Cost-Effective Production of L-DOPA by Tyrosinase-Immobilized Polyhydroxyalkanoate Nanogranules in Engineered Halomonas bluephagenesis TD01
Abstract
:1. Introduction
2. Results
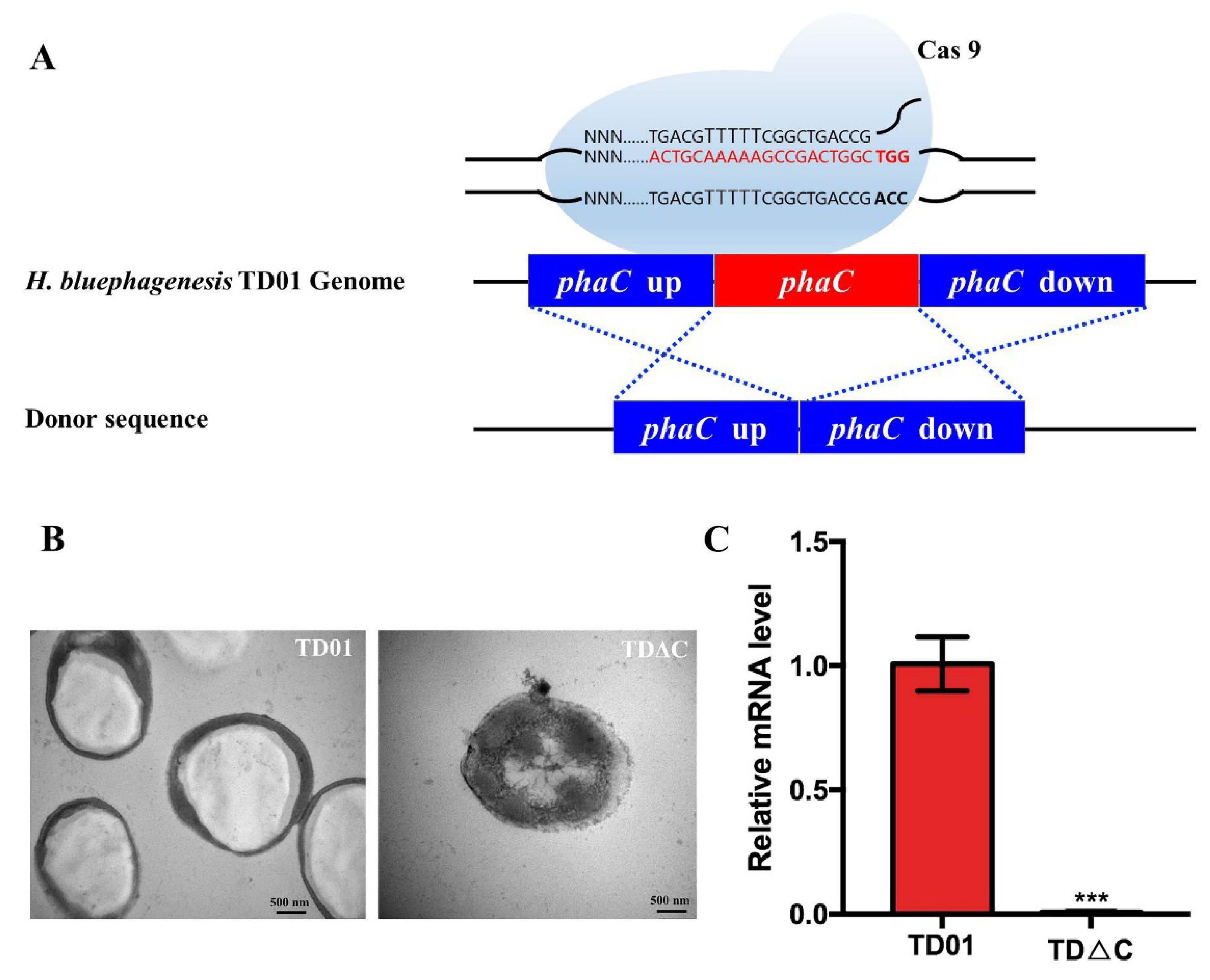
2.1. Construction and Identification of PhaCHb Knockout Mutant H. bluephagenesis TDΔC
2.2. Expression of Free Tyrosinase TyrVs in H. bluephagenesis TD Strain and Determination of Its L-DOPA Productivity
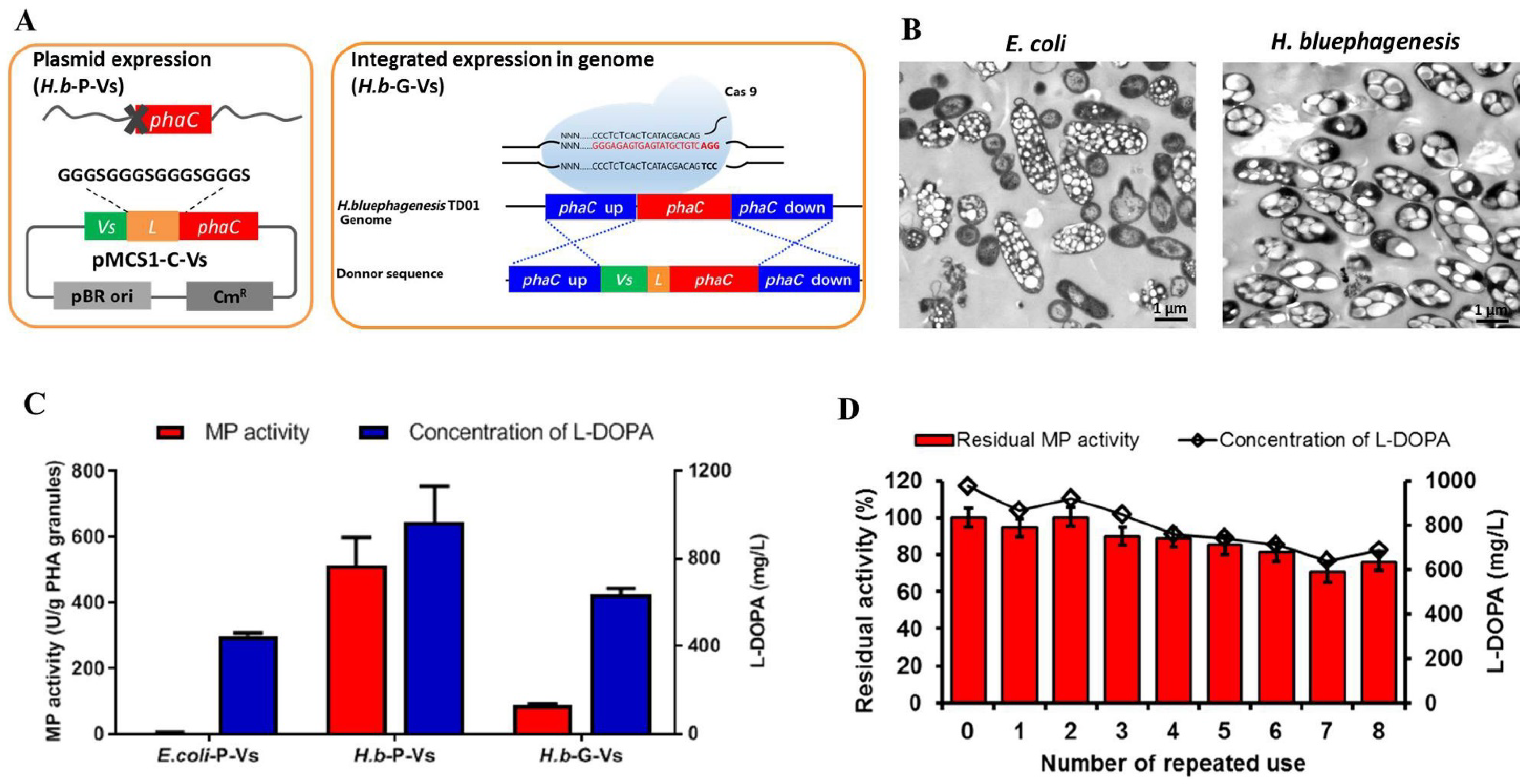
2.3. One-Step Production and Identification of PHA-TyrVs Nanogranules by H. bluephagenesis
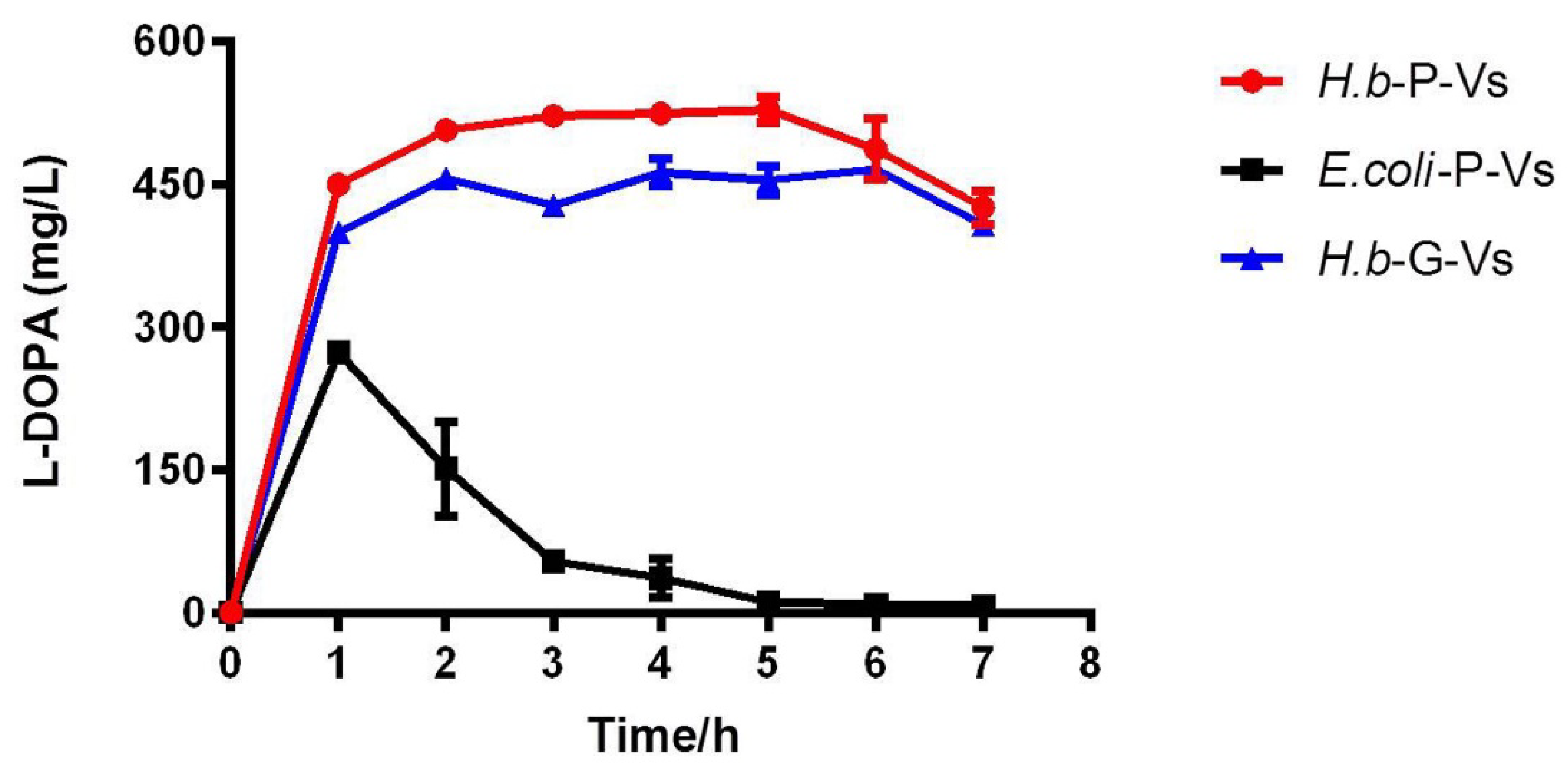
2.4. L-DOPA Production and Reusability of PHA-TyrVs Nanogranules Produced by H. bluephagenesis
2.5. Robust L-DOPA Production by Cell Lysates Containing PHA-TyrVs Nanogranules from Three Hosts
3. Discussions
4. Materials and Methods
4.1. Microorganisms, Plasmids, and Culture Conditions
4.2. Molecular Manipulations of H. bluephagenesis TD Strain
4.2.1. Construction of Expression Vectors for Free and Immobilized TyrVs in H. bluephagenesis TD Strain
4.2.2. Genome Editing by Adapted CRISPR/Cas9 in H. bluephagenesis TD01
4.3. Production and Purification of PHA-TyrVs Nanogranules
4.4. Characterizations of PHA-TyrVs Nanogranules
4.4.1. PHA Content Determination and Granule Visualization
4.4.2. Identification and Absolute Quantification of Immobilized TyrVs on PHA Nanogranules via MS-Based Quantitative Proteomic
4.5. Production of L-DOPA
4.5.1. Production of L-DOPA In Vivo
4.5.2. Production of L-DOPA In Vitro by Cell Lysates or Purified PHA-TyrVs Nanogranules
4.6. Determination of L-DOPA Concentration and the Specific Monophenolase (MP) Activity of Tyrosinase
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Etemadi, F.; Hashemi, M.; Autio, W.R.; Mangan, F.X.; Omidreza, Z. Yield and accumulation trend of biomass and L-dopa in different parts of eight faba bean cultivars. Crop Sci. 2018, 58, 2020–2028. [Google Scholar] [CrossRef]
- Surwase, S.N.; Jadhav, J.P. Bioconversion of L-tyrosine to L-DOPA by a novel bacterium Bacillus sp. JPJ. Amino Acids 2011, 41, 495–506. [Google Scholar] [CrossRef]
- Fordjour, E.; Adipah, F.K.; Zhou, S.H.; Du, G.C.; Zhou, J.W. Metabolic engineering of Escherichia coli BL21 (DE3) for de novo production of L-DOPA from D-glucose. Microb. Cell Factories 2019, 18, 74. [Google Scholar] [CrossRef]
- Patil, S.A.; Apine, O.A.; Surwase, S.N.; Jadhav, J.P. Biological sources of L-DOPA: An alternative approach. Adv. Parkinson’s Dis. 2013, 2, 81–87. [Google Scholar] [CrossRef]
- Tan, X.; Song, W.; Chen, X.; Liu, L.; Wu, J. Recent advances in biocatalytic derivatization of L-tyrosine. Appl. Microbiol. Biotechnol. 2020, 104, 9907–9920. [Google Scholar] [CrossRef]
- Bulduk, I. Optimization of extraction techniques and RP-HPLC analysis of anti-parkinson drug levodopa from flowers of Vicia faba L. Acta Chromatogr. 2020, 32, 281–288. [Google Scholar] [CrossRef] [Green Version]
- Min, K.; Park, K.; Park, D.H.; Yoo, Y.J. Overview on the biotechnological production of L-DOPA. Appl. Microbiol. Biotechnol. 2015, 99, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Faccio, G.; Kruus, K.; Saloheimo, M.; Thony-Meyer, L. Bacterial tyrosinases and their applications. Process Biochem. 2012, 47, 1749–1760. [Google Scholar] [CrossRef]
- Yuan, W.; Zhong, S.; Xiao, Y.M.; Wang, Z.; Sun, J. Efficient biocatalyst of L-DOPA with Escherichia coli expressing a tyrosine phenol-lyase mutant from Kluyvera intermedia. Appl. Biochem. Biotechnol. 2020, 190, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Munoz, A.J.; Hernandez-Chavez, G.; de Anda, R.; Martinez, A.; Bolivar, F.; Gosset, G. Metabolic engineering of Escherichia coli for improving L-3,4-dihydroxyphenylalanine (L-DOPA) synthesis from glucose. J. Ind. Microbiol. Biotechnol. 2011, 38, 1845–1852. [Google Scholar] [CrossRef]
- Ali, S.; Shultz, J.L. Ikram-ul-Haq. High performance microbiological transformation of L-tyrosine to L-dopa by Yarrowia lipolytica NRRL-143. BMC Biotechnol. 2007, 7, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koyanagi, T.; Katayama, T.; Suzuki, H.; Nakazawa, H.; Yokozeki, K.; Kumagai, H. Effective production of 3,4-dihydroxyphenyl-L-alanine (L-DOPA) with Erwinia herbicola cells carrying a mutant transcriptional regulator TyrR. J. Biotechnol. 2005, 115, 303–306. [Google Scholar] [CrossRef] [PubMed]
- Fairhead, M.; Thony-Meyer, L. Role of the C-terminal extension in a bacterial tyrosinase. FEBS J. 2010, 277, 2083–2095. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Borron, J.C.; Solano, F. Molecular anatomy of tyrosinase and its related proteins: Beyond the histidine-bound metal catalytic center. Pigment Cell Res. 2002, 15, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Capecchi, E.; Piccinino, D.; Delfino, I.; Bollella, P.; Antiochia, R.; Saladino, R. Functionalized tyrosinase-lignin nanoparticles as sustainable catalysts for the oxidation of phenols. Nanomaterials 2018, 8, 438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pialis, P.; Saville, B.A. Production of L-DOPA from tyrosinase immobilized on nylon 6,6: Enzyme stability and scaleup. Enzym. Microb. Technol. 1998, 22, 261–268. [Google Scholar] [CrossRef]
- Seetharam, G.; Saville, B.A. L-DOPA production from tyrosinase immobilized on zeolite. Enzym. Microb. Technol. 2002, 31, 747–753. [Google Scholar] [CrossRef]
- Tan, D.; Zhao, J.P.; Ran, G.Q.; Zhu, X.L.; Lu, X.Y. Highly efficient biocatalytic synthesis of L-DOPA using in situ immobilized Verrucomicrobium spinosum tyrosinase on polyhydroxyalkanoate nano-granules. Appl. Microbiol. Biot. 2019, 1033, 5663–5678. [Google Scholar] [CrossRef]
- Bresan, S.; Sznajder, A.; Hauf, W.; Forchhammer, K.; Pfeiffer, D.; Jendrossek, D. Polyhydroxyalkanoate (PHA) granules have no phospholipids. Sci. Rep. 2016, 6, 26612. [Google Scholar] [CrossRef] [Green Version]
- Jendrossek, D. Polyhydroxyalkanoate Granules are complex subcellular organelles (Carbonosomes). J. Bacteriol. 2009, 191, 3195–3202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.Q.; Jiang, X.R. Engineering microorganisms for improving polyhydroxyalkanoate biosynthesis. Curr. Opin. Biotechnol. 2018, 53, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.; Wang, Y.; Tong, Y.; Chen, G.Q.J.T.i.B. Grand challenges for industrializing polyhydroxyalkanoates (PHAs). Trends Biotechnol. 2021. [Google Scholar] [CrossRef]
- Ye, J.W.; Hu, D.K.; Yin, J.; Huang, W.Z.; Xiang, R.J.; Zhang, L.Z.; Wang, X.; Han, J.N.; Chen, G.Q. Stimulus response-based fine-tuning of polyhydroxyalkanoate pathway in Halomonas. Metab. Eng. 2020, 57, 85–95. [Google Scholar] [CrossRef]
- Tan, D.; Xue, Y.S.; Aibaidula, G.; Chen, G.Q. Unsterile and continuous production of polyhydroxybutyrate by Halomonas TD01. Bioresour. Technol. 2011, 102, 8130–8136. [Google Scholar] [CrossRef]
- Qin, Q.; Ling, C.; Zhao, Y.; Yang, T.; Yin, J.; Guo, Y.; Chen, G.Q. CRISPR/Cas9 editing genome of extremophile Halomonas spp. Metab. Eng. 2018, 47, 219–229. [Google Scholar] [CrossRef]
- Jiang, X.R.; Yao, Z.H.; Chen, G.Q. Controlling cell volume for efficient PHB production by Halomonas. Metab. Eng. 2017, 44, 30–37. [Google Scholar] [CrossRef]
- Cai, L.; Tan, D.; Aibaidula, G.; Dong, X.R.; Chen, J.C.; Tian, W.D.; Chen, G.Q. Comparative genomics study of polyhydroxyalkanoates (PHA) and ectoine relevant genes from Halomonas sp TD01 revealed extensive horizontal gene transfer events and co-evolutionary relationships. Microb. Cell Factories 2011, 10, 88. [Google Scholar] [CrossRef] [Green Version]
- Pastor, J.M.; Salvador, M.; Argandona, M.; Bernal, V.; Reina-Bueno, M.; Csonka, L.N.; Iborra, J.L.; Vargas, C.; Nieto, J.J.; Canovas, M. Ectoines in cell stress protection: Uses and biotechnological production. Biotechnol. Adv. 2010, 28, 782–801. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, L.H. Ectoine improves yield of biodiesel catalyzed by immobilized lipase. J. Mol. Catal. B Enzym. 2010, 62, 91–96. [Google Scholar] [CrossRef]
- Min, K.; Park, G.W.; Yoo, Y.J.; Lee, J.S. A perspective on the biotechnological applications of the versatile tyrosinase. Bioresour. Technol. 2019, 289, 121730. [Google Scholar] [CrossRef] [PubMed]
- Fairhead, M.; Thony-Meyer, L. Bacterial tyrosinases: Old enzymes with new relevance to biotechnology. New Biotechnol. 2012, 29, 183–191. [Google Scholar] [CrossRef]
- Kovach, M.E.; Phillips, R.W.; Elzer, P.H.; Roop, R.M.; Peterson, K.M. pBBR1MCS: A broad-host-range cloning vector. Biotechniques 1994, 16, 800. [Google Scholar] [CrossRef]
- Simon, R.; Priefer, U.; Puhler, A. A broad host mobilization system for in vivo genetic engineering: Transposon mutagenesis in Gram-negative bacteria. Bio/Technolgy 1983, 1, 37–45. [Google Scholar] [CrossRef]
- Tan, D.; Wu, Q.; Chen, J.C.; Chen, G.Q. Engineering Halomonas TD01 for the low-cost production of polyhydroxyalkanoates. Metab. Eng. 2014, 26, 34–47. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Engler, C.; Gruetzner, R.; Kandzia, R.; Marillonnet, S. Golden Gate Shuffling: A one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS ONE 2009, 4, e5553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, X.Z.; Tan, D.; Aibaidula, G.; Wu, Q.; Chen, J.C.; Chen, G.Q. Development of Halomonas TD01 as a host for open production of chemicals. Metab. Eng. 2014, 23, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Jahns, A.C.; Haverkamp, R.G.; Rehm, B.H.A. Multifunctional inorganic-binding beads self-assembled inside engineered bacteria. Bioconj. Chem. 2008, 19, 2072–2080. [Google Scholar] [CrossRef]
- Denner, E.; Mcgenity, T.J.; Busse, H.; Grant, W.D.; Wanner, G. Halococcus salifodinae sp. nov., an archaeal isolate from an Austrian salt mine. Int. J. Syst. Bacteriol. 1994, 44, 774–780. [Google Scholar] [CrossRef] [Green Version]
- Ran, G.Q.; Tan, D.; Dai, W.E.; Zhu, X.L.; Zhao, J.P.; Ma, Q.; Lu, X.Y. Immobilization of alkaline polygalacturonate lyase from Bacillus subtilis on the surface of bacterial polyhydroxyalkanoate nano-granules. Appl. Microbiol. Biot. 2017, 101, 3247–3258. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Arnow, L.E. Colorimetric determination of the components of 3,4-dihydroxyphenylalanine tyrosine mixtures. J. Biol. Chem. 1937, 118, 531–537. [Google Scholar] [CrossRef]


| Source of PHA-TyrVs Nanogranules | Plasmid Expression in E. coli (E.coli-P-Vs) | Plasmid Expression in H. bluephagenesis TD (H.b-P-Vs) | Genome Expression in H. bluephagenesis TD (H.b-G-Vs) |
|---|---|---|---|
| CDW (g/L) | 1.14 ± 0.05 | 2.50 ± 0.17 | 9.64 ± 0.53 |
| Content of PHA nanogranules in cell (wt%) | 22.28 ± 1.25 | 23.22 ± 0.40 | 56.10 ± 2.60 |
| Theoretical concentration of PHA nanogranules (g/L) | 0.25 ± 0.01 | 0.58 ± 0.06 | 5.41 ± 1.12 |
| MS quantification of immobilized content of TyrVs on PHA nanogranules | 0.06% | 6.85% | 3.48% |
| Strains/Plasmids | Descriptions | Sources/References |
|---|---|---|
| Strains | ||
| E. coli DH5α | F-φ-5dlacZΔlac Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK+,mk+) phoA supE44 λ-thi-1 gyrA96 relA1 | Takara |
| E. coli BL21 (DE3) | F- ompT hsdSB (rB- mB-) gal dcm (DE3) | Novagen |
| E. coli S17-1 pir | TpR SmR recA, thi, pro, hsdR-M+RP4: 2-Tc:Mu: Km Tn7 λpir | [33] |
| H. bluephagenesis TD01 | H. bluephagenesis TD01 wild type, isolated from a salt lake in China | [24] |
| H. bluephagenesis TDΔC | H. bluephagenesis TD01 with phaCHb gene deleted | This study |
| H. bluephagenesis H.b-G-Vs | H. bluephagenesis TD01 with tyrVs-phaCHb fusion gene knocked in | This study |
| Plasmids | ||
| pQ08 | pSEVA321 derivative, Streptococcus pyogenes cas9, CmR | [25] |
| pQ31 | pSEVA241 derivative, PJ23119-sgRNA template, KmR and SpeR | [25] |
| pQ31-C | pQ31 derivative, PJ23119-sgRNA template, 1000 bp donor DNA for phaCHb knockout, KmR and SpeR | This study |
| pQ31-VsC | pQ31 derivative, PJ23119-sgRNA template, 3001 bp donor DNA for tyrVs-phaCHb knockin, KmR and SpeR | This study |
| pBBR1MCS-1 | A broad host plasmid for H. bluephagenesis TD01, KmR | [32] |
| pMCS1-Vs | pBBR1MCS-1 derivative, Pporin promoter, free tyrosinase TyrVs expression vector in H. bluephagenesis TD, CmR | This study |
| pMCS1-C-Vs | pBBR1MCS-1 derivative, Pporin promoter, expression vector for PHA-TyrVs nanogranules in H. bluephagenesis TD, CmR | This study |
| pCDF-Vs | Expression vector for free tyrosinase from Verrucomicrobium spinosum TyrVs in E. coli, SmR | [18] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Ran, G.; Xu, M.; Lu, X.; Tan, D. Cost-Effective Production of L-DOPA by Tyrosinase-Immobilized Polyhydroxyalkanoate Nanogranules in Engineered Halomonas bluephagenesis TD01. Molecules 2021, 26, 3778. https://doi.org/10.3390/molecules26133778
Zhao J, Ran G, Xu M, Lu X, Tan D. Cost-Effective Production of L-DOPA by Tyrosinase-Immobilized Polyhydroxyalkanoate Nanogranules in Engineered Halomonas bluephagenesis TD01. Molecules. 2021; 26(13):3778. https://doi.org/10.3390/molecules26133778
Chicago/Turabian StyleZhao, Jiping, Ganqiao Ran, Mengmeng Xu, Xiaoyun Lu, and Dan Tan. 2021. "Cost-Effective Production of L-DOPA by Tyrosinase-Immobilized Polyhydroxyalkanoate Nanogranules in Engineered Halomonas bluephagenesis TD01" Molecules 26, no. 13: 3778. https://doi.org/10.3390/molecules26133778
APA StyleZhao, J., Ran, G., Xu, M., Lu, X., & Tan, D. (2021). Cost-Effective Production of L-DOPA by Tyrosinase-Immobilized Polyhydroxyalkanoate Nanogranules in Engineered Halomonas bluephagenesis TD01. Molecules, 26(13), 3778. https://doi.org/10.3390/molecules26133778






