Structure of Nanobody Nb23
Abstract
1. Introduction
2. Results
2.1. Nb23 Sequence Inferences
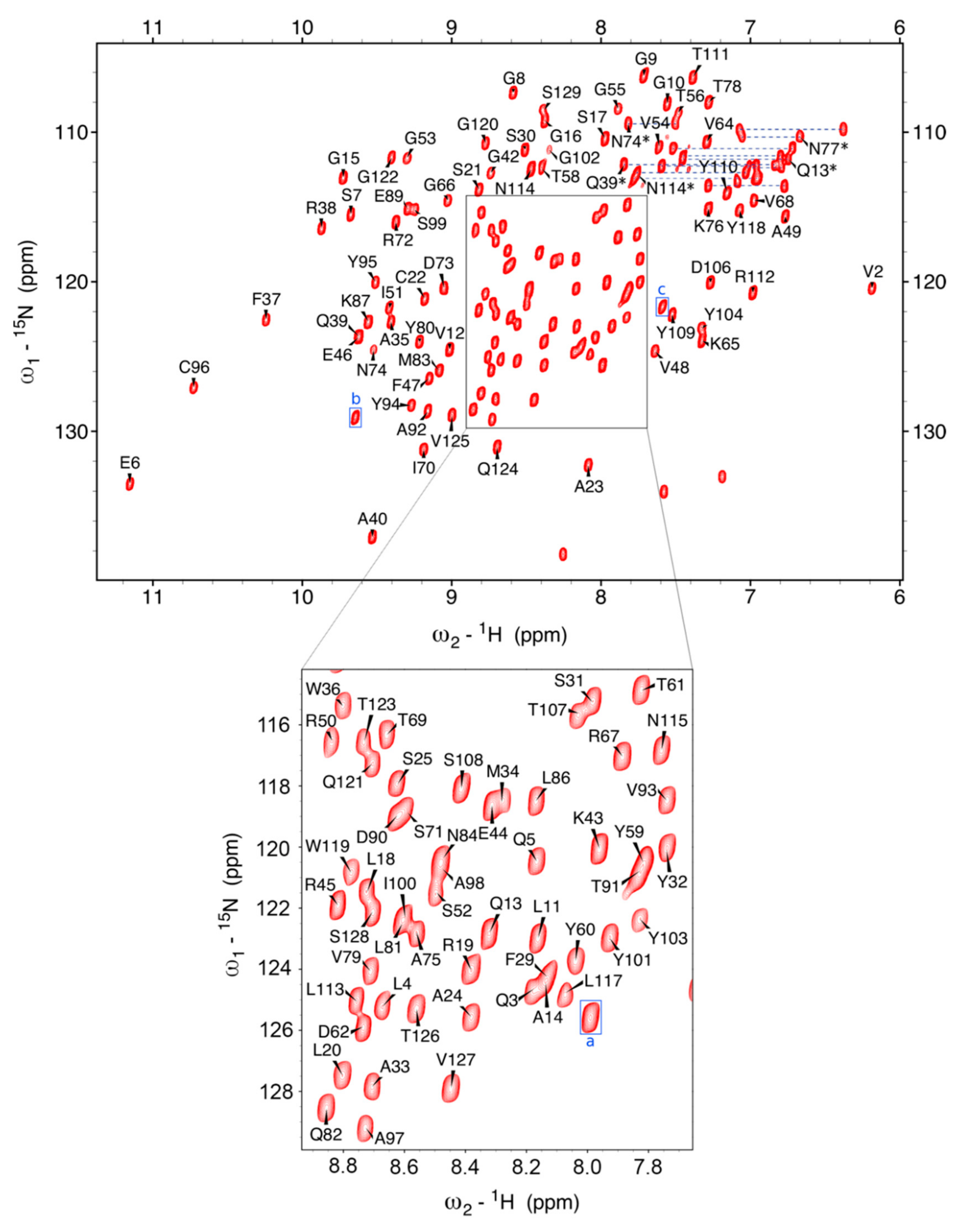
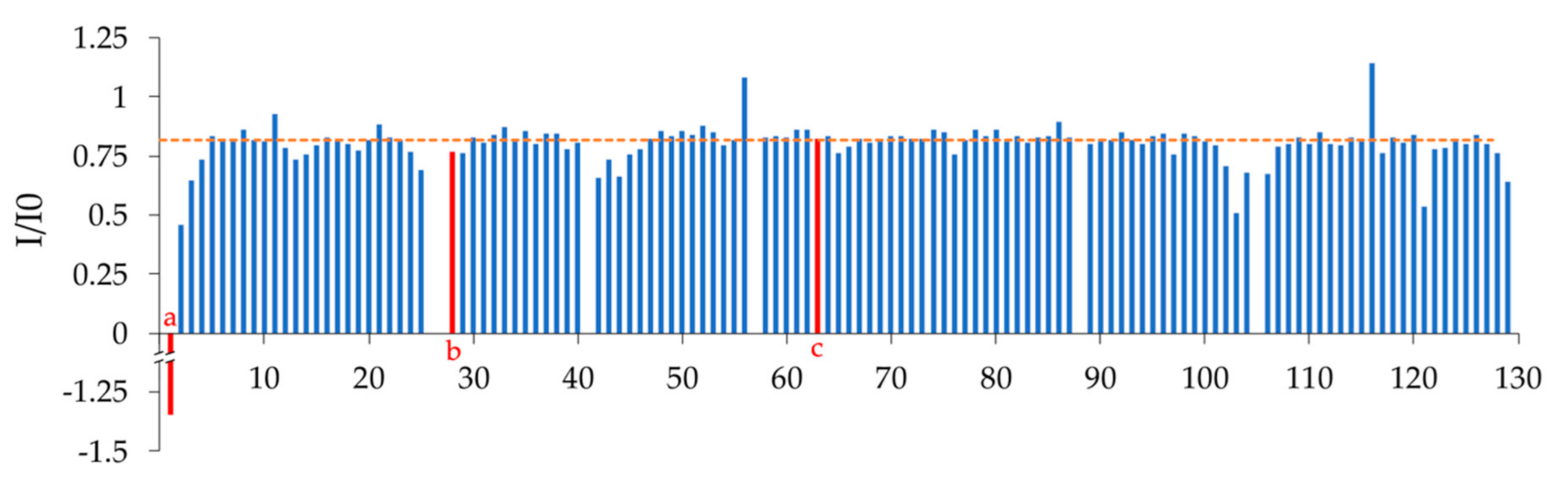
2.2. NMR Spectroscopy Results and Chemical Shift Assignment Completeness
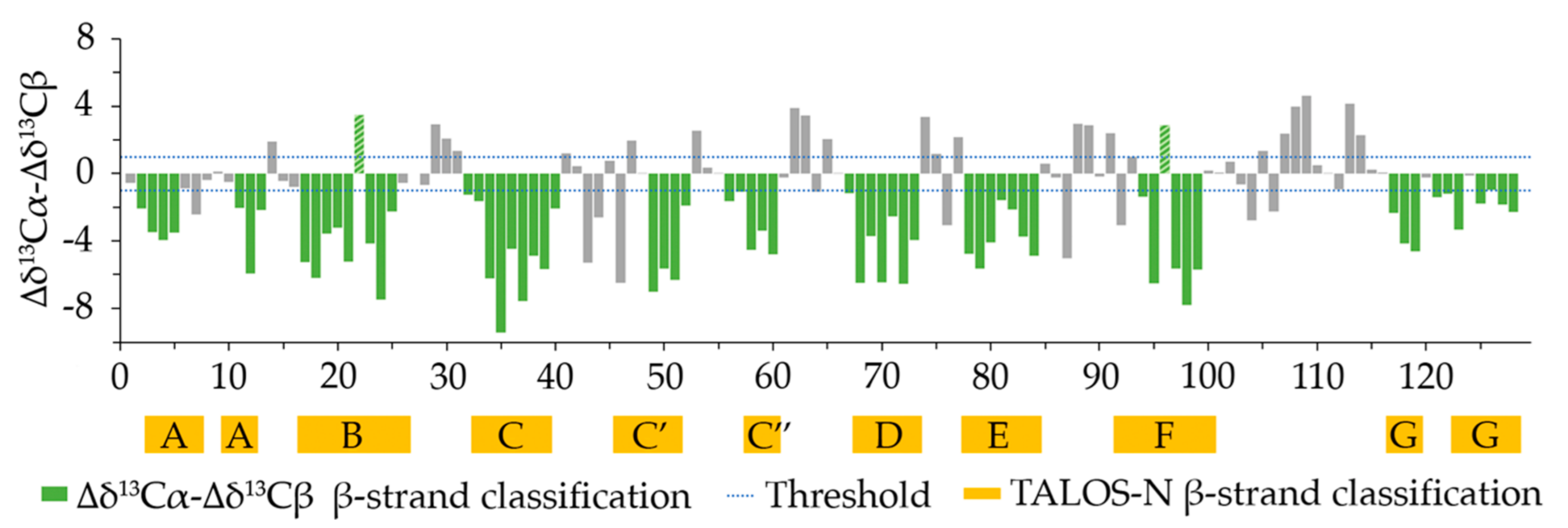
2.3. Secondary Structure Content Assessment
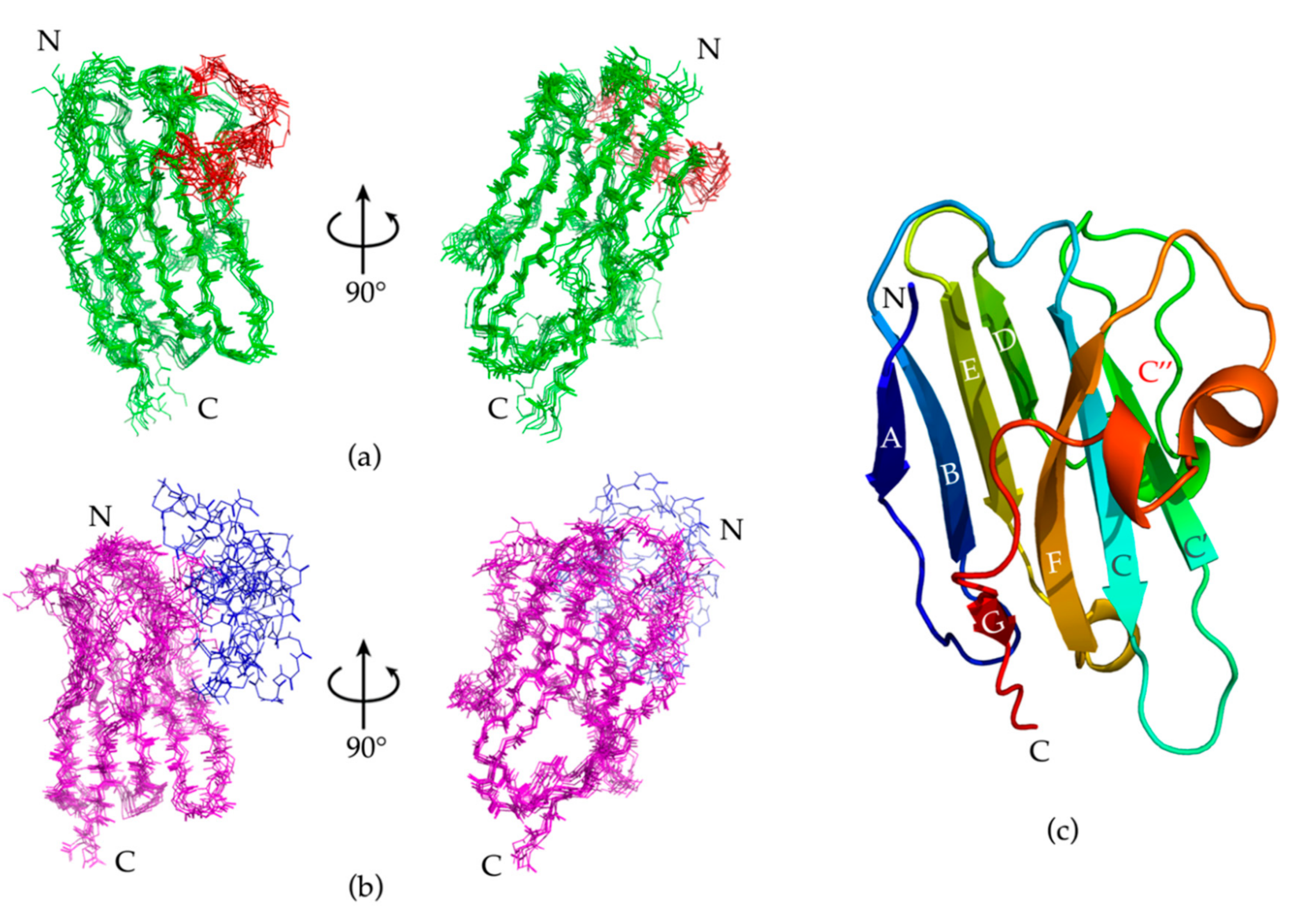
2.4. Constraints and Nb23 Structure Calculation
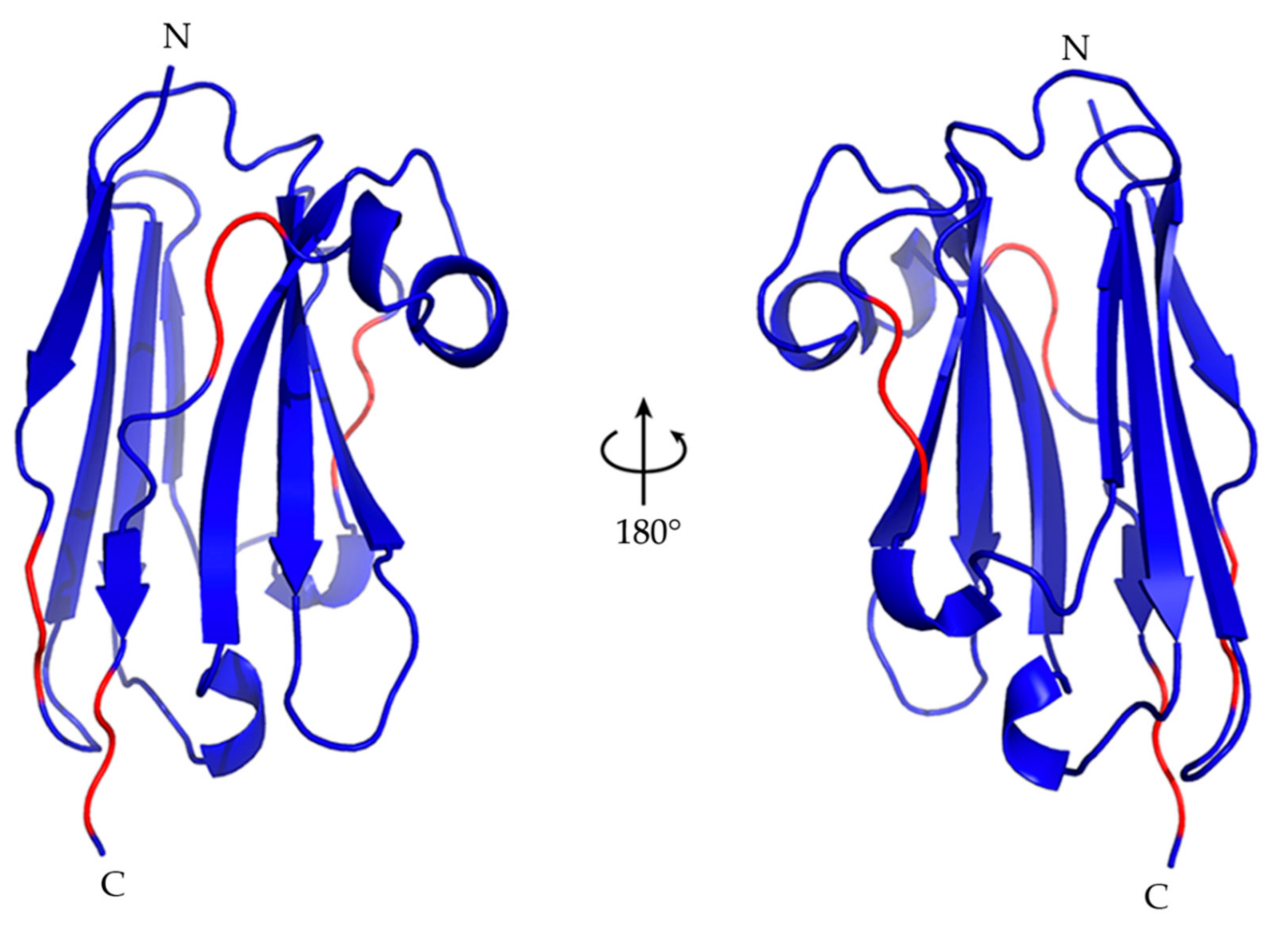
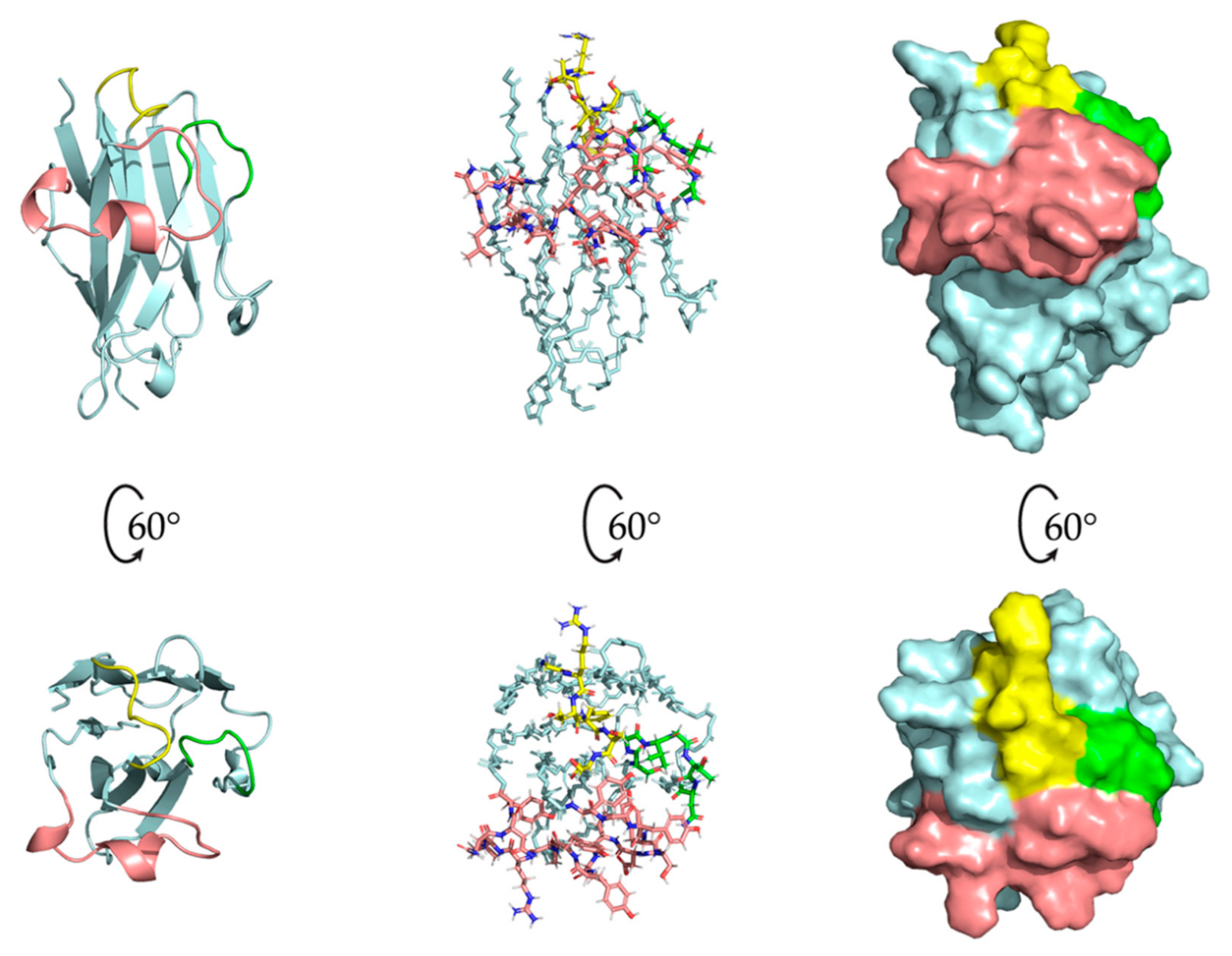
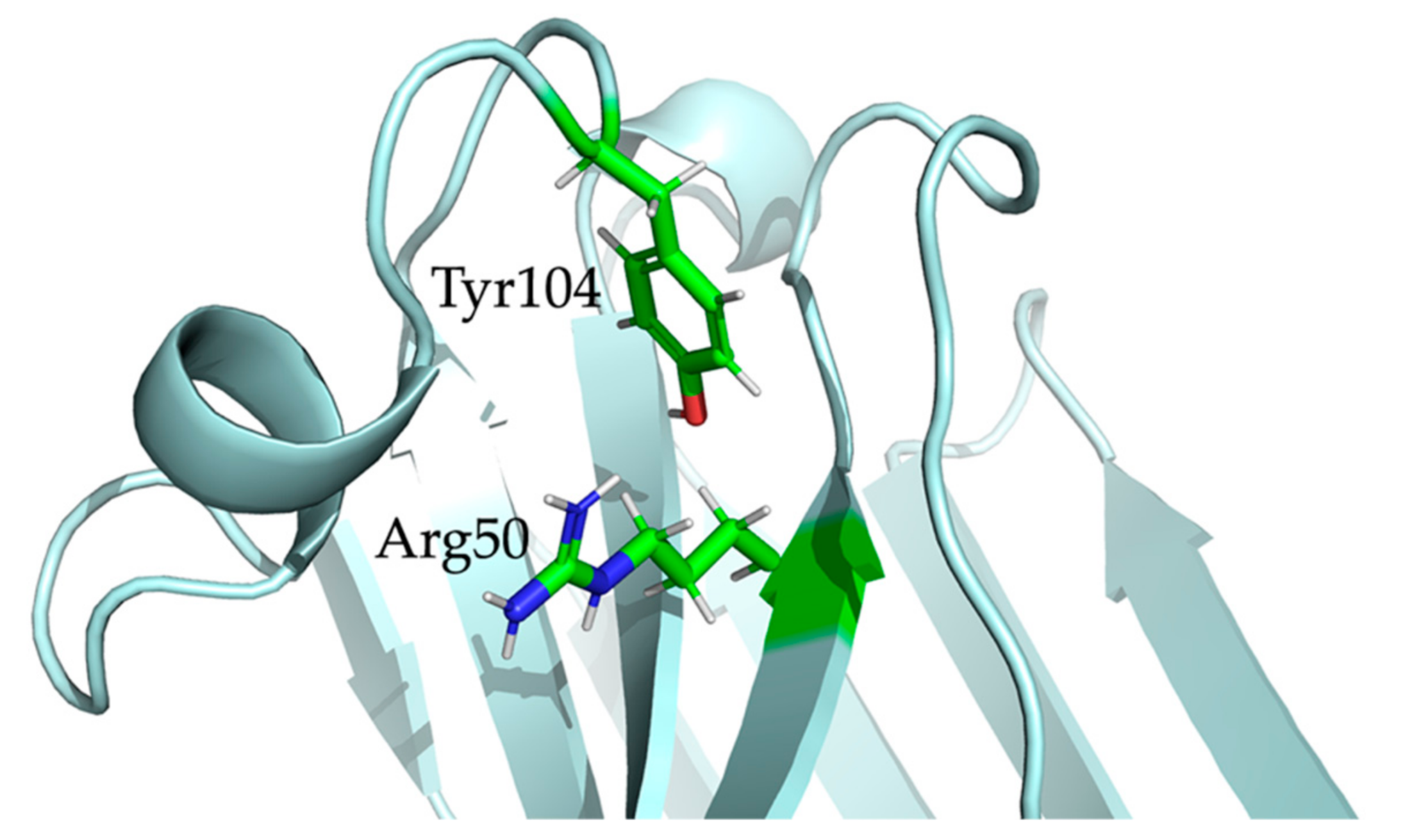
2.5. Nb23 Structural Features
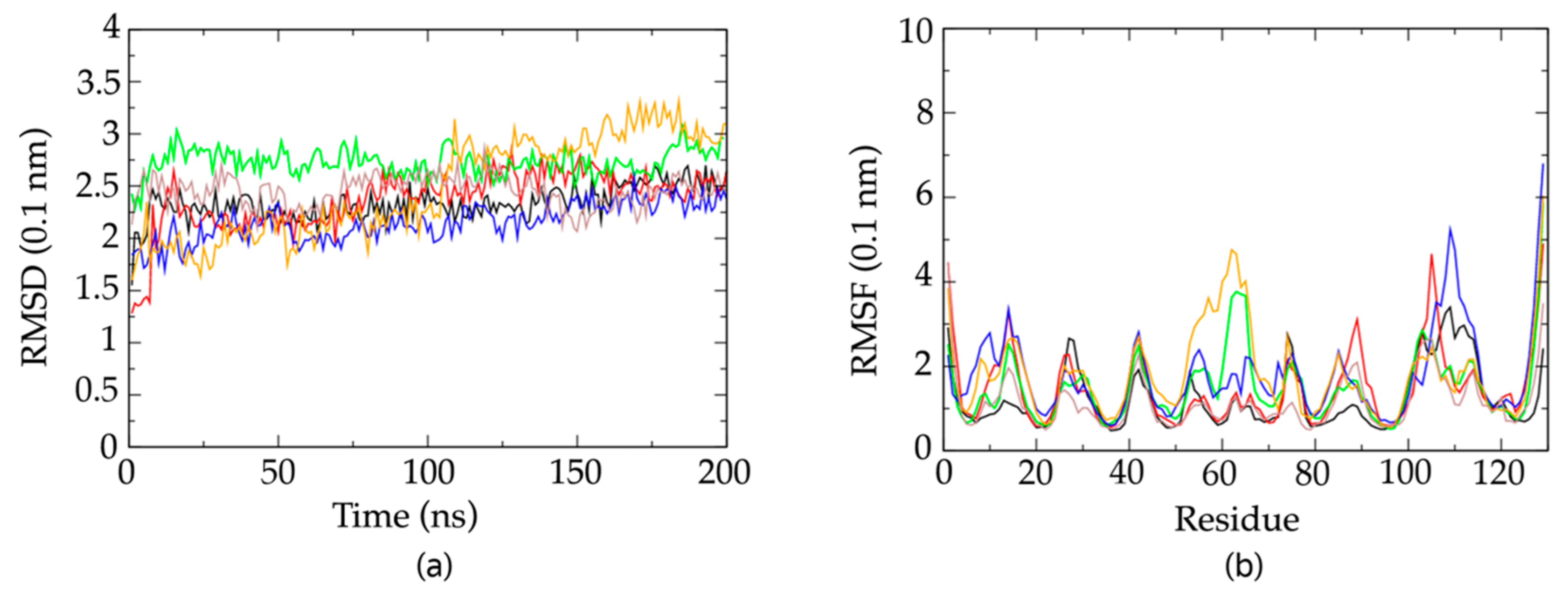
2.6. Molecular Dynamics Simulations
3. Discussion
4. Materials and Methods
4.1. Nb23 Expression and Labeling
4.2. Nb23 Sample Preparation, NMR Data Acquisition, and Peak Assignment
4.3. Restrained Modeling
4.4. Energy Minimization
4.5. Molecular Dynamics Simulations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Muyldermans, S. Nanobodies: Natural Single-Domain Antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef] [PubMed]
- Jovčevska, I.; Muyldermans, S. The Therapeutic Potential of Nanobodies. BioDrugs 2020, 34, 11–26. [Google Scholar] [CrossRef]
- Boulenouar, H.; Amar, Y.; Bouchoutrouch, N.; Faouzi, M.E.A.; Cherrah, Y.; Sefrioui, H. Nanobodies and Their Medical Applications. Genet. Mol. Res. 2020, 19, 1–12. [Google Scholar] [CrossRef]
- Hassanzadeh-Ghassabeh, G.; Devoogdt, N.; De Pauw, P.; Vincke, C.; Muyldermans, S. Nanobodies and their potential applications. Nanomedicine 2013, 8, 1013–1026. [Google Scholar] [CrossRef]
- Bannas, P.; Hambach, J.; Koch-Nolte, F. Nanobodies and Nanobody-Based Human Heavy Chain Antibodies As Antitumor Therapeutics. Front. Immunol. 2017, 8, 1603. [Google Scholar] [CrossRef]
- Lafaye, P.; Achour, I.; England, P.; Duyckaerts, C.; Rougeon, F. Single-domain antibodies recognize selectively small oligomeric forms of amyloid beta, prevent Abeta-induced neurotoxicity and inhibit fibril formation. Mol. Immunol. 2009, 46, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Gejyo, F.; Yamada, T.; Odani, S.; Nakagawa, Y.; Arakawa, M.; Kunitomo, T.; Kataoka, H.; Suzuki, M.; Hirasawa, Y.; Shirahama, T.; et al. A new form of amyloid protein associated with chronic hemodialysis was identified as beta 2-microglobulin. Biochem. Biophys. Res. Commun. 1985, 129, 701–706. [Google Scholar] [CrossRef]
- Esposito, G.; Michelutti, R.; Verdone, G.; Viglino, P.; Hernández, H.; Robinson, C.V.; Amoresano, A.; Dal Piaz, F.; Monti, M.; Pucci, P.; et al. Removal of the N-terminal hexapeptide from human beta2-microglobulin facilitates protein aggregation and fibril formation. Protein Sci. 2000, 9, 831–845. [Google Scholar] [CrossRef] [PubMed]
- Valleix, S.; Gillmore, J.D.; Bridoux, F.; Mangione, P.P.; Dogan, A.; Nedelec, B.; Boimard, M.; Touchard, G.; Goujon, J.M.; Lacombe, C.; et al. Hereditary systemic amyloidosis due to Asp76Asn variant β2-microglobulin. N. Engl. J. Med. 2012, 366, 2276–2283. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, S.; Porcari, R.; Mangione, P.P.; Verona, G.; Marcoux, J.; Giorgetti, S.; Taylor, G.W.; Ellmerich, S.; Ballico, M.; Zanini, S.; et al. A specific nanobody prevents amyloidogenesis of D76N β(2)-microglobulin in vitro and modifies its tissue distribution in vivo. Sci. Rep. 2017, 7, 46711. [Google Scholar] [CrossRef]
- Domanska, K.; Vanderhaegen, S.; Srinivasan, V.; Pardon, E.; Dupeux, F.; Marquez, J.A.; Giorgetti, S.; Stoppini, M.; Wyns, L.; Bellotti, V.; et al. Atomic structure of a nanobody-trapped domain-swapped dimer of an amyloidogenic beta2-microglobulin variant. Proc. Natl. Acad. Sci. USA 2011, 108, 1314–1319. [Google Scholar] [CrossRef] [PubMed]
- Vanderhaegen, S.; Fislage, M.; Domanska, K.; Versees, W.; Pardon, E.; Bellotti, V.; Steyaert, J. Structure of an early native-like intermediate of β2-microglobulin amyloidogenesis. Protein Sci. 2013, 22, 1349–1357. [Google Scholar] [CrossRef] [PubMed]
- Kinjo, A.R.; Nishikawa, K. Eigenvalue analysis of amino acid substitution matrices reveals a sharp transition of the mode of sequence conservation in proteins. Bioinformatics 2004, 20, 2504–2508. [Google Scholar] [CrossRef] [PubMed]
- Schulte-Herbrüggen, T.; Sørensen, O.W. Clean TROSY: Compensation for Relaxation-Induced Artifacts. J. Magn. Reason. 2000, 144, 123–128. [Google Scholar] [CrossRef]
- Salzmann, M.; Pervushin, K.; Wider, G.; Senn, H.; Wüthrich, K. TROSY in triple-resonance experiments: New perspectives for sequential NMR assignment of large proteins. Proc. Natl. Acad. Sci. USA 1998, 95, 13585. [Google Scholar] [CrossRef]
- Yamazaki, T.; Forman-Kay, J.D.; Kay, L.E. Two-dimensional NMR experiments for correlating carbon-13 beta. and proton.delta./.epsilon. chemical shifts of aromatic residues in 13C-labeled proteins via scalar couplings. J. Am. Chem. Soc. 1993, 115, 11054–11055. [Google Scholar] [CrossRef]
- Hafsa, N.E.; Arndt, D.; Wishart, D.S. CSI 3.0: A web server for identifying secondary and super-secondary structure in proteins using NMR chemical shifts. Nucleic Acids Res. 2015, 43, W370–W377. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Sykes, B.D. The 13C chemical-shift index: A simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR 1994, 4, 171–180. [Google Scholar] [CrossRef]
- Shen, Y.; Bax, A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 2013, 56, 227–241. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Bulyáki, É.; Kun, J.; Moussong, É.; Lee, Y.H.; Goto, Y.; Réfrégiers, M.; Kardos, J. BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018, 46, W315–W322. [Google Scholar] [CrossRef]
- Shen, Y.; Lange, O.; Delaglio, F.; Rossi, P.; Aramini, J.M.; Liu, G.; Eletsky, A.; Wu, Y.; Singarapu, K.K.; Lemak, A.; et al. Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl. Acad. Sci. USA 2008, 105, 4685–4690. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Kim, J.H.; Westler, W.M.; Markley, J.L. PONDEROSA, an automated 3D-NOESY peak picking program, enables automated protein structure determination. Bioinformatics 2011, 27, 1727–1728. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Stark, J.L.; Markley, J.L. PONDEROSA-C/S: Client-server based software package for automated protein 3D structure determination. J. Biomol. NMR 2014, 60, 73–75. [Google Scholar] [CrossRef]
- Wüthrich, K.; Billeter, M.; Braun, W. Polypeptide secondary structure determination by nuclear magnetic resonance observation of short proton-proton distances. J. Mol. Biol. 1984, 180, 715–740. [Google Scholar] [CrossRef]
- Englander, S.W.; Kallenbach, N.R. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q. Rev. Biophys. 1983, 16, 521–655. [Google Scholar] [CrossRef] [PubMed]
- Gore, S.; Sanz García, E.; Hendrickx, P.M.S.; Gutmanas, A.; Westbrook, J.D.; Yang, H.; Feng, Z.; Baskaran, K.; Berrisford, J.M.; Hudson, B.P.; et al. Validation of Structures in the Protein Data Bank. Structure 2017, 25, 1916–1927. [Google Scholar] [CrossRef] [PubMed]
- Davis, I.W.; Leaver-Fay, A.; Chen, V.B.; Block, J.N.; Kapral, G.J.; Wang, X.; Murray, L.W.; Arendall, W.B., 3rd; Snoeyink, J.; Richardson, J.S.; et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007, 35, W375–W383. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef] [PubMed]
- Fowler, N.J.; Sljoka, A.; Williamson, M.P. A method for validating the accuracy of NMR protein structures. Nature Commun. 2020, 11, 6321. [Google Scholar] [CrossRef]
- Gallivan, J.P.; Dougherty, D.A. Cation-pi interactions in structural biology. Proc. Natl. Acad. Sci. USA 1999, 96, 9459–9464. [Google Scholar] [CrossRef]
- Palmer, A.G.; Cavanagh, J.; Wright, P.E.; Rance, M. Sensitivity improvement in proton-detected two-dimensional heteronuclear correlation NMR spectroscopy. J. Magn. Reason. 1991, 93, 151–170. [Google Scholar] [CrossRef]
- States, D.J.; Haberkorn, R.A.; Ruben, D.J. A two-dimensional nuclear overhauser experiment with pure absorption phase in four quadrants. J. Magn. Reason. 1982, 48, 286–292. [Google Scholar] [CrossRef]
- Marion, D.; Wüthrich, K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem. Biophys. Res. Commun. 1983, 113, 967–974. [Google Scholar] [CrossRef]
- Grzesiek, S.; Bax, A. The importance of not saturating water in protein NMR. Application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc. 1993, 115, 12593–12594. [Google Scholar] [CrossRef]
- Piotto, M.; Saudek, V.; Sklenár, V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 1992, 2, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Hwang, T.L.; Shaka, A.J. Water Suppression That Works. Excitation Sculpting Using Arbitrary Wave-Forms and Pulsed-Field Gradients. J. Magn. Reason. Series A 1995, 112, 275–279. [Google Scholar] [CrossRef]
- Orekhov, V.Y.; Ibraghimov, I.; Billeter, M. Optimizing resolution in multidimensional NMR by three-way decomposition. J. Biomol. NMR 2003, 27, 165–173. [Google Scholar] [CrossRef]
- Lee, W.; Tonelli, M.; Markley, J.L. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 2015, 31, 1325–1327. [Google Scholar] [CrossRef]
- Kay, L.; Keifer, P.; Saarinen, T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 1992, 114, 10663–10665. [Google Scholar] [CrossRef]
- Nietlispach, D. Suppression of anti-TROSY lines in a sensitivity enhanced gradient selection TROSY scheme. J. Biomol. NMR 2005, 31, 161–166. [Google Scholar] [CrossRef]
- Grzesiek, S.; Bax, A. An efficient experiment for sequential backbone assignment of medium-sized isotopically enriched proteins. J. Magn. Reason. 1992, 99, 201–207. [Google Scholar] [CrossRef]
- Grzesiek, S.; Bax, A. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 1992, 114, 6291–6293. [Google Scholar] [CrossRef]
- Grzesiek, S.; Anglister, J.; Bax, A. Correlation of Backbone Amide and Aliphatic Side-Chain Resonances in 13C/15N-Enriched Proteins by Isotropic Mixing of 13C Magnetization. J. Magn. Reason. Ser. B 1993, 101, 114–119. [Google Scholar] [CrossRef]
- Grzesiek, S.; Bax, A. Amino acid type determination in the sequential assignment procedure of uniformly 13C/15N-enriched proteins. J. Biomol. NMR 1993, 3, 185–204. [Google Scholar] [CrossRef]
- Salzmann, M.; Wider, G.; Pervushin, K.; Senn, H.; Wüthrich, K. TROSY-type Triple-Resonance Experiments for Sequential NMR Assignments of Large Proteins. J. Am. Chem. Soc. 1999, 121, 844–848. [Google Scholar] [CrossRef]
- Shaka, A.J.; Lee, C.J.; Pines, A. Iterative schemes for bilinear operators; application to spin decoupling. J. Magn. Reason. 1988, 77, 274–293. [Google Scholar] [CrossRef]
- Bax, A.; Clore, G.M.; Gronenborn, A.M. 1H-1H correlation via isotropic mixing of 13C magnetization, a new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J. Magn. Reason. 1990, 88, 425–431. [Google Scholar] [CrossRef]
- Schleucher, J.; Schwendinger, M.; Sattler, M.; Schmidt, P.; Schedletzky, O.; Glaser, S.J.; Sørensen, O.W.; Griesinger, C. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J. Biomol. NMR 1994, 4, 301–306. [Google Scholar] [CrossRef]
- Kumar, A.; Ernst, R.R.; Wüthrich, K. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 1980, 95, 1–6. [Google Scholar] [CrossRef]
- Lee, W.; Petit, C.M.; Cornilescu, G.; Stark, J.L.; Markley, J.L. The AUDANA algorithm for automated protein 3D structure determination from NMR NOE data. J. Biomol. NMR 2016, 65, 51–57. [Google Scholar] [CrossRef]
- Kalé, L.; Skeel, R.; Bhdarkar, M.; Brunner, R.; Gursoy, A.; Krawetz, N.; Phillips, J.; Shinozaki, A.; Varadarajan, K.; Schulten, K. NAMD2: Greater Scalability for Parallel Molecular Dynamics. J. Comput. Phys. 1999, 151, 283–312. [Google Scholar] [CrossRef]
- Onufriev, A.; Bashford, D.; Case, D.A. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins 2004, 55, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 2010, 78, 1950–1958. [Google Scholar] [CrossRef]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Feller, S.E.; Zhang, Y.; Pastor, R.W.; Brooks, B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
| Total | 1H | 13C | 15N | |
|---|---|---|---|---|
| Backbone | 95% | 96% | 94% | 96% |
| Sidechain | 67% | 73% | 69% | 0% |
| Aromatic | 50% | 68% | 32% | 0% |
| Overall | 77% | 80% | 75% | 71% |
| Nb23 CS-Rosetta (10 Structures) | |
| Clashes | |
| van der Waals clashes | 8 (0.8 clashes/structure) |
| Average clash | 0.48 ± 0.05 Å |
| Ramachandran plot distribution | |
| Residues in favored regions | 97% |
| Residues in allowed regions | 2% |
| Outliers | 1% |
| χ outliers per structure | 0.1 |
| Cα-RMSD 1–129 w.r.t. lowest energy structure * | 3.442 ± 2.212 Å |
| Cα-RMSD 1–102, 117–122 w.r.t. lowest energy structure * | 1.531 ± 0.994 Å |
| Nb23 NOE-Restrained (20 Structures) | |
|---|---|
| Distance Constraints | |
| Short-range | 417 |
| Medium-range | 16 |
| Long-range | 186 |
| Hydrogen bonds | 18 |
| Total | 637 |
| Violations | |
| Distance constraint violations | 115 (5.75 violations/structure) |
| Short-range | 4 |
| Medium-range | 6 |
| Long-range | 76 |
| Hydrogen bonds | 29 |
| Average violation | 1.13 ± 0.61 Å |
| Clashes | |
| van der Waals clashes | 189 (9.45 clashes/structure) |
| Average clash | 0.48 ± 0.08 Å |
| Ramachandran Plot Distribution | |
| Residues in favored regions | 93% |
| Residues in allowed regions | 6% |
| Outliers | 1% |
| χ outliers per structure | 2.45 |
| Cα-RMSD 1–129 w.r.t. least violation structure | 1.98 ± 0.68 Å |
| Cα-RMSD 3–100, 118–128 [Å] w.r.t least violation structure | 1.70 ± 0.68 Å |
| Nb23 NOE-Restrained Energy-Minimized (10 Structures) | |
| Clashes | |
| van der Waals clashes | 0 |
| Ramachandran Plot Distribution | |
| Residues in favored regions | 96% |
| Residues in allowed regions | 4% |
| Outliers | 0% |
| χ outliers per structure | 0.6 |
| Cα-RMSD 1–129 w.r.t least violation structure * | 1.57 ± 0.32 Å |
| Cα-RMSD 3–100, 118–128 w.r.t least violation structure * | 1.23 ± 0.30 Å |
| β-Strand | A | A * | B | C | C’ | C’’ | D | E | F | G | G * |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Structure 1 | 3–7 | - | 17–25 | 32–39 | 46–51 | - | 69–73 | 77–84 | 92–100 | - | 123–125 |
| Structure 2 | 3–7 | - | 17–25 | 32–39 | 46–51 | - | 69–73 | 77–84 | 92–100 | 117–119 | - |
| Structure 3 | 3–7 | - | 17–25 | 32–39 | 46–51 | - | 69–73 | 77–84 | 92–100 | 117–119 | 123–125 |
| Structure 4 | 3–7 | - | 17–25 | 32–39 | 46–51 | - | 69–73 | 77–84 | 93–100 | - | - |
| Structure 5 | 3–7 | - | 17–25 | 32–38 | 46–51 | - | 69–73 | 77–84 | 93–99 | - | - |
| Structure 6 | 3–7 | - | 17–25 | 32–39 | 46–52 | - | 69–73 | 77–84 | 93–100 | 117–119 | - |
| Structure 7 | 3–7 | - | 17–25 | 32–39 | 46–51 | - | 69–73 | 77–84 | 93–101 | 117–119 | - |
| Structure 8 | 3–7 | - | 17–26 | 32–39 | 46–51 | 59–61 | 69–73 | 77–84 | 92–100 | 117–119 | 123–125 |
| Structure 9 | 3–7 | - | 17–25 | 32–39 | 46–51 | - | 69–73 | 77–84 | 93–100 | 117–119 | - |
| Structure 10 | 3–7 | - | 17–25 | 32–39 | 46–51 | - | 69–73 | 77–84 | 93–100 | 117–119 | - |
| Spectrum | Time Domain Dimensions | Transients (NS) | Carrier (ppm) | Spectral Width (ppm) | References |
|---|---|---|---|---|---|
| 2D 15N-1H HSQC | t2 (1H): 2048 t1 (15N): 128 | 8, 16 | t2 (1H): 4.7 t1 (15N): 118 | t2 (1H): 16 t1 (15N): 50 | [39] |
| 2D tr-15N-1H HSQC | t2 (1H): 2048 t1 (15N): 80 | 16 | t2 (1H): 4.7 t1 (15N): 118 | t2 (1H): 16 t1 (15N): 50 | [40] |
| 3D tr-CBCANH | t3 (1H): 1024 t2 (15N): 50 t1 (13C): 128 | 576 | t3 (1H): 4.7 t2 (15N): 118 t1 (13C): 43 | t3 (1H): 14 t2 (15N): 50 t1 (13C): 80 | [14,41] |
| 3D tr-CBCA(CO)NH | t3 (1H): 1024 t2 (15N): 50 t1 (13C): 128 | 96 | t3 (1H): 4.7 t2 (15N): 118 t1 (13C): 43 | t3 (1H): 14 t2 (15N): 50 t1 (13C): 80 | [14,42] |
| 3D tr-HNCA | t3 (1H): 1024 t2 (15N): 50 t1 (13C): 96 | 32 | t3 (1H): 4.7 t2 (15N): 118 t1 (13C): 54 | t3 (1H): 18 t2 (15N): 50 t1 (13C): 80 | [15] |
| 3D tr-CC(CO)NH | t3 (1H): 1024 t2 (15N): 50 t1 (13C): 128 | 256 | t3 (1H): 4.7 t2 (15N): 118 t1 (13C): 43 | t3 (1H): 14 t2 (15N): 50 t1 (13C): 80 | [14,43] |
| 3D tr-H(CCO)NH | t3 (1H): 1024 t2 (15N): 50 t1 (1H): 128 | 256 | t3 (1H): 4.7 t2 (15N): 118 1H: 4.7 | t3 (1H): 14 t2 (15N): 50 1H: 14 | [14,43] |
| 3D tr-HBHA(CO)NH | t3 (1H): 1024 t2 (15N): 50 t1 (1H): 128 | 96 | t3 (1H): 4.7 t2 (15N): 118 t1 (1H): 4.7 | t3 (1H): 14 t2 (15N): 50 t1 (1H): 8 | [14,44] |
| 3D tr-HNCO | t3 (1H): 1024 t2 (15N): 50 t1 (13C): 96 | 32 | t3 (1H): 4.7 t2 (15N): 118 t1 (13C): 173.5 | t3 (1H): 14 t2 (15N): 50 t1 (13C): 22 | [15] |
| 3D tr-HN(CA)CO | t3 (1H): 1024 t2 (15N): 50 t1 (13C): 96 | 96 | t3 (1H): 4.7 t2 (15N): 118 t1 (13C): 173.5 | t3 (1H): 14 t2 (15N): 50 t1 (13C): 22 | [45] |
| 2D 1H-1H TOCSY | t2 (1H): 4096 t1 (1H): 768 | 192 | t2 (1H): 4.7 t1 (1H): 4.7 | t2 (1H): 14.4 t1 (1H): 14.4 | [36,46,47] |
| 3D 15N-1H NOESY HSQC | t3 (1H): 1024 t2 (15N): 50 t1 (1H): 400 | 96 | t3 (1H): 4.7 t2 (15N): 122 t1 (1H): 4.7 | t3 (1H): 14 t2 (15N): 42 t1 (1H): 14 | [31,39,48] |
| 2D CBHD (D2O) | t2 (1H): 2048 t1 (13C): 98 | 1024 | t2 (1H): 4.7 t1 (13C): 36 | t2 (1H): 16 t1 (13C): 28 | [16] |
| 2D CBHE (D2O) | t2 (1H): 2048 t1 (13C): 98 | 1024 | t2 (1H): 4.7 t1 (13C): 36 | t2 (1H): 16 t1 (13C): 28 | [16] |
| 2D 1H-1H NOESY (D2O) | t2 (1H): 4096 t1 (1H): 400 | 192 | t2 (1H): 4.7 t1 (1H): 4.7 | t2 (1H): 14.4 t1 (1H): 14.4 | [36,46,49] |
| 3D 13C-1H NOESY HSQC aliphatic (D2O) | t3 (1H): 1024 t2 (13C): 80 t1 (1H): 160 | 96 | t3 (1H): 4.7 t2 (13C): 43 t1 (1H): 4.7 | t3 (1H): 14 t2 (13C): 80 t1 (1H): 14 | [31,39,48] |
| 3D 13C-1H NOESY HSQC aromatic (D2O) | t3 (1H): 1024 t2 (13C): 80 t1 (1H): 160 | 96 | t3 (1H): 4.7 t2 (13C): 105 t1 (1H): 4.7 | t3 (1H): 14 t2 (13C): 80 t1 (1H): 14 | [31,39,48] |
| 2D 13C-1H HSQC | t2 (1H): 1024 t1 (13C): 196 | 32 | t2 (1H): 4.7 t1 (13C): 72 | t2 (1H): 16 t1 (13C): 165 | [39] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Percipalle, M.; Hunashal, Y.; Steyaert, J.; Fogolari, F.; Esposito, G. Structure of Nanobody Nb23. Molecules 2021, 26, 3567. https://doi.org/10.3390/molecules26123567
Percipalle M, Hunashal Y, Steyaert J, Fogolari F, Esposito G. Structure of Nanobody Nb23. Molecules. 2021; 26(12):3567. https://doi.org/10.3390/molecules26123567
Chicago/Turabian StylePercipalle, Mathias, Yamanappa Hunashal, Jan Steyaert, Federico Fogolari, and Gennaro Esposito. 2021. "Structure of Nanobody Nb23" Molecules 26, no. 12: 3567. https://doi.org/10.3390/molecules26123567
APA StylePercipalle, M., Hunashal, Y., Steyaert, J., Fogolari, F., & Esposito, G. (2021). Structure of Nanobody Nb23. Molecules, 26(12), 3567. https://doi.org/10.3390/molecules26123567







