 146563. [Google Scholar] [CrossRef]
- Yamamoto, T.; Miyazaki, R.; Yamada, T. Intracerebroventricular administration of 26RFa produces an analgesic effect in the rat formalin test. Peptides 2009, 30, 1683–1688. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Miyazaki, R.; Yamada, T.; Shinozaki, T. Anti-allodynic effects of intrathecally and intracerebroventricularly administered 26RFa, an intrinsic agonist for GRP103, in the rat partial sciatic nerve ligation model. Peptides 2011, 32, 1262–1269. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Nonaka, T.; Nakamura, S.; Araki, M.; Yamamoto, T. Microinjection of 26RFa, an endogenous ligand for the glutamine RF-amide peptide receptor (QRFP receptor), into the rostral ventromedial medulla (RVM), locus coelureus (LC), and periaqueductal grey (PAG) produces an analgesic effect in rats. Peptides 2019, 115, 1–7. [Google Scholar] [CrossRef]
- Gouardères, C.; Mazarguil, H.; Mollereau, C.; Chartrel, N.; Leprince, J.; Vaudry, H.; Zajac, J.-M. Functional differences between NPFF1 and NPFF2 receptor coupling: High intrinsic activities of RFamide-related peptides on stimulation of [35S]GTPγS binding. Neuropharmacology 2007, 52, 376–386. [Google Scholar] [CrossRef] [Green Version]
- Chartrel, N.; Alonzeau, J.; Alexandre, D.; Jeandel, L.; Alvear-Perez, R.; Leprince, J.; Boutin, J.; Vaudry, H.; Anouar, Y.; Llorens-Cortes, C. The RFamide neuropeptide 26RFa and its role in the control of neuroendocrine functions. Front. Neuroendocrinol. 2011, 32, 387–397. [Google Scholar] [CrossRef]
- Prévost, G.; Jeandel, L.; Arabo, A.; Coëffier, M.; El Ouahli, M.; Picot, M.; Alexandre, D.; Gobet, F.; Leprince, J.; Berrahmoune, H.; et al. Hypothalamic neuropeptide 26RFa acts as an incretin to regulate glucose homeostasis. Diabetes 2015, 64, 2805–2816. [Google Scholar] [CrossRef] [Green Version]
- Prévost, G.; Arabo, A.; Le Solliec, M.-A.; Bons, J.; Picot, M.; Maucotel, J.; Berrahmoune, H.; El Mehdi, M.; Cherifi, S.; Benani, A.; et al. Neuropeptide 26RFa (QRFP) is a key regulator of glucose homeostasis and its activity is markedly altered in obese/hyperglycemic mice. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E147–E157. [Google Scholar] [CrossRef]
- Prévost, G.; Picot, M.; Le Solliec, M.-A.; Arabo, A.; Berrahmoune, H.; El Mehdi, M.; Cherifi, S.; Benani, A.; Nédélec, E.; Gobet, F.; et al. The neuropeptide 26RFa in the human gut and pancreas: Potential involvement in glucose homeostasis. Endocr. Connect. 2019, 8, 941–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Mehdi, M.; Takhlidjt, S.; Khiar, F.; Prévost, G.; Do Rego, J.-L.; Do Rego, J.-C.; Benani, A.; Nedelec, E.; Godefroy, D.; Arabo, A.; et al. Glucose homeostasis is impaired in mice deficient in the neuropeptide 26RFa (QRFP). BMJ Open Diabetes Res. Care 2020, 8, e000942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quillet, R.; Ayachi, S.; Bihel, F.; Elhabazi, K.; Ilien, B.; Simonin, F. RF-amide neuropeptides and their receptors in Mammals: Pharmacological properties, drug development and main physiological functions. Pharmacol. Ther. 2016, 160, 84–132. [Google Scholar] [CrossRef]
- Mazarguil, H.; Gouardères, C.; Tafani, J.A.; Marcus, D.; Kotani, M.; Mollereau, C.; Roumy, M.; Zajac, J.M. Structure-activity relationships of neuropeptide FF: Role of C-terminal regions. Peptides 2001, 22, 1471–1478. [Google Scholar] [CrossRef]
- Boyle, R.G.; Downham, R.; Ganguly, T.; Humphries, J.; Smith, J.; Travers, S. Structure–activity studies on prolactin-releasing peptide (PrRP). Analogues of PrRP-(19–31)-peptide. J. Pept. Sci. 2005, 11, 161–165. [Google Scholar] [CrossRef]
- Orsini, M.J.; Klein, M.A.; Beavers, M.P.; Connolly, P.J.; Middleton, S.A.; Mayo, K.H. Metastin (KiSS-1) mimetics identified from peptide structure–activity relationship-derived pharmacophores and directed small molecule database screening. J. Med. Chem. 2007, 50, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Gicquel, S.; Mazarguil, H.; Desprat, C.; Allard, M.; Devillers, J.P.; Simonnet, G.; Zajac, J.M. Structure-activity study of neuropeptide FF: Contribution of N-terminal regions to affinity and activity. J. Med. Chem. 1994, 37, 3477–3481. [Google Scholar] [CrossRef]
- Mazarguil, H.; Mollereau, C.; Czaplicki, G.; Zajac, J.M. Study of the N-terminal part of peptidic selective NPFF2 agonists. Peptides 2012, 37, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Le Marec, O.; Neveu, C.; Lefranc, B.; Dubessy, C.; Boutin, J.A.; Do-Régo, J.-C.; Costentin, J.; Tonon, M.-C.; Tena-Sempere, M.; Vaudry, H.; et al. Structure–activity relationships of a series of analogues of the RFamide-related peptide 26RFa. J. Med. Chem. 2011, 54, 4806–4814. [Google Scholar] [CrossRef]
- Neveu, C.; Lefranc, B.; Tasseau, O.; Do Rego, J.-C.; Bourmaud, A.; Chan, P.; Bauchat, P.; Le Marec, O.; Chuquet, J.; Guilhaudis, L.; et al. Rational design of a low molecular weight, stable, potent, and long-lasting GPR103 aza-β3-pseudopeptide agonist. J. Med. Chem. 2012, 55, 7516–7524. [Google Scholar] [CrossRef]
- Alim, K.; Lefranc, B.; Sopkova-de Oliveira Santos, J.; Dubessy, C.; Picot, M.; Boutin, J.A.; Vaudry, H.; Chartrel, N.; Vaudry, D.; Chuquet, J.; et al. Design, synthesis, molecular dynamics simulation and functional evaluation of a novel series of 26RFa peptide analogues containing a mono- or polyalkyl guanidino arginine derivative. J. Med. Chem. 2018, 61, 10185–10197. [Google Scholar] [CrossRef]
- Pierry, C.; Couve-Bonnaire, S.; Guilhaudis, L.; Neveu, C.; Marotte, A.; Lefranc, B.; Cahard, D.; Segalas-Milazzo, I.; Leprince, J.; Pannecoucke, X. Fluorinated pseudopeptide analogues of neuropeptide 26RFa: Synthesis, biological and structural studies. ChemBioChem. 2013, 14, 1620–1633. [Google Scholar] [CrossRef]
- Rouméas, L.; Humbert, J.P.; Schneider, S.; Doebelin, C.; Bertin, I.; Schmitt, M.; Bourguignon, J.J.; Simonin, F.; Bihel, F. Effects of systematic N-terminus deletions and benzoylations of endogenous RF-amide peptides on NPFF1R, NPFF2R, GPR10, GPR54 and GPR103. Peptides 2015, 71, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Anjana, R.; Vaishnavi, M.K.; Sherlin, D.; Kumar, S.P.; Naveen, K.; Kanth, P.S.; Sekar, K. Aromatic-aromatic interactions in structures of proteins and protein-DNA complexes: A study based on orientation and distance. Bioinformation 2012, 8, 1220–1224. [Google Scholar] [CrossRef]
- Makwana, K.M.; Mahalakshmi, R. Implications of aromatic-aromatic interactions: From protein structures to peptide models. Protein Sci. 2015, 24, 1920–1933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Freitas, R.F.; Schapira, M. A systematic analysis of atomic protein-ligand interactions in the PDB. MedChemComm 2017, 8, 1970–1981. [Google Scholar] [CrossRef] [Green Version]
- Meyer, E.A.; Castellano, R.K.; Diederich, F. Interactions with aromatic rings in chemical and biological recognition. Angew. Chem. Int. Ed. Engl. 2003, 42, 1210–1250. [Google Scholar] [CrossRef] [PubMed]
- Neveu, C.; Dulin, F.; Lefranc, B.; Galas, L.; Calbrix, C.; Bureau, R.; Rault, S.; Chuquet, J.; Boutin, J.A.; Guilhaudis, L.; et al. Molecular basis of agonist docking in a human GPR103 homology model by site-directed mutagenesis and structure-activity relationship studies. Br. J. Pharmacol. 2014, 171, 4425–4439. [Google Scholar] [CrossRef] [Green Version]
- Kotha, S.; Deodhar, D.; Khedkar, P. Diversity-oriented synthesis of medicinally important 1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid (Tic) derivatives and higher analogs. Org. Biomol. Chem. 2014, 12, 9054–9091. [Google Scholar] [CrossRef]
- Williams, P.G.; Yoshida, W.Y.; Moore, R.E.; Paul, V.J. The isolation and structure elucidation of Tasiamide B, a 4-amino-3-hydroxy-5-phenylpentanoic acid containing peptide from the marine Cyanobacterium symploca sp. J. Nat. Prod. 2003, 66, 1006–1009. [Google Scholar] [CrossRef] [PubMed]
- Fauchère, J.L.; Thurieau, C. Evaluation of the stability of peptides and pseudopeptides as a tool in peptide drug design. Adv. Drug Res. 1992, 23, 127–158. [Google Scholar]
- Thuau, R.; Guilhaudis, L.; Ségalas-Milazzo, I.; Chartrel, N.; Oulyadi, H.; Boivin, S.; Fournier, A.; Leprince, J.; Davoust, D.; Vaudry, H. Structural studies on 26RFa, a novel human RFamide-related peptide with orexigenic activity. Peptides 2005, 26, 779–789. [Google Scholar] [CrossRef]
- Butterfield, S.M.; Patel, P.R.; Waters, M.L. Contribution of aromatic interactions to α-helix stability. J. Am. Chem. Soc. 2002, 124, 9751–9755. [Google Scholar] [CrossRef]
- Mosberg, H.I.; Hurst, R.; Hruby, V.J.; Gee, K.; Yamamura, H.I.; Galligan, J.J.; Burks, T.F. Bis-penicillamine enkephalins possess highly improved specificity toward delta opioid receptors. Proc. Natl. Acad. Sci. USA 1983, 80, 5871–5874. [Google Scholar] [CrossRef] [Green Version]
- Marcotte, I.; Separovic, F.; Auger, M.; Gagné, S.M. A multidimensional 1H NMR investigation of the conformation of methionine-enkephalin in fast-tumbling bicelles. Biophys. J. 2004, 86, 1587–1600. [Google Scholar] [CrossRef] [Green Version]
- Datta, S.; Shamala, N.; Banerjee, A.; Balaram, P. Conformational variability of Gly-Gly segments in peptides: A comparison of the crystal structures of an acyclic pentapeptide and an octapeptide. Biopolymers 1997, 41, 331–336. [Google Scholar] [CrossRef]
- Touchard, A.; Aili, S.R.; Téné, N.; Barassé, V.; Klopp, C.; Dejean, A.; Kini, R.M.; Mrinalini; Coquet, L.; Jouenne, T.; et al. Venom peptide repertoire of the European myrmicine ant Manica rubida: Identification of insecticidal toxins. J. Proteome Res. 2020, 19, 1800–1811. [Google Scholar] [CrossRef] [PubMed]
- Dubessy, C.; Cartier, D.; Lectez, B.; Bucharles, C.; Chartrel, N.; Montero-Hadjadje, M.; Bizet, P.; Chatenet, D.; Tostivint, H.; Scalbert, E.; et al. Characterization of urotensin II, distribution of urotensin II, urotensin II-related peptide and UT receptor mRNAs in mouse: Evidence of urotensin II at the neuromuscular junction. J. Neurochem. 2008, 17, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Pascual, E.; Leprince, J.; Martínez-Fuentes, A.J.; Ségalas-Milazzo, I.; Pineda, R.; Roa, J.; Duran-Prado, M.; Guilhaudis, L.; Desperrois, E.; Lebreton, A.; et al. In vivo and in vitro structure-activity relationships and structural conformation of kisspeptin-10-related peptides. Mol. Pharmacol. 2009, 76, 58–67. [Google Scholar] [CrossRef] [Green Version]
- Alexander, S.P.H.; Christopoulos, A.; Davenport, A.P.; Kelly, E.; Mathie, A.; Peters, J.A.; Veale, E.L.; Armstrong, J.F.; Faccenda, E.; Harding, S.D.; et al. The concise guide to pharmacology 2019/20: G protein-coupled receptors. Br. J. Pharmacol. 2019, 176, S21–S141. [Google Scholar] [CrossRef] [PubMed] [Green Version]

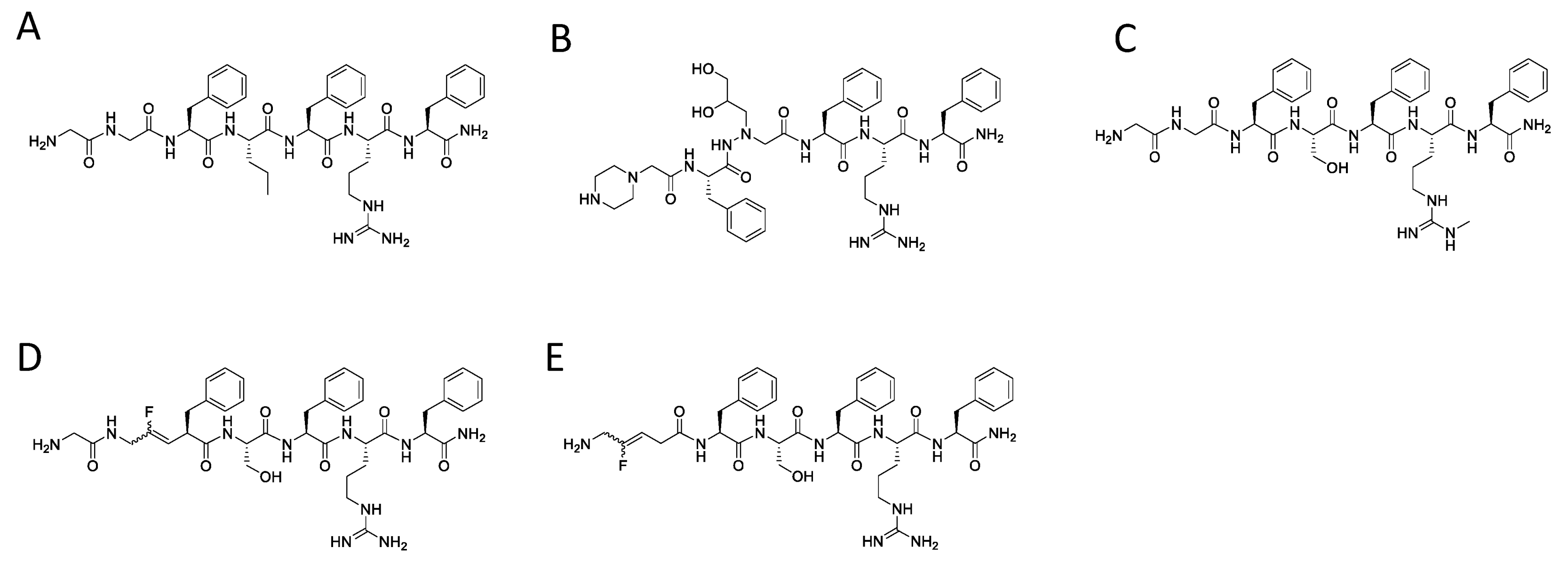

| Peptide | Structure of Residue 22, 24, 26 | Code | HPLC | MS | |||
|---|---|---|---|---|---|---|---|
| Rt (min) a | Purity (%) | Calcd b | Obsd c | ||||
| 1 | 26RFa(20–26) | LV-2021 | 18.0 | 99.9 | 815.41 | 816.53 | |
| 2 | [Thi22]26RFa(20–26) |  | LV-2050 | 18.6 | 99.9 | 821.36 | 822.33 |
| 3 | [Thi24]26RFa(20–26) | LV-2051 | 18.6 | 99.9 | 821.36 | 822.37 | |
| 4 | [Thi26]26RFa(20–26) | LV-2052 | 18.6 | 99.9 | 821.36 | 822.38 | |
| 5 | [2Nal22]26RFa(20–26) | 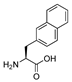 | LV-2065 | 20.4 | 99.9 | 865.42 | 866.45 |
| 6 | [2Nal24]26RFa(20–26) | LV-2213 | 20.8 | 99.9 | 865.42 | 866.46 | |
| 7 | [2Nal26]26RFa(20–26) | LV-2191 | 20.6 | 99.9 | 865.42 | 866.57 | |
| 8 | [Trp22]26RFa(20–26) | 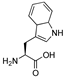 | LV-2210 | 18.9 | 99.9 | 854.42 | 855.27 |
| 9 | [Trp24]26RFa(20–26) | LV-2204 | 18.6 | 99.9 | 854.42 | 855.43 | |
| 10 | [Trp26]26RFa(20–26) | LV-2187 | 19.3 | 99.9 | 854.42 | 855.48 | |
| 11 | [ptBuPhe22]26RFa(20–26) | 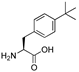 | LV-2068 | 22.1 | 99.9 | 871.47 | 872.49 |
| 12 | [ptBuPhe24]26RFa(20–26) | LV-2235 | 22.3 | 99.9 | 871.47 | 872.45 | |
| 13 | [ptBuPhe26]26RFa(20–26) | LV-2238 | 22.4 | 99.9 | 871.47 | 872.55 | |
| 14 | [Pcp22]26RFa(20–26) | 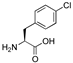 | LV-2069 | 19.9 | 99.9 | 849.37 | 850.30 |
| 15 | [Pcp24]26RFa(20–26) | LV-2232 | 20.5 | 99.9 | 849.37 | 850.41 | |
| 16 | [Pcp26]26RFa(20–26) | LV-2193 | 20.0 | 99.9 | 849.37 | 850.42 | |
| 17 | [pNO2Phe22]26RFa(20–26) |  | LV-2214 | 19.9 | 99.9 | 860.39 | 861.53 |
| 18 | [pNO2Phe24]26RFa(20–26) | LV-2236 | 19.0 | 99.9 | 860.39 | 861.46 | |
| 19 | [pNO2Phe26]26RFa(20–26) | LV-2194 | 18.9 | 99.9 | 860.39 | 861.56 | |
| 20 | [Phg22]26RFa(20–26) dia 2 | 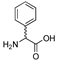 | LV-2053 | 24.2 | 97.8 | 801.39 | 802.40 |
| 21 | [Phg24]26RFa(20–26) dia 1 | LV-2054 | 21.1 | 99.9 | 801.39 | 802.40 | |
| 22 | [Phg24]26RFa(20–26) dia 2 | LV-2055 | 22.2 | 98.3 | 801.39 | 802.40 | |
| 23 | [Phg26]26RFa(20–26) dia 1 | LV-2056 | 21.0 | 99.9 | 801.39 | 802.70 | |
| 24 | [Phg26]26RFa(20–26) dia 2 | LV-2057 | 21.6 | 97.3 | 801.39 | 802.62 | |
| 25 | [NMePhe22]26RFa(20–26) |  | LV-2066 | 18.9 | 99.9 | 829.42 | 830.53 |
| 26 | [NMePhe24]26RFa(20–26) | LV-2233 | 19.2 | 98.9 | 829.42 | 830.48 | |
| 27 | [NMePhe26]26RFa(20–26) | LV-2242 | 18.8 | 99.9 | 829.42 | 830.48 | |
| 28 | [Tic22]26RFa(20–26) | 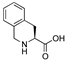 | LV-2211 | 20.3 | 99.9 | 827.41 | 828.42 |
| 29 | [Tic24]26RFa(20–26) | LV-2205 | 17.7 | 99.9 | 827.41 | 828.46 | |
| 30 | [Tic26]26RFa(20–26) | LV-2189 | 18.5 | 99.9 | 827.41 | 828.53 | |
| 31 | [NPhe22]26RFa(20–26) | 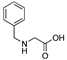 | LV-2058 | 21.1 | 99.9 | 815.41 | 816.23 |
| 32 | [NPhe24]26RFa(20–26) | LV-2060 | 17.9 | 99.9 | 815.41 | 816.56 | |
| 33 | [NPhe26]26RFa(20–26) | LV-2061 | 18.6 | 99.9 | 815.41 | 816.40 | |
| 34 | [AHPPA22]26RFa(20–26) | 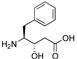 | LV-2062 | 17.8 | 99.9 | 859.43 | 860.30 |
| 35 | [AHPPA24]26RFa(20–26) | LV-2063 | 17.7 | 99.9 | 859.43 | 860.40 | |
| 36 | [AHPPA26]26RFa(20–26) | LV-2064 | 18.1 | 99.9 | 859.43 | 860.55 | |
| 37 | [DTrp22]26RFa(20–26) | 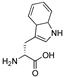 | LV-2237 | 18.2 | 99.9 | 854.42 | 855.56 |
| 38 | [DTrp24]26RFa(20–26) | LV-2070 | 17.7 | 99.9 | 854.42 | 855.53 | |
| 39 | [DTrp26]26RFa(20–26) | LV-2188 | 18.2 | 99.9 | 854.42 | 855.39 | |
| 40 | [DTic22]26RFa(20–26) |  | LV-2212 | 18.4 | 99.9 | 827.41 | 828.49 |
| 41 | [DTic24]26RFa(20–26) | LV-2215 | 18.2 | 99.9 | 827.41 | 828.36 | |
| 42 | [DTic26]26RFa(20–26) | LV-2190 | 18.3 | 99.9 | 827.41 | 828.41 | |
| 43 | [Oic22]26RFa(20–26) | 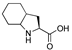 | LV-2067 | 18.6 | 99.9 | 819.44 | 820.56 |
| 44 | [Oic24]26RFa(20–26) | LV-2234 | 18.3 | 99.9 | 819.44 | 820.36 | |
| 45 | [Oic26]26RFa(20–26) | LV-2241 | 18.1 | 99.9 | 819.44 | 820.45 | |
| 46 | [Cys22,24]26RFa(20–26) |  | LV-2105 | 11.5 | 99.9 | 725.27 | 726.28 |
| 47 | [Cys22,26]26RFa(20–26) | LV-2107 | 6.7 | 99.9 | 725.27 | 726.37 | |
| 48 | [Cys24,26]26RFa(20–26) | LV-2106 | 11.8 | 99.9 | 725.27 | 726.32 | |
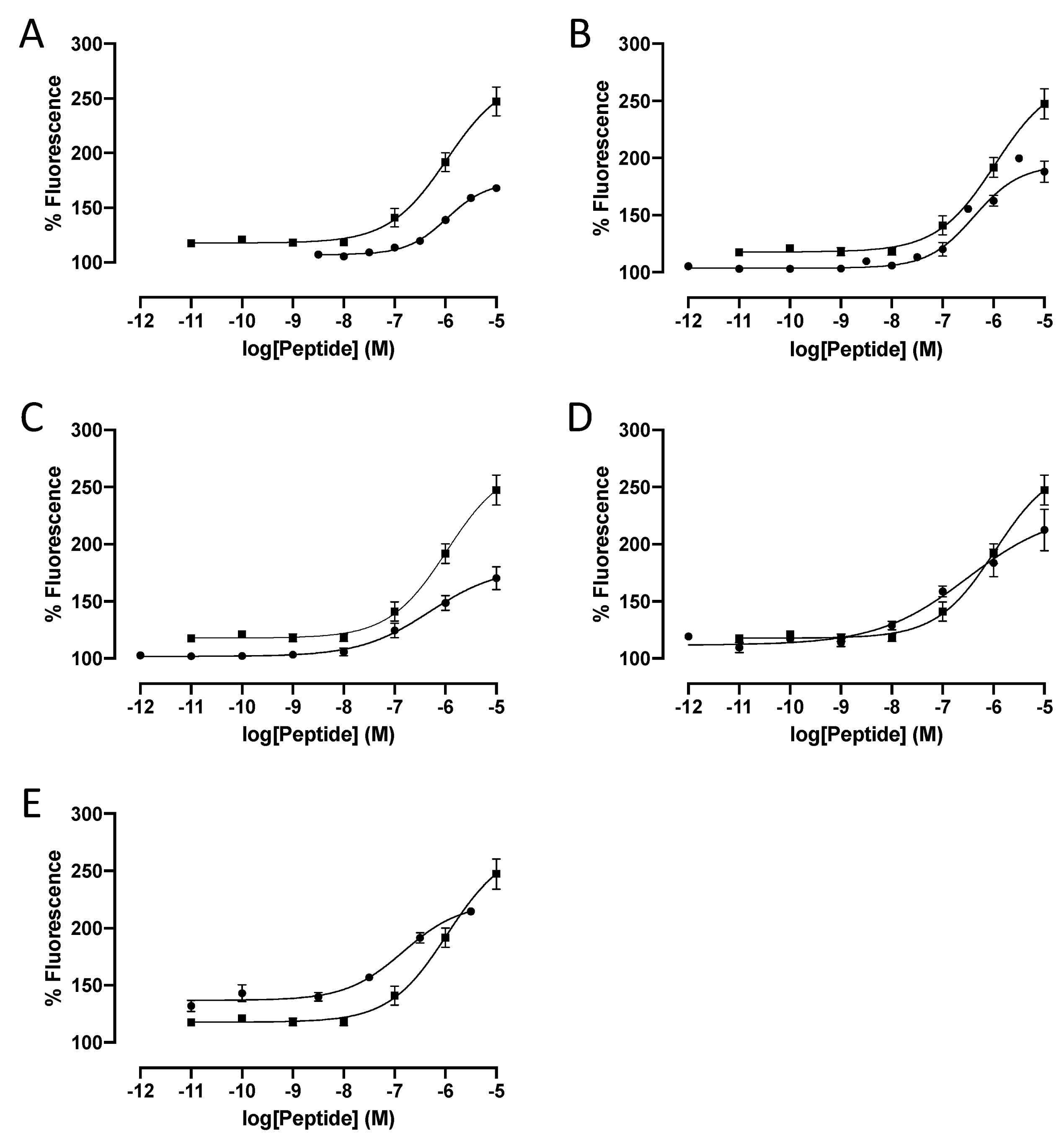
| Peptide | Code | EC50 | IC50 | Imax | |||||
|---|---|---|---|---|---|---|---|---|---|
| (nM) | (nM) | (%) a | |||||||
| 1 | 26RFa(20–26) | LV-2021 | 1640 | ± | 259 | - | |||
| 2 | [Thi22]26RFa(20–26) | LV-2050 | 3160 | ± | 1110 | - | |||
| 3 | [Thi24]26RFa(20–26) | LV-2051 | 2530 | ± | 1020 | - | |||
| 4 | [Thi26]26RFa(20–26) | LV-2052 | 7227 | ± | 96 | - | |||
| 5 | [2Nal22]26RFa(20–26) | LV-2065 | 1150 | ± | 170 | - | |||
| 6 | [2Nal24]26RFa(20–26) | LV-2213 | 15,300 | ± | 5400 | - | |||
| 7 | [2Nal26]26RFa(20–26) | LV-2191 | 3790 | ± | 1800 | - | |||
| 8 | [Trp22]26RFa(20–26) | LV-2210 | 1980 | ± | 880 | - | |||
| 9 | [Trp24]26RFa(20–26) | LV-2204 | 2040 | ± | 1600 | - | |||
| 10 | [Trp26]26RFa(20–26) | LV-2187 | 5060 | ± | 1300 | - | |||
| 11 | [ptBuPhe22]26RFa(20–26) | LV-2068 | >10,000 | 6220 | ± | 2500 | 45 | ||
| 12 | [ptBuPhe24]26RFa(20–26) | LV-2235 | >10,000 | ND | |||||
| 13 | [ptBuPhe26]26RFa(20–26) | LV-2238 | 1040 | ± | 25 ** | - | |||
| 14 | [Pcp22]26RFa(20–26) | LV-2069 | 5000 | ± | 4500 | - | |||
| 15 | [Pcp24]26RFa(20–26) | LV-2232 | 850 | ± | 240 NS | - | |||
| 16 | [Pcp26]26RFa(20–26) | LV-2193 | 457 | ± | 71 ** | - | |||
| 17 | [pNO2Phe22]26RFa(20–26) | LV-2214 | 2130 | ± | 1100 | - | |||
| 18 | [pNO2Phe24]26RFa(20–26) | LV-2236 | >10,000 | ND | |||||
| 19 | [pNO2Phe26]26RFa(20–26) | LV-2194 | 491 | ± | 33 ** | - | |||
| 20 | [Phg22]26RFa(20–26) dia 2 | LV-2053 | 5010 | ± | 3600 | - | |||
| 21 | [Phg24]26RFa(20–26) dia 1 | LV-2054 | >10,000 | ND | |||||
| 22 | [Phg24]26RFa(20–26) dia 2 | LV-2055 | >10,000 | ND | |||||
| 23 | [Phg26]26RFa(20–26) dia 1 | LV-2056 | >10,000 | ND | |||||
| 24 | [Phg26]26RFa(20–26) dia 2 | LV-2057 | >10,000 | ND | |||||
| 25 | [NMePhe22]26RFa(20–26) | LV-2066 | 15,100 | ± | 4200 | - | |||
| 26 | [NMePhe24]26RFa(20–26) | LV-2233 | 1360 | ± | 750 | - | |||
| 27 | [NMePhe26]26RFa(20–26) | LV-2242 | >10,000 | ND | |||||
| 28 | [Tic22]26RFa(20–26) | LV-2211 | 327 | ± | 170 * | - | |||
| 29 | [Tic24]26RFa(20–26) | LV-2205 | >10,000 | ND | |||||
| 30 | [Tic26]26RFa(20–26) | LV-2189 | 7700 | ± | 4400 | - | |||
| 31 | [NPhe22]26RFa(20–26) | LV-2058 | 578 | ± | 110 * | - | |||
| 32 | [NPhe24]26RFa(20–26) | LV-2060 | >10,000 | ND | |||||
| 33 | [NPhe26]26RFa(20–26) | LV-2061 | >10,000 | ND | |||||
| 34 | [AHPPA22]26RFa(20–26) | LV-2062 | 2850 | ± | 380 | - | |||
| 35 | [AHPPA24]26RFa(20–26) | LV-2063 | >10,000 | ND | |||||
| 36 | [AHPPA26]26RFa(20–26) | LV-2064 | >10,000 | ND | |||||
| 37 | [DTrp22]26RFa(20–26) | LV-2237 | 6160 | ± | 2400 | - | |||
| 38 | [DTrp24]26RFa(20–26) | LV-2070 | >10,000 | ND | |||||
| 39 | [DTrp26]26RFa(20–26) | LV-2188 | >10,000 | >10,000 | 46 | ||||
| 40 | [DTic22]26RFa(20–26) | LV-2212 | 4990 | ± | 2100 | - | |||
| 41 | [DTic24]26RFa(20–26) | LV-2215 | 6200 | ± | 2600 | - | |||
| 42 | [DTic26]26RFa(20–26) | LV-2190 | >10,000 | ND | |||||
| 43 | [Oic22]26RFa(20–26) | LV-2067 | >10,000 | ND | |||||
| 44 | [Oic24]26RFa(20–26) | LV-2234 | >10,000 | ND | |||||
| 45 | [Oic26]26RFa(20–26) | LV-2241 | >10,000 | ND | |||||
| 46 | [Cys22,24]26RFa(20–26) | LV-2105 | >10,000 | ND | |||||
| 47 | [Cys22,26]26RFa(20–26) | LV-2107 | >10,000 | ND | |||||
| 48 | [Cys24,26]26RFa(20–26) | LV-2106 | >10,000 | ND | |||||
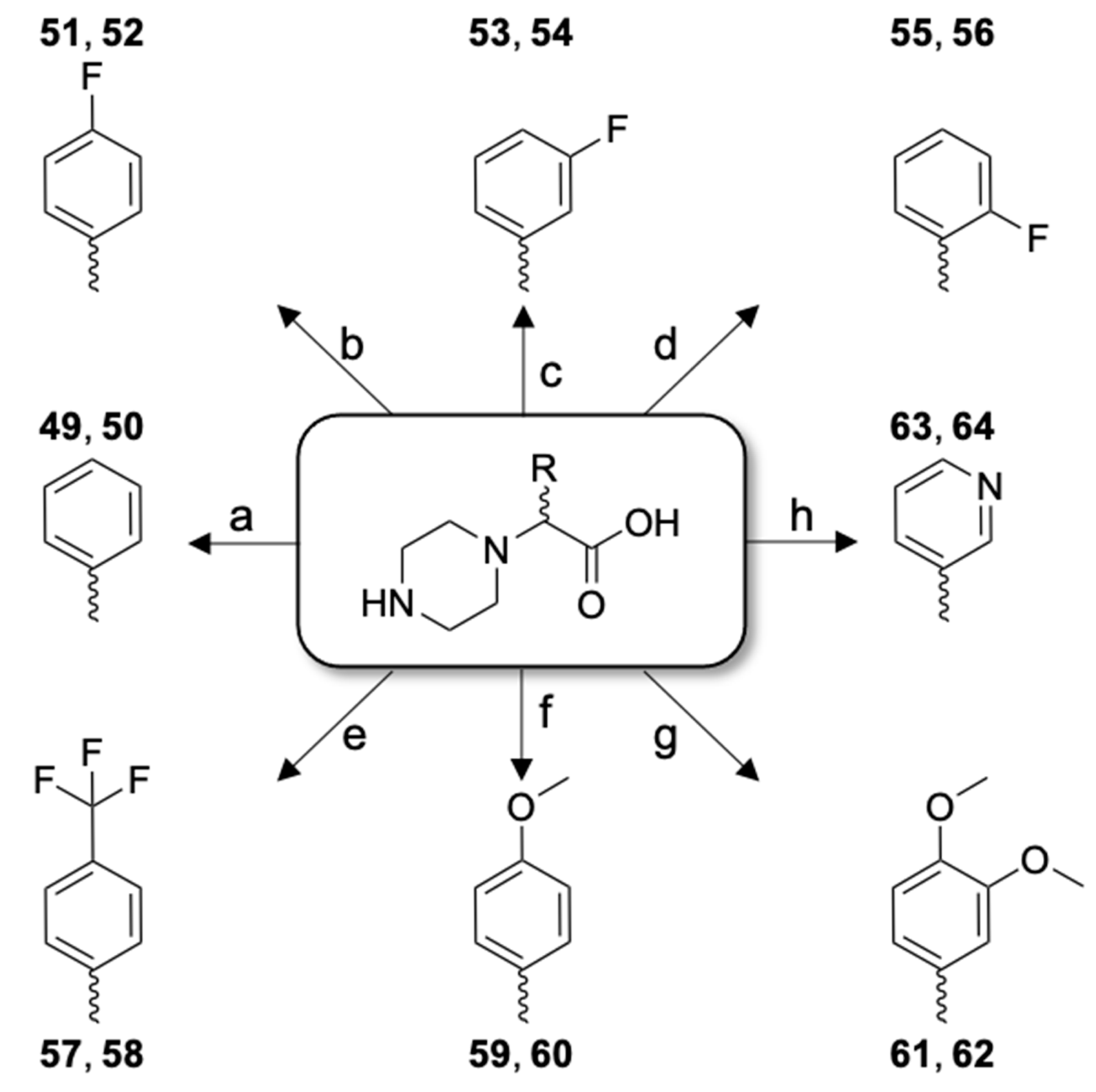
| Peptide Derivative | Code | HPLC | MS | |||
|---|---|---|---|---|---|---|
| Rt (min) a | Purity (%) | Calcd b | Obsd c | |||
| 1 | 26RFa(20–26) | LV-2021 | 18.0 | 99.9 | 815.41 | 816.53 |
| 49 | [Cmpi(Phg)22]26RFa(22–26) dia 1 | LV-2099 | 12.8 | 99.9 | 756.41 | 757.46 |
| 50 | [Cmpi(Phg)22]26RFa(22–26) dia 2 | LV-2100 | 12.9 | 99.9 | 756.41 | 757.46 |
| 51 | [Cmpi(4-FPhg)22]26RFa(22–26) dia 1 | LV-2101 | 15.3 | 99.9 | 774.40 | 775.37 |
| 52 | [Cmpi(4-FPhg)22]26RFa(22–26) dia 2 | LV-2102 | 15.7 | 99.9 | 774.40 | 775.39 |
| 53 | [Cmpi(3-FPhg)22]26RFa(22–26) dia 1 | LV-2103 | 15.3 | 99.9 | 774.40 | 775.42 |
| 54 | [Cmpi(3-FPhg)22]26RFa(22–26) dia 2 | LV-2104 | 15.8 | 99.9 | 774.40 | 775.41 |
| 55 | [Cmpi(2-FPhg)22]26RFa(22–26) dia 1 | LV-2175 | 13.9 | 98.4 | 774.40 | 775.47 |
| 56 | [Cmpi(2-FPhg)22]26RFa(22–26) dia 2 | LV-2176 | 14.7 | 99.9 | 774.40 | 775.53 |
| 57 | [Cmpi(4-TfmPhg)22]26RFa(22–26) dia 1 | LV-2177 | 25.3 | 99.9 | 824.39 | 825.47 |
| 58 | [Cmpi(4-TfmPhg)22]26RFa(22–26) dia 2 | LV-2178 | 26.2 | 99.9 | 824.39 | 825.39 |
| 59 | [Cmpi(4-MeOPhg)22]26RFa(22–26) dia 1 | LV-2179 | 13.2 | 99.9 | 786.42 | 787.47 |
| 60 | [Cmpi(4-MeOPhg)22]26RFa(22–26) dia 2 | LV-2180 | 13.4 | 99.9 | 786.42 | 787.41 |
| 61 | [Cmpi(3,4-diMeOPhg)22]26RFa(22–26) dia 1 | LV-2181 | 10.9 | 99.9 | 816.43 | 817.37 |
| 62 | [Cmpi(3,4-diMeOPhg)22]26RFa(22–26) dia 2 | LV-2182 | 11.5 | 99.9 | 816.43 | 817.49 |
| 63 | [Cmpi(3-Pyg)22]26RFa(22–26) dia 1 | LV-2183 | 19.5 | 99.9 | 757.40 | 758.40 |
| 64 | [Cmpi(3-Pyg)22]26RFa(22–26) dia 2 | LV-2184 | 20.3 | 99.9 | 757.40 | 758.31 |
| Peptide Derivative | Code | EC50 | IC50 | |||
|---|---|---|---|---|---|---|
| (nM) | (nM) | |||||
| 1 | 26RFa(20–26) | LV-2021 | 1640 | ± | 259 | - |
| 49 | [Cmpi(Phg)22]26RFa(22–26) dia 1 | LV-2099 | >10,000 | ND | ||
| 50 | [Cmpi(Phg)22]26RFa(22–26) dia 2 | LV-2100 | >10,000 | ND | ||
| 51 | [Cmpi(4-FPhg)22]26RFa(22–26) dia 1 | LV-2101 | 4050 | ± | 1200 | - |
| 52 | [Cmpi(4-FPhg)22]26RFa(22–26) dia 2 | LV-2102 | 1660 | ± | 788 | - |
| 53 | [Cmpi(3-FPhg)22]26RFa(22–26) dia 1 | LV-2103 | 3560 | ± | 440 | - |
| 54 | [Cmpi(3-FPhg)22]26RFa(22–26) dia 2 | LV-2104 | 2710 | ± | 1000 | - |
| 55 | [Cmpi(2-FPhg)22]26RFa(22–26) dia 1 | LV-2175 | 2480 | ± | 1113 | - |
| 56 | [Cmpi(2-FPhg)22]26RFa(22–26) dia 2 | LV-2176 | >10,000 | ND | ||
| 57 | [Cmpi(4-TfmPhg)22]26RFa(22–26) dia 1 | LV-2177 | >10,000 | ND | ||
| 58 | [Cmpi(4-TfmPhg)22]26RFa(22–26) dia 2 | LV-2178 | >10,000 | ND | ||
| 59 | [Cmpi(4-MeOPhg)22]26RFa(22–26) dia 1 | LV-2179 | >10,000 | ND | ||
| 60 | [Cmpi(4-MeOPhg)22]26RFa(22–26) dia 2 | LV-2180 | >10,000 | ND | ||
| 61 | [Cmpi(3,4-diMeOPhg)22]26RFa(22–26) dia 1 | LV-2181 | >10,000 | ND | ||
| 62 | [Cmpi(3,4-diMeOPhg)22]26RFa(22–26) dia 2 | LV-2182 | >10,000 | ND | ||
| 63 | [Cmpi(3-Pyg)22]26RFa(22–26) dia 1 | LV-2183 | 4990 | ± | 2800 | - |
| 64 | [Cmpi(3-Pyg)22]26RFa(22–26) dia 2 | LV-2184 | >10,000 | ND | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lefranc, B.; Alim, K.; Neveu, C.; Le Marec, O.; Dubessy, C.; Boutin, J.A.; Chuquet, J.; Vaudry, D.; Prévost, G.; Picot, M.; et al. Point-Substitution of Phenylalanine Residues of 26RFa Neuropeptide: A Structure-Activity Relationship Study. Molecules 2021, 26, 4312. https://doi.org/10.3390/molecules26144312
Lefranc B, Alim K, Neveu C, Le Marec O, Dubessy C, Boutin JA, Chuquet J, Vaudry D, Prévost G, Picot M, et al. Point-Substitution of Phenylalanine Residues of 26RFa Neuropeptide: A Structure-Activity Relationship Study. Molecules. 2021; 26(14):4312. https://doi.org/10.3390/molecules26144312
Chicago/Turabian StyleLefranc, Benjamin, Karima Alim, Cindy Neveu, Olivier Le Marec, Christophe Dubessy, Jean A. Boutin, Julien Chuquet, David Vaudry, Gaëtan Prévost, Marie Picot, and et al. 2021. "Point-Substitution of Phenylalanine Residues of 26RFa Neuropeptide: A Structure-Activity Relationship Study" Molecules 26, no. 14: 4312. https://doi.org/10.3390/molecules26144312
APA StyleLefranc, B., Alim, K., Neveu, C., Le Marec, O., Dubessy, C., Boutin, J. A., Chuquet, J., Vaudry, D., Prévost, G., Picot, M., Vaudry, H., Chartrel, N., & Leprince, J. (2021). Point-Substitution of Phenylalanine Residues of 26RFa Neuropeptide: A Structure-Activity Relationship Study. Molecules, 26(14), 4312. https://doi.org/10.3390/molecules26144312








