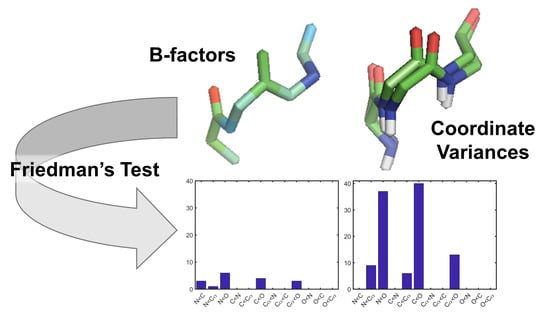
Patterns in Protein Flexibility: A Comparison of NMR “Ensembles”, MD Trajectories, and Crystallographic B-Factors
Abstract
1. Introduction
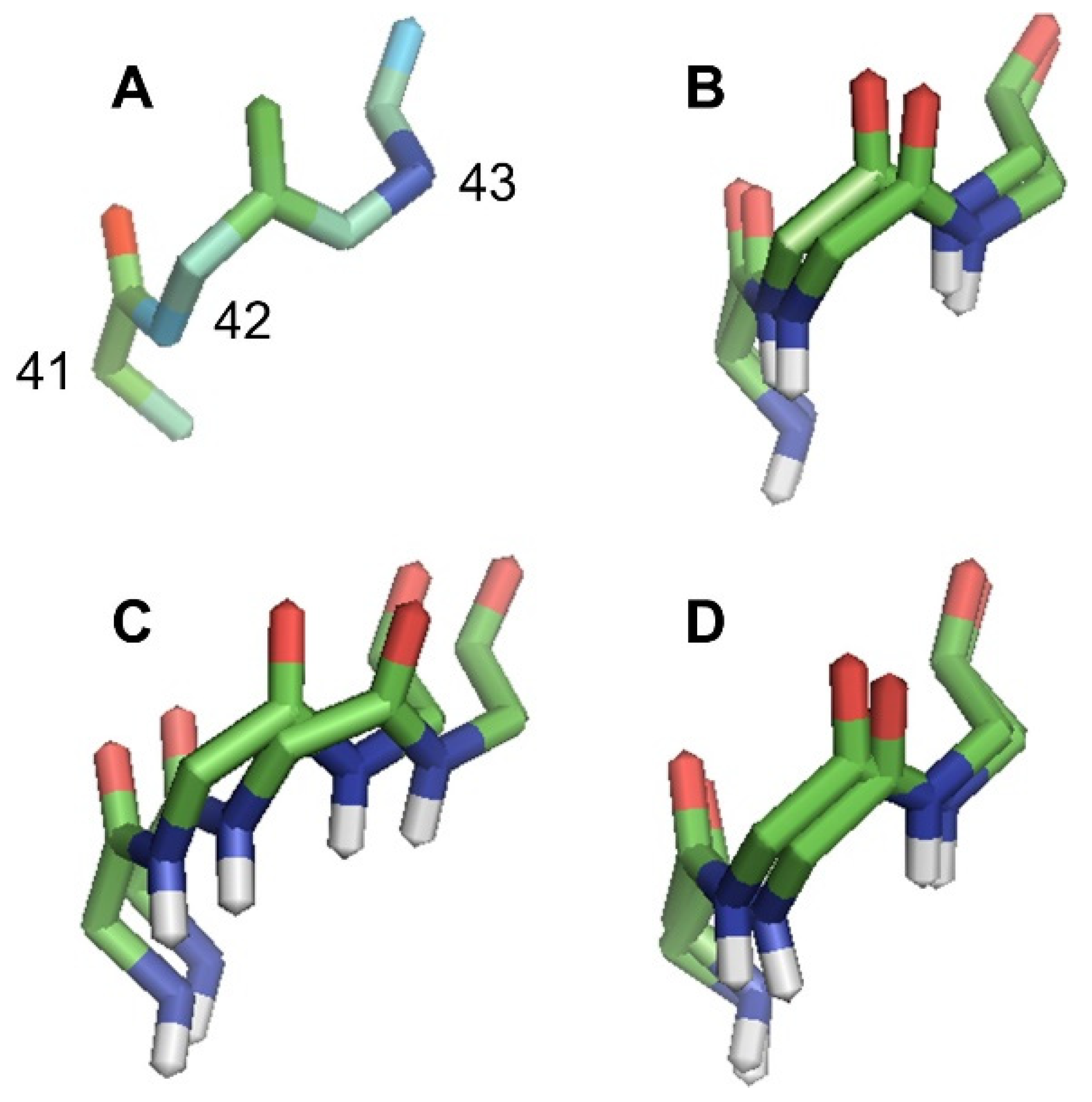
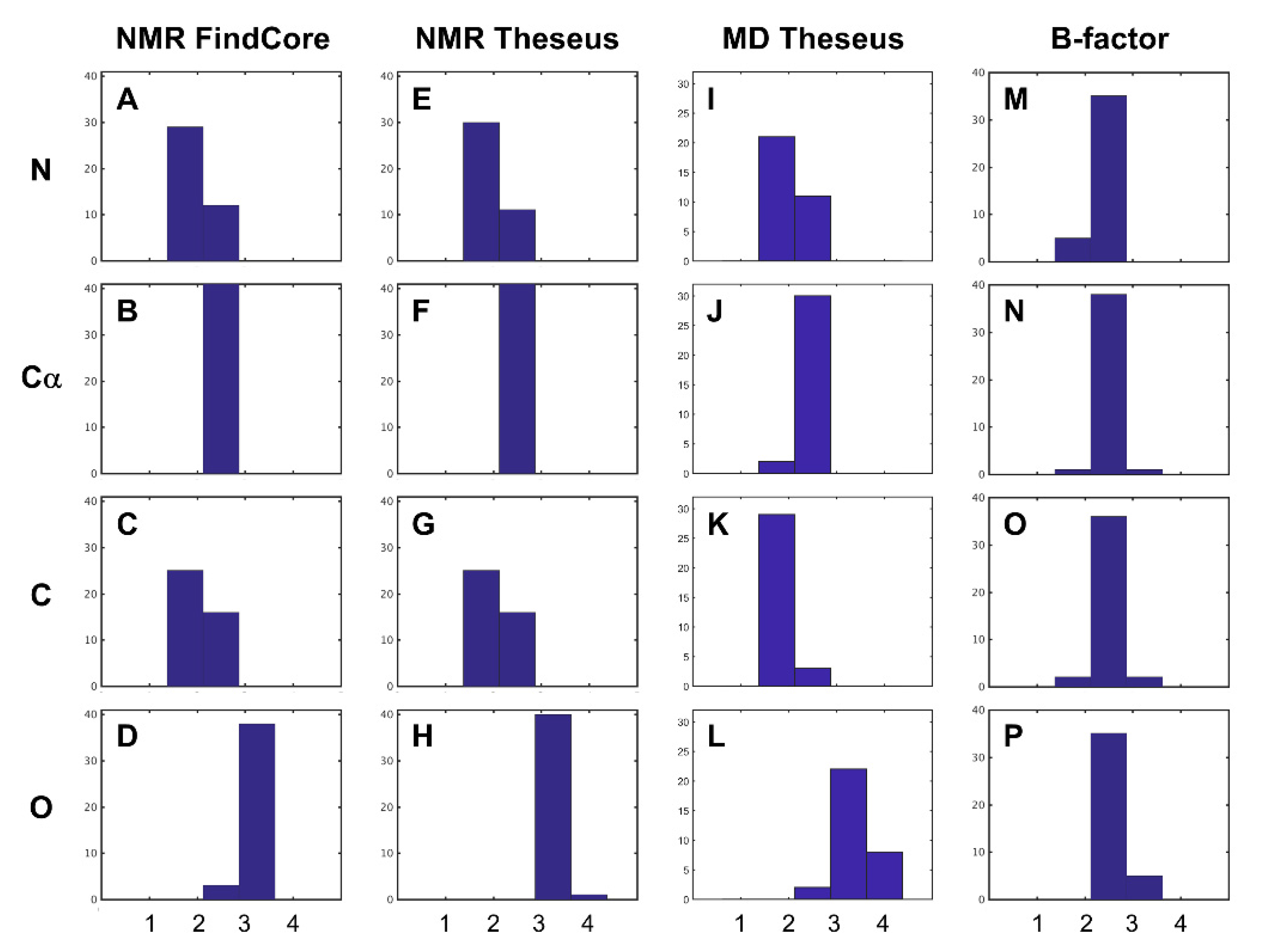
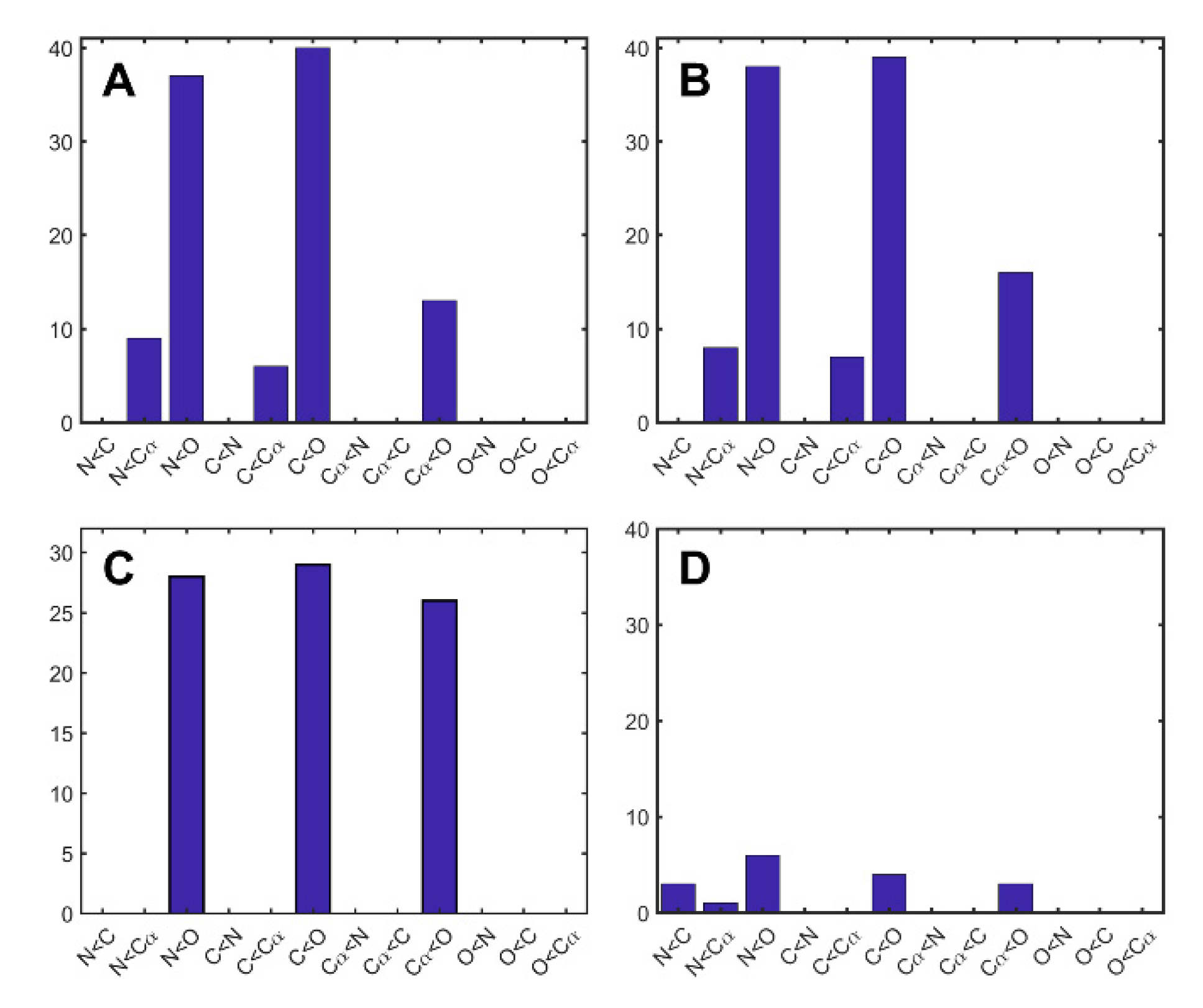
2. Results and Discussion
3. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Rashin, A.A.; Rashin, A.H.; Jernigan, R.L. Protein flexibility: Coordinate uncertainties and interpretation of structural differences. Acta Crystallogr. D Biol. Crystallogr. 2009, 65, 1140–1161. [Google Scholar] [CrossRef]
- Fenwick, R.B.; Bedem, H.V.D.; Fraser, J.S.; Wright, P.E. Integrated description of protein dynamics from room-temperature X-ray crystallography and NMR. Proc. Natl. Acad. Sci. USA 2014, 111, E445–E454. [Google Scholar] [CrossRef] [PubMed]
- Karplus, M.; McCammon, J.A. Molecular dynamics simulations of biomolecules. Nat. Genet. 2002, 9, 646–652. [Google Scholar] [CrossRef]
- Sapienza, P.J.; Lee, A.L. Using NMR to study fast dynamics in proteins: Methods and applications. Curr. Opin. Pharmacol. 2010, 10, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Wand, A.J. The dark energy of proteins comes to light: Conformational entropy and its role in protein function revealed by NMR relaxation. Curr. Opin. Struct. Biol. 2013, 23, 75–81. [Google Scholar] [CrossRef]
- Lipari, G.; Szabo, A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 1982, 104, 4546–4559. [Google Scholar] [CrossRef]
- Lipari, G.; Szabo, A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J. Am. Chem. Soc. 1982, 104, 4559–4570. [Google Scholar] [CrossRef]
- Robustelli, P.; Stafford, K.A.; Palmer, A.G. Interpreting Protein Structural Dynamics from NMR Chemical Shifts. J. Am. Chem. Soc. 2012, 134, 6365–6374. [Google Scholar] [CrossRef]
- Berjanskii, M.V.; Wishart, D.S. A Simple Method to Measure Protein Side-Chain Mobility Using NMR Chemical Shifts. J. Am. Chem. Soc. 2013, 135, 14536–14539. [Google Scholar] [CrossRef]
- Eyal, E.; Gerzon, S.; Potapov, V.; Edelman, M.; Sobolev, V. The Limit of Accuracy of Protein Modeling: Influence of Crystal Packing on Protein Structure. J. Mol. Biol. 2005, 351, 431–442. [Google Scholar] [CrossRef]
- Read, R.J. Structure-factor probabilities for related structures. Acta Crystallogr. Sect. A Found. Crystallogr. 1990, 46, 900–912. [Google Scholar] [CrossRef]
- Li, D.-W.; Brüschweiler, R. Protocol to Make Protein NMR Structures Amenable to Stable Long Time Scale Molecular Dynamics Simulations. J. Chem. Theory Comput. 2014, 10, 1781–1787. [Google Scholar] [CrossRef]
- Showalter, S.A.; Brüschweiler, R. Validation of molecular dynamics simulations of biomolecules using NMR spin relaxation as benchmarks: Application to the AMBER99SB force field. J. Chem. Theory Comput. 2007, 3, 961–975. [Google Scholar] [CrossRef] [PubMed]
- Brüschweiler, R. Certification of Molecular Dynamics Trajectories with NMR Chemical Shifts. J. Phys. Chem. Lett. 2009, 1, 246–248. [Google Scholar] [CrossRef]
- Rueda, M.; Ferrer-Costa, C.; Meyer, T.; Pérez, A.; Camps, J.; Gelpí, J.L.; Orozco, M. A consensus view of protein dynamics. Proc. Natl. Acad. Sci. USA 2007, 104, 796–801. [Google Scholar] [CrossRef]
- Showalter, S.A.; Johnson, E.; Rance, M.; Brüschweiler, R. Toward Quantitative Interpretation of Methyl Side-Chain Dynamics from NMR by Molecular Dynamics Simulations. J. Am. Chem. Soc. 2007, 129, 14146–14147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Brüschweiler, R. Contact Model for the Prediction of NMR N−H Order Parameters in Globular Proteins. J. Am. Chem. Soc. 2002, 124, 12654–12655. [Google Scholar] [CrossRef]
- Ming, D.; Brüschweiler, R. Reorientational Contact-Weighted Elastic Network Model for the Prediction of Protein Dynamics: Comparison with NMR Relaxation. Biophys. J. 2006, 90, 3382–3388. [Google Scholar] [CrossRef]
- Rieping, W.; Nilges, M.; Habeck, M. ISD: A software package for Bayesian NMR structure calculation. Bioinformatics 2008, 24, 1104–1105. [Google Scholar] [CrossRef]
- Carstens, S.; Nilges, M.; Habeck, M. Inferential Structure Determination of Chromosomes from Single-Cell Hi-C Data. PLoS Comput. Biol. 2016, 12, e1005292. [Google Scholar] [CrossRef] [PubMed]
- Richter, B.; Gsponer, J.; Várnai, P.; Salvatella, X.; Vendruscolo, M. The MUMO (minimal under-restraining minimal over-restraining) method for the determination of native state ensembles of proteins. J. Biomol. NMR 2007, 37, 117–135. [Google Scholar] [CrossRef] [PubMed]
- Snyder, D.A.; Grullon, J.; Huang, Y.J.; Tejero, R.; Montelione, G.T. The expanded FindCore method for identification of a core atom set for assessment of protein structure prediction. Proteins Struct. Funct. Bioinform. 2013, 82, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Tejero, R.; Bassolino-Klimas, D.; Bruccoleri, R.E.; Montelione, G.T. Simulated annealing with restrained molecular dynamics using CONGEN: Energy refinement of the NMR solution structures of epidermal and type-α transforming growth factors. Protein Sci. 1996, 5, 578–592. [Google Scholar] [CrossRef] [PubMed]
- Salvatella, X. Understanding Protein Dynamics Using Conformational Ensembles; Springer International Publishing: New York, NY, USA, 2013; Volume 805, pp. 67–85. [Google Scholar]
- Jamroz, M.; Kolinski, A.; Kmiecik, S. CABS-flex predictions of protein flexibility compared with NMR ensembles. Bioinformatics 2014, 30, 2150–2154. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, D.K.; Güntert, P. Objective identification of residue ranges for the superposition of protein structures. BMC Bioinform. 2011, 12, 170. [Google Scholar] [CrossRef] [PubMed]
- Snyder, D.A.; Montelione, G.T. Clustering algorithms for identifying core atom sets and for assessing the precision of protein structure ensembles. Proteins Struct. Funct. Bioinform. 2005, 59, 673–686. [Google Scholar] [CrossRef]
- Hyberts, S.G.; Goldberg, M.S.; Havel, T.F.; Wagner, G. The solution structure of eglin c based on measurements of many NOEs and coupling constants and its comparison with X-ray structures. Protein Sci. 1992, 1, 736–751. [Google Scholar] [CrossRef]
- Kelley, L.A.; Gardner, S.P.; Sutcliffe, M.J. An automated approach for defining core atoms and domains in an ensemble of NMR-derived protein structures. Protein Eng. 1997, 10, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Theobald, D.L.; Wuttke, D.S. THESEUS: Maximum likelihood superpositioning and analysis of macromolecular structures. Bioinformatics 2006, 22, 2171–2172. [Google Scholar] [CrossRef]
- Theobald, D.L.; Wuttke, D.S. Accurate structural correlations from maximum likelihood superpositions. PLoS Comput. Biol. 2008, 4, e43. [Google Scholar] [CrossRef] [PubMed]
- Moult, J.; Fidelis, K.; Kryshtafovych, A.; Tramontano, A. Critical assessment of methods of protein structure prediction (CASP)-round IX. Proteins Struct. Funct. Bioinform. 2011, 79, 1–5. [Google Scholar] [CrossRef]
- Kryshtafovych, A.; Moult, J.; Bales, P.; Bazan, J.F.; Biasini, M.; Burgin, A.; Chen, C.; Cochran, F.V.; Craig, T.K.; Das, R.; et al. Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10. Proteins Struct. Funct. Bioinform. 2013, 82, 26–42. [Google Scholar] [CrossRef] [PubMed]
- Mao, B.; Tejero, R.; Baker, D.; Montelione, G.T. Protein NMR Structures Refined with Rosetta Have Higher Accuracy Relative to Corresponding X-ray Crystal Structures. J. Am. Chem. Soc. 2014, 136, 1893–1906. [Google Scholar] [CrossRef]
- Everett, J.K.; Tejero, R.; Murthy, S.B.K.; Acton, T.B.; Aramini, J.M.; Baran, M.C.; Benach, J.; Cort, J.R.; Eletsky, A.; Forouhar, F.; et al. A community resource of experimental data for NMR/X-ray crystal structure pairs. Protein Sci. 2015, 25, 30–45. [Google Scholar] [CrossRef] [PubMed]
- Hollander, M.; Wolfe, D.A.; Chicken, E. Nonparametric Statistical Methods, 3rd ed.; Wiley: Hoboken, NY, USA, 2015; pp. 1–10. [Google Scholar]
- Brunger, A.T. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2007, 2, 2728–2733. [Google Scholar] [CrossRef] [PubMed]
- Brunger, A.T.; Adams, P.D.; Clore, G.M.; Delano, W.L.; Gros, P.; Grosse-Kunstleve, R.W.; Jiang, J.-S.; Kuszewski, J.; Nilges, M.; Pannu, N.S.; et al. Crystallography & NMR System: A New Software Suite for Macromolecular Structure Determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998, 54, 905–921. [Google Scholar] [CrossRef]
- Schwieters, C.D.; Kuszewski, J.J.; Tjandra, N.; Clore, G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003, 160, 65–73. [Google Scholar] [CrossRef]
- Jain, A.; Purohit, C.S.; Verma, S.; Sankararamakrishnan, R. Close Contacts between Carbonyl Oxygen Atoms and Aromatic Centers in Protein Structures: π⋅⋅⋅π or Lone-Pair⋅⋅⋅π Interactions? J. Phys. Chem. B 2007, 111, 8680–8683. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Das, A. The n → π* interaction: A rapidly emerging non-covalent interaction. Phys. Chem. Chem. Phys. 2015, 17, 9596–9612. [Google Scholar] [CrossRef]
- Lange, O.F.; Van Der Spoel, D.; De Groot, B.L. Scrutinizing Molecular Mechanics Force Fields on the Submicrosecond Timescale with NMR Data. Biophys. J. 2010, 99, 647–655. [Google Scholar] [CrossRef]
- Pace, C.N.; Fu, H.; Lee Fryar, K.; Landua, J.; Trevino, S.R.; Schell, D.; Thurlkill, R.L.; Imura, S.; Scholtz, J.M.; Gajiwala, K.; et al. Contribution of hydrogen bonds to protein stability. Protein Sci. 2014, 23, 652–661. [Google Scholar] [CrossRef]
- Reichert, D.; Zinkevich, T.; Saalwächter, K.; Krushelnitsky, A. The relation of the X-ray B-factor to protein dynamics: Insights from recent dynamic solid-state NMR data. J. Biomol. Struct. Dyn. 2012, 30, 617–627. [Google Scholar] [CrossRef]
- Kuzmanic, A.; Pannu, N.S.; Zagrovic, B. X-ray refinement significantly underestimates the level of microscopic heterogeneity in biomolecular crystals. Nat. Commun. 2014, 5, 3220. [Google Scholar] [CrossRef] [PubMed]
- Shivakumar, D.; Williams, J.; Wu, Y.; Damm, W.; Shelley, J.; Sherman, W. Prediction of Absolute Solvation Free Energies using Molecular Dynamics Free Energy Perturbation and the OPLS Force Field. J. Chem. Theory Comput. 2010, 6, 1509–1519. [Google Scholar] [CrossRef] [PubMed]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins Struct. Funct. Bioinform. 2006, 65, 712–725. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Bowers, K.J.; Chow, D.E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, New York, NY, USA, 11 November 2006; p. 43. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- MATLAB, version 9.2.0.538062 (R2017a); The MathWorks Inc.: Natick, MA, USA, 2017.

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reinknecht, C.; Riga, A.; Rivera, J.; Snyder, D.A. Patterns in Protein Flexibility: A Comparison of NMR “Ensembles”, MD Trajectories, and Crystallographic B-Factors. Molecules 2021, 26, 1484. https://doi.org/10.3390/molecules26051484
Reinknecht C, Riga A, Rivera J, Snyder DA. Patterns in Protein Flexibility: A Comparison of NMR “Ensembles”, MD Trajectories, and Crystallographic B-Factors. Molecules. 2021; 26(5):1484. https://doi.org/10.3390/molecules26051484
Chicago/Turabian StyleReinknecht, Christopher, Anthony Riga, Jasmin Rivera, and David A. Snyder. 2021. "Patterns in Protein Flexibility: A Comparison of NMR “Ensembles”, MD Trajectories, and Crystallographic B-Factors" Molecules 26, no. 5: 1484. https://doi.org/10.3390/molecules26051484
APA StyleReinknecht, C., Riga, A., Rivera, J., & Snyder, D. A. (2021). Patterns in Protein Flexibility: A Comparison of NMR “Ensembles”, MD Trajectories, and Crystallographic B-Factors. Molecules, 26(5), 1484. https://doi.org/10.3390/molecules26051484