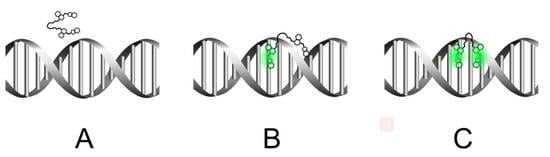
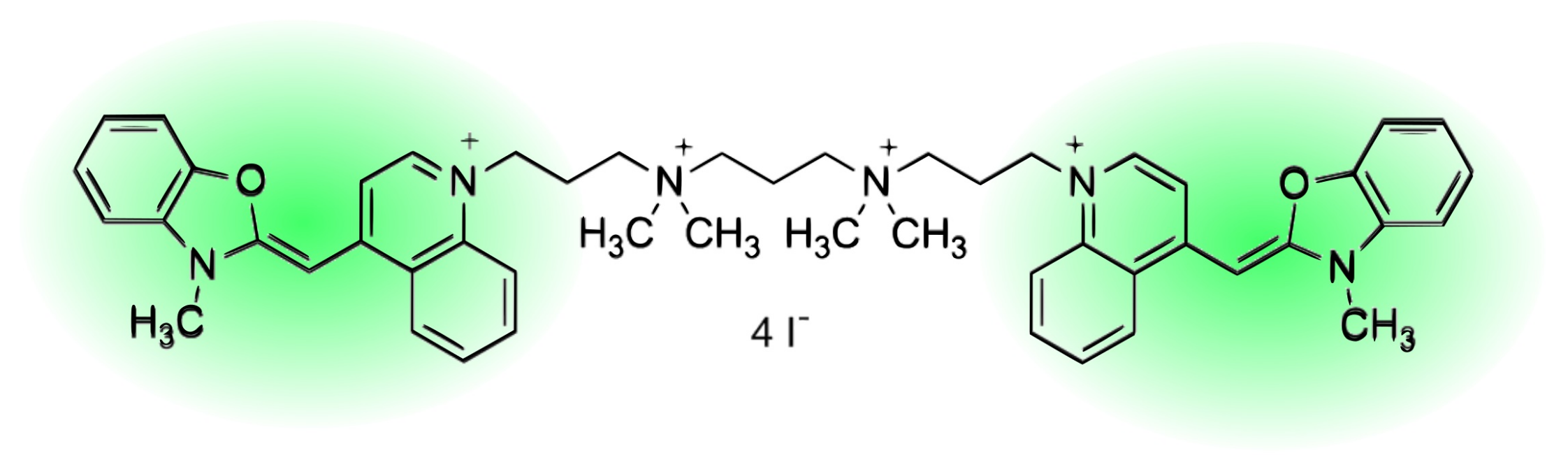
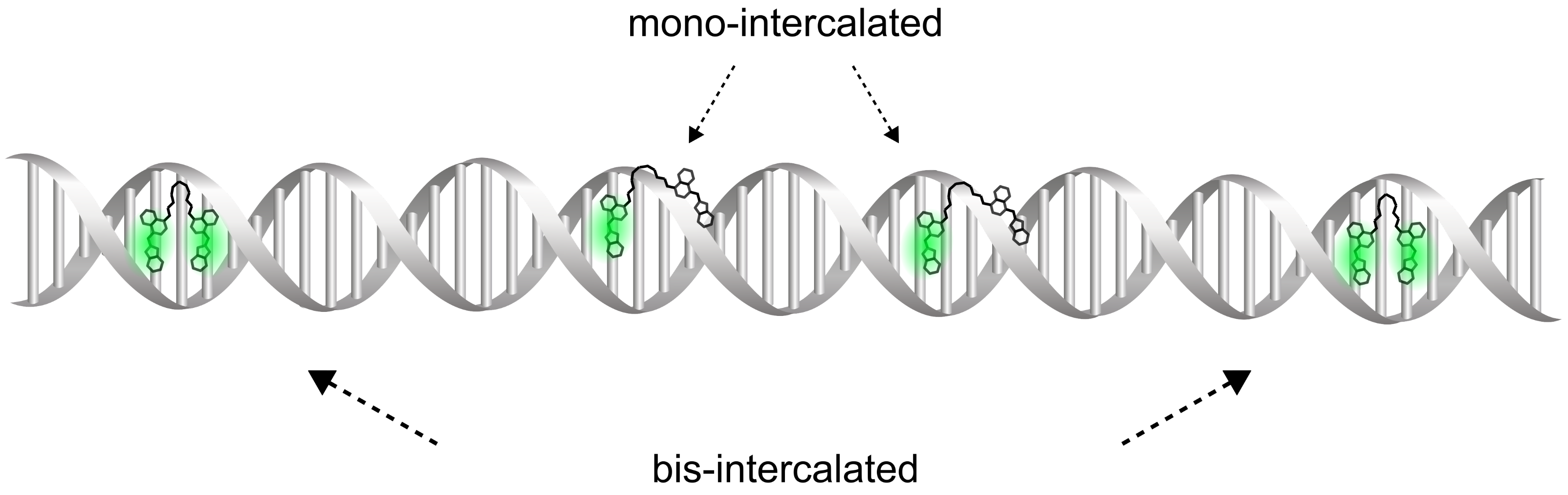
Two Intercalation Mechanisms of Oxazole Yellow Dimer (YOYO-1) into DNA
Abstract
1. Introduction
2. Results
2.1. The Validation of DNA-YOYO-1 Binding Models
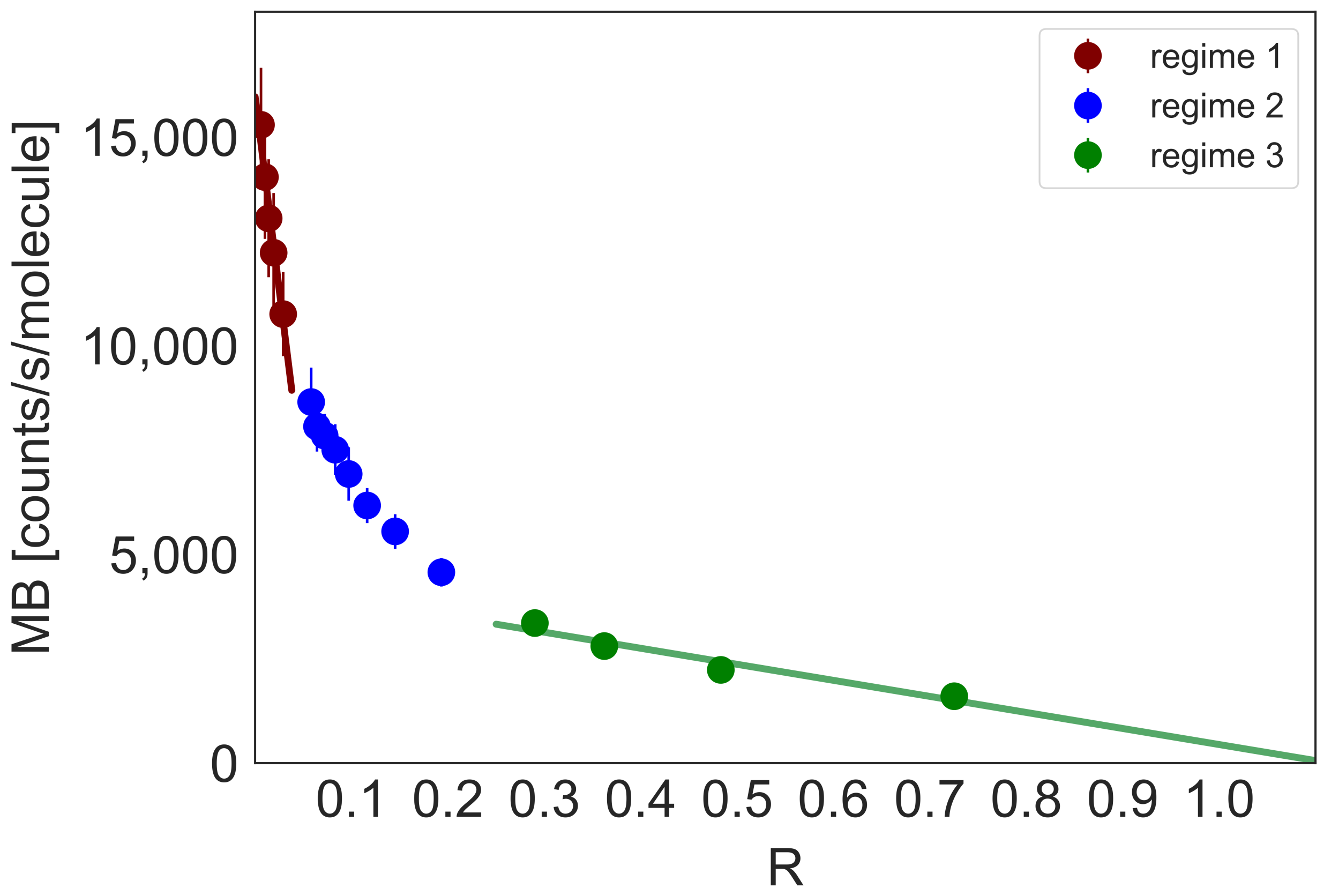
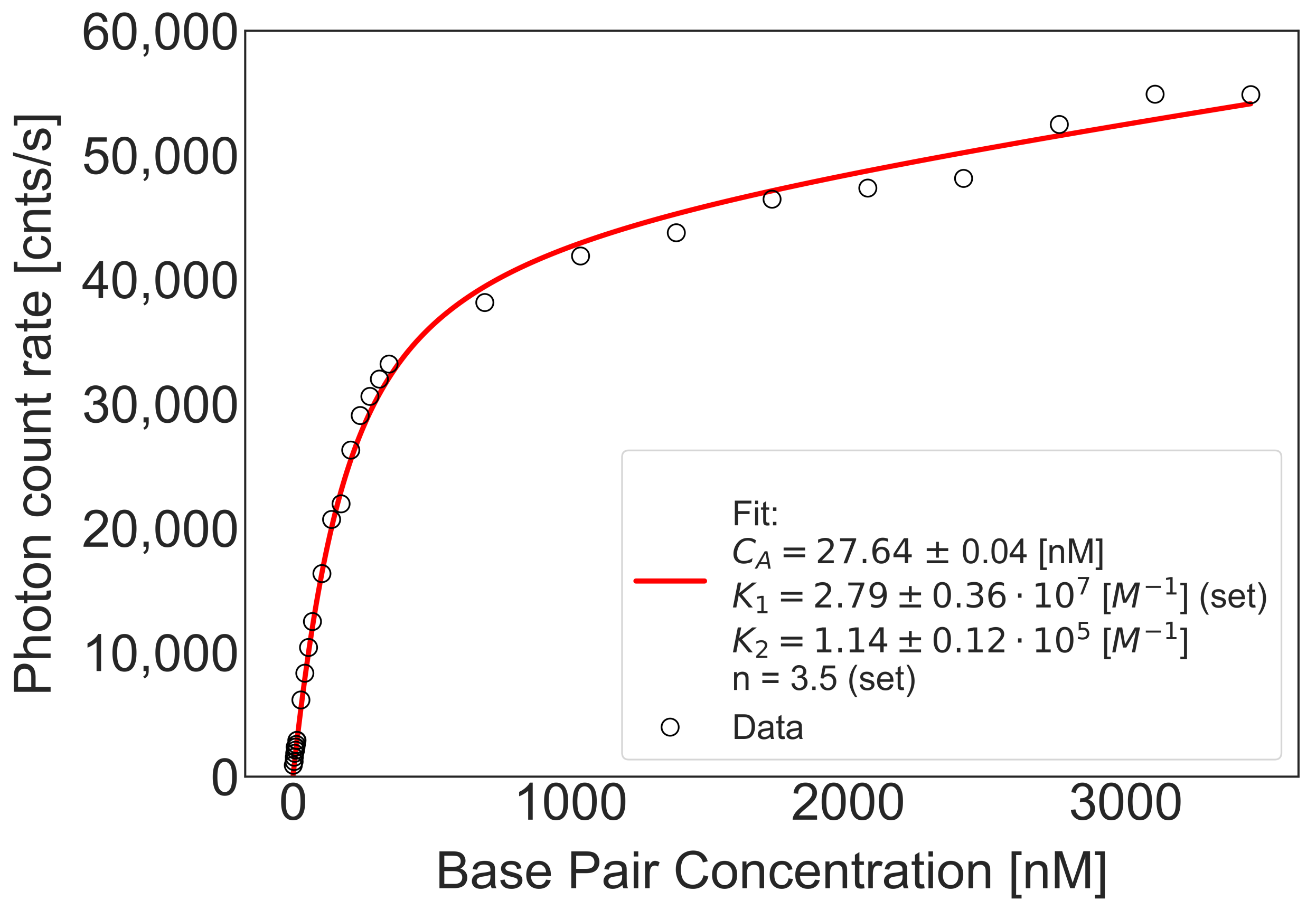
2.2. Equilibrium Constants of DNA-YOYO-1 Complexes Formation
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. FCS Set-Up
4.3. FCS Measurements
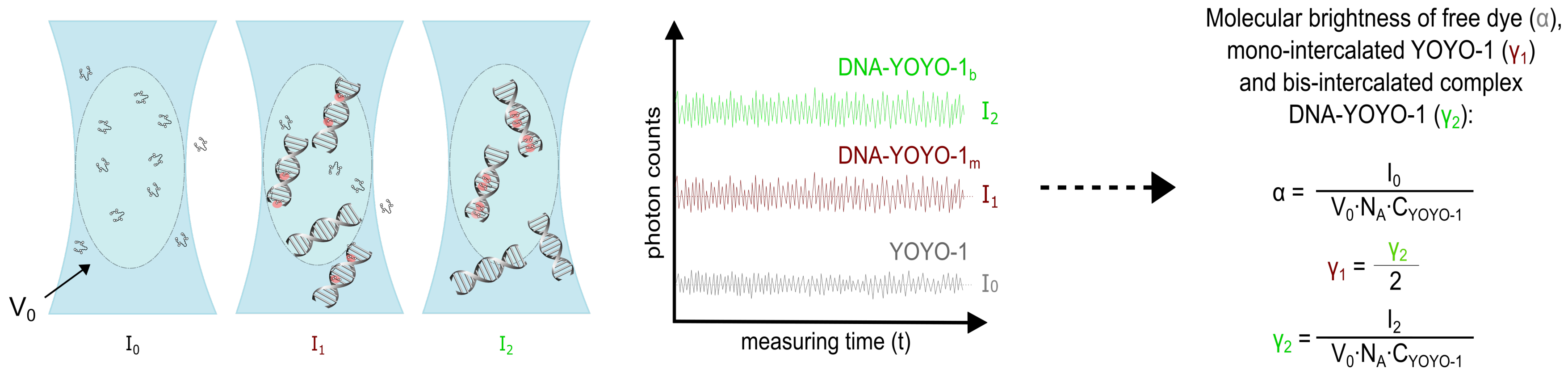
4.4. Brightness Method
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Schwille, P. Fluorescence correlation spectroscopy and its potential for intracellular applications. Cell Biochem. Biophys. 2001, 34, 383–408. [Google Scholar] [CrossRef]
- Plum, G.E.; Breslauer, K.J. Fluorescence energy transfer monitored competitive equilibria of nucleic acids: Applications in thermodynamics and screening. Biopolymers 2002, 61, 214–223. [Google Scholar] [CrossRef]
- Thompson, N.L.; Drake, A.W.; Chen, L.; Vanden Broek, W. Equilibrium, kinetics, diffusion and self-association of proteins at membrane surfaces: Measurement by total internal reflection fluorescence microscopy. Photochem. Photobiol. 1997, 65, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.; Myong, S. Protein induced fluorescence enhancement (PIFE) for probing protein-nucleic acid interactions. Chem. Soc. Rev. 2014, 43, 1221–1229. [Google Scholar] [CrossRef]
- Bielec, K.; Bubak, G.; Kalwarczyk, T.; Hołyst, R. Analysis of Brightness of a Single Fluorophore for Quantitative Characterization of Biochemical Reactions. J. Phys. Chem. B 2020, 124, 1941–1948. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Bielec, K.; Pasitsuparoad, P.; Hołyst, R. Single-molecule brightness analysis for the determination of anticancer drug interactions with DNA. Analyst 2020, 145, 6600–6606. [Google Scholar] [CrossRef] [PubMed]
- Dixit, A.B.; Ray, K.; Black, L.W. A viral small terminase subunit (TerS) twin ring Pac Synapsis DNA Packag. Model Is Support fluorescent Fusion Proteins. Virology 2019, 536, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; He, D.; Li, H.W. A fluorometric assay of thrombin using magnetic nanoparticles and enzyme-free hybridization chain reaction. Microchim. Acta 2020, 187, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Murade, C.; Subramaniam, V.; Otto, C.; Bennink, M. Interaction of Oxazole Yellow Dyes with DNA Studied with Hybrid Optical Tweezers and Fluorescence Microscopy. Biophys. J. 2009, 97, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sischka, A.; Walhorn, V.; Tönsing, K.; Anselmetti, D. Nanomechanics of Fluorescent DNA Dyes on DNA Investigated by Magnetic Tweezers. Biophys. J. 2016, 111, 1604–1611. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rye, H.; Yue, S.; Wemmer, D.; Quesada, M.; Haugland, R.; Mathies, R.; Glazer, G. Stable fluorescent complexes of double-stranded DNA with bis-intercalating asymmetric cyanine dyes: Properties and applications. Nucleic Acids Res. 1992, 20, 2803–2812. [Google Scholar] [CrossRef] [PubMed]
- Netzel, T.; Nafisi, K.; Zhao, M.; Lenhard, J.; Johnson, I. Base-Content Dependence of Emission Enhancements, Quantum Yields, and Lifetimes for Cyanine Dyes Bound to Double-Strand DNA: Photophysical Properties of Monomeric and Bichromophoric DNA Stains. J. Phys. Chem. 1995, 99, 17936–17947. [Google Scholar] [CrossRef]
- Öz, R.; Sriram, K.K.; Westerlund, F. A nanofluidic device for real-time visualization of DNA–protein interactions on the single DNA molecule level. Nanoscale 2019, 11, 2071–2078. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, Y.; Lim, S.; Jo, K. Single-molecule visualization of ROS-induced DNA damage in large DNA molecules. Analyst 2016, 141, 847–852. [Google Scholar] [CrossRef] [PubMed]
- Larsson, A.; Carlsson, C.; Jonsson, M.; Albinsson, B. Characterization of the Binding of the Fluorescent Dyes YO and YOYO to DNA by Polarized Light Spectroscopy. J. Am. Chem. Soc. 1994, 116, 8459–8465. [Google Scholar] [CrossRef]
- Paik, D.; Perkins, T. Dynamics and Multiple Stable Binding Modes of DNA Intercalators Revealed by Single-Molecule Force Spectroscopy. Angew. Chem. 2012, 51, 1811–1815. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Sierra, M.; Quiñones, E. Assays for the determination of the activity of DNA nucleases based on the fluorometric properties of the YOYO dye. Arch. Biochem. Biophys. 2015, 570, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Günter, K.; Mertig, M.; Seidel, R. Mechanical and structural properties of YOYO-1 complexed DNA. Nucleic Acids Res. 2010, 38, 6526–6532. [Google Scholar] [CrossRef] [PubMed]
- Bielec, K.; Sozański, K.; Seynen, M.; Dziekan, Z.; ten Wolde, P.R.; Hołyst, R. Kinetics and equilibrium constants of oligonucleotides at low concentrations. Hybridization and melting study. Phys. Chem. Chem. Phys. 2019, 21, 10798–10807. [Google Scholar] [CrossRef] [PubMed]
Samples of the compounds are not available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kucharska, K.; Pilz, M.; Bielec, K.; Kalwarczyk, T.; Kuźma, P.; Hołyst, R. Two Intercalation Mechanisms of Oxazole Yellow Dimer (YOYO-1) into DNA. Molecules 2021, 26, 3748. https://doi.org/10.3390/molecules26123748
Kucharska K, Pilz M, Bielec K, Kalwarczyk T, Kuźma P, Hołyst R. Two Intercalation Mechanisms of Oxazole Yellow Dimer (YOYO-1) into DNA. Molecules. 2021; 26(12):3748. https://doi.org/10.3390/molecules26123748
Chicago/Turabian StyleKucharska, Karolina, Marta Pilz, Krzysztof Bielec, Tomasz Kalwarczyk, Patrycja Kuźma, and Robert Hołyst. 2021. "Two Intercalation Mechanisms of Oxazole Yellow Dimer (YOYO-1) into DNA" Molecules 26, no. 12: 3748. https://doi.org/10.3390/molecules26123748
APA StyleKucharska, K., Pilz, M., Bielec, K., Kalwarczyk, T., Kuźma, P., & Hołyst, R. (2021). Two Intercalation Mechanisms of Oxazole Yellow Dimer (YOYO-1) into DNA. Molecules, 26(12), 3748. https://doi.org/10.3390/molecules26123748