Abstract
In this paper, peptide conjugates were designed and synthesized by incorporating the antimicrobial undecapeptide BP16 at the C- or N-terminus of the plant defense elicitor peptide flg15, leading to BP358 and BP359, respectively. The evaluation of their in vitro activity against six plant pathogenic bacteria revealed that BP358 displayed MIC values between 1.6 and 12.5 μM, being more active than flg15, BP16, BP359, and an equimolar mixture of BP16 and flg15. Moreover, BP358 was neither hemolytic nor toxic to tobacco leaves. BP358 triggered the overexpression of 6 out of the 11 plant defense-related genes tested. Interestingly, BP358 inhibited Erwinia amylovora infections in pear plants, showing slightly higher efficacy than the mixture of BP16 and flg15, and both treatments were as effective as the antibiotic kasugamycin. Thus, the bifunctional peptide conjugate BP358 is a promising agent to control fire blight and possibly other plant bacterial diseases.
1. Introduction
Fire blight caused by Erwinia amylovora is one of the main diseases that affect plants of the family Rosaceae, causing substantial production losses worldwide in important fruit crops, such as apple and pear trees [1,2,3,4]. Management of fire blight relies on a combination of strategies that include cultural practices and/or the use of tolerant cultivars and of preventive bactericide sprays [3]. Preventive applications of bactericides, such as antibiotics and copper compounds, for the control of fire blight, have several drawbacks. In particular, repeated antibiotic treatments have led to the emergence of resistance within the pathogen population which limits their efficacy, and the use of antibiotics is not authorized in many countries [5,6,7,8,9,10]. In the case of copper compounds, they are not always effective, even after multiple applications, and they may cause phytotoxicity by increasing fruit russeting [8]. These limitations entail an impending need to find safer and more effective methods to combat fire blight.
Antimicrobial peptides, found in the defense mechanisms of most living organisms, have been reported as promising compounds for fire blight management [11,12,13,14,15]. These peptides are cationic sequences of 2 to 50 amino acids, able to adopt an amphipathic structure [16,17,18]. In addition to complying with safety standards, they have some advantages over other treatments. In particular, they are biodegradable, and it is difficult for bacteria to develop resistance to them because their mechanism of action mainly involves the disruption of the bacterial cell membrane and, moreover, they are selective for bacterial cells over plant or mammal tissues [16,17,19,20,21].
Plant defense inducers are also viewed as an interesting complement to strategies using bactericides in plant protection. Plants have an immune system able to activate defense mechanisms by innate immunity, and systemic induced resistance (ISR), or/and systemic acquired resistance (SAR) [22,23]. The recognition of different molecular patterns leads to the activation of transcriptional regulators and defense-related genes. Microbe-associated molecular patterns (MAMPs) are able to cause defense priming that triggers the ISR mediated by jasmonic acid and ethylene [23]. Other molecular patterns such as pathogen or damage-associated (PAMPs or DAMPs) induce the pattern-triggered immunity (PTI) followed, in a second phase, by the effector-triggered immunity (ETI). The initiation of ETI results in a local resistance reaction of the infected tissue represented by localized programmed cell death (PCD) called the hypersensitive response (HR) and, subsequently, induces SAR mediated by salicylic acid. Compounds, such as bion, acibenzolar-S-methyl, or harpins, have been described as acting as defense elicitors in apple or pear plants, being effective in the control of fire blight [3,24]. Moreover, peptides flg22 or flg15, derived from bacterial flagellin, have been reported to elicit immune responses in a broad variety of plants, including tomatoes or apples [25,26]. Therefore, they could be useful as agents to control fire blight.
The conjugation of two different peptides constitutes an efficient approach to obtain novel compounds with improved antimicrobial activity compared to the individual sequences [27,28,29]. In fact, the conjugation of sequences with antimicrobial activity has been extensively reported [30,31,32,33,34,35]. In this context, we conjugated the antimicrobial peptide BP100 with fragments of natural antimicrobial peptides to obtain peptide conjugates with improved biological activity against plant pathogens than their corresponding monomers [13]. By contrast, peptide conjugates resulting from the combination of a peptide with antimicrobial activity with another peptide with a different activity are limited [36,37,38]. To the best of our knowledge, the conjugation of an antimicrobial peptide and a plant defense elicitor sequence has only been described by the group of G.-Y. Chen, who conjugated the active domains of cecropin A and melittin to the elicitor harpin Hpa1 to obtain a chimeric protein that conserved the biological activities of the parent monomers [39]. In addition, few papers describing the design of new peptide conjugates have evaluated the effect on the activity of the order of the monomers in the sequence of these conjugates [13,40,41,42,43]. Results from this evaluation point out that the biological activity of the peptide conjugates is significantly sensitive to this structural modification.
Studies on antimicrobial peptides are almost solely focused on the use of individual peptides. However, another attractive approach relies on mixtures of different peptides or mixtures of peptides and conventional antibiotics [18,44]. Concerning the combination of two antimicrobial peptides, these mixtures have been reported to cause a higher antimicrobial effect than the individual components [18,45,46,47,48]. In this context, the differences in biological activity between the conjugation of two peptides in a single sequence and a mixture of the two individual peptides have been scarcely analyzed [43,49,50,51,52]. In one of these studies, it was concluded that a hybrid peptide derived from magainin 2 and PGLa showed similar or even stronger antimicrobial activity than an equimolar mixture of magainin and PGLa [49]. Similarly, the hybrid antimicrobial peptide LFchimera, incorporating the sequences of bovine lactoferrampin (265–284) and lactoferricin (17–30), exhibited significantly higher antimicrobial and candidacidal activity than the individual peptides or an equimolar mixture of both of them [51,52]. Recently, Wade et al. demonstrated that peptide conjugates derived from parasin were more active than cocktails of the corresponding individual sequences, while hybrid peptides derived from magainin 2 were at least as active as the mixtures of the corresponding monomers [43].
The aim of the present study was to obtain efficient agents for the management of fire blight by conjugating the antimicrobial peptide BP16 at the N- or at the C-terminus of the plant defense elicitor flg15. The linear undecapeptide BP16 was previously identified to display moderate antimicrobial activity against E. amylovora with low toxicity [2,11,20]. We evaluated the resulting peptide conjugates to determine the antimicrobial activity against several bacterial phytopathogens. Furthermore, the capacity of the best peptide conjugate to induce the expression of defense-related genes on tomato plants was studied. Finally, the effectiveness of this peptide conjugate was assessed in whole plants against fire blight disease in controlled greenhouse environmental conditions. The activity of the peptide conjugates was compared to that of a mixture of both monomers BP16 and flg15.
2. Results
2.1. Design and Solid-Phase Synthesis of the Peptides
Peptide conjugates were designed by combining the antimicrobial peptide BP16 and the plant defense elicitor peptide flg15 (Table 1). With the aim of studying the influence of the order of the monomers on the biological activity, BP16 was conjugated at the C- or at the N-terminus of flg15, resulting in peptide conjugates BP358 (flg15-BP16) and BP359 (BP16-flg15), respectively.

Table 1.
Sequences, retention times, HPLC purities, and mass spectrometry data of the peptides.
The synthesis was performed in solid-phase following a standard 9-fluorenylmethoxycarbonyl (Fmoc)/tert-butyl (tBu) strategy to yield a C-terminal amide for BP16 and BP358 and a C-terminal carboxylic acid in the case of flg15 and BP359. After purification, peptides were obtained at >99% HPLC purity and were characterized by HRMS (Table 1) (Supplementary Materials for characterization of BP16, flg15, BP358 and BP359).
2.2. In Vitro Biological Activity of Peptides
The antibacterial activity of the peptide conjugates BP358 and BP359 was assayed against E. amylovora, Pseudomonas syringae pv. syringae, Pseudomonas syringae pv. actinidiae, Xanthomonas arboricola pv. pruni, Xanthomonas fragariae, and Xanthomonas axonopodis pv. vesicatoria at 1.6, 3.1, 6.2, 12.5, 25, 50, and 100 μM (Table 2). The monomers BP16 and flg15 were included for comparison purposes. In order to analyze the effect of the conjugation, assays were also performed using an equimolar mixture of these two peptides.

Table 2.
Antimicrobial activity (MIC), hemolysis, and phytotoxicity of the peptides.
BP16 was active against the above bacteria with MIC values ranging from 3.1 to 50 μM. In contrast, as expected, flg15 did not display antibacterial activity (MIC > 100 μM). The peptide conjugate BP358 (flg15-BP16) was considerably more active (MIC values between 1.6 and 12.5 μM) than both BP359 (BP16-flg15, MIC of 3.1 to > 50 μM) and BP16 (MIC of 3.1 to 50 μM). In particular, BP358 showed lower MIC values than BP16 against E. amylovora, X. arboricola pv. pruni, X. fragariae, and X. axonopodis pv. vesicatoria. The mixture of the monomers BP16 and flg15 exhibited similar antibacterial activity to that of BP16 (MIC of 3.1 to 50 μM). In addition, the reference antibiotic kanamycin sulfate displayed a similar MIC than the peptides against E. amylovora (MIC of 6.2 to 12.5 μM).
Bactericidal activity was evaluated by comparing the time course to kill mid-logarithmic-phase culture suspensions of E. amylovora. As shown in Figure 1, at the concentration tested, BP358, BP16, and the mixture of BP16 and flg15 showed a bactericidal effect. They reduced E. amylovora population 3–4 log after a 180 min exposure period; BP358 was the peptide that showed faster bactericidal activity.
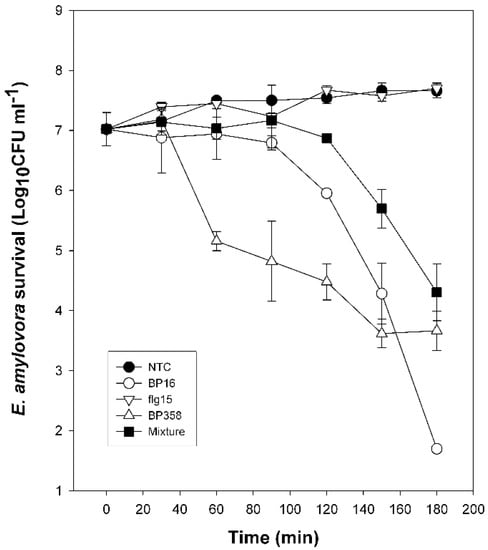
Figure 1.
Kinetics of survival of E. amylovora in the presence of peptides. Bacterial suspensions were untreated (NTC) or treated with BP16 (50 µM), flg15 (100 µM), BP358 (6.2 µM), or a mixture of flg15 and BP16 at equal molar quantities (25 μM) (Mixture). Viable cells were determined at different time intervals.
The toxicity of these peptides to eukaryotic cells was determined as the ability to lyse erythrocytes in comparison to melittin (100% hemolysis). All the peptides, as well as the mixture of BP16 and flg15, displayed low hemolytic activity with percentages of hemolysis ranging from 0% to 5% at 375 μM (Table 2). These peptides were also evaluated for their effect on tobacco leaves. Melittin was used as a positive control, which caused a brown necrotic area of around 2 cm diameter at 250 μM. In contrast, the monomers (BP16 and flg15) and the conjugates (BP358 and BP359) had a considerably less toxic effect at this concentration, causing a lesion diameter of 0.26, 0, 0.42, and 0.30 cm, respectively. The phytotoxicity of the mixture of BP16 and flg15 was similar to that of the monomers, causing a lesion diameter of 0.30 cm at 250 μM.
2.3. Effect of Peptides on Defense Gene Expression of Tomato Plants
The capacity to induce the expression of genes related to defense response in tomato plants was evaluated for the peptide conjugate BP358, the corresponding monomers BP16 and flg15, and an equimolar mixture of BP16 and flg15 (Table 3). Acibenzolar-S-methyl was included as positive control. The relative quantification for the expression of the selected genes was performed using the ΔΔCt method [53]. A fold induction above 2 was considered overexpression.

Table 3.
Expression of genes related to defense response in tomato after the treatment with the reference compound acibenzolar-S-methyl (ASM), peptides flg15, BP16, and BP358, and a mixture of BP16 and flg15 (BP16 + flg15). Fold induction above 2 was considered overexpression in the relative quantification using the ΔΔCt method. Significant values are indicated in bold.
Tobacco plants treated with acibenzolar-S-methyl showed overexpression of 9 out of the 11 genes analyzed (Harp, PR1, GluA, PPO, LOX, PinII, Sub1, ERT3, and BCB). The monomer flg15 caused overexpression of all tested genes except for LOX, whereas BP16 only induced the overexpression of Tas14. Notably, the peptide conjugate BP358 promoted the overexpression of 6 genes: Harp, GluA, Sub1, and BCB, with a relative overexpression of more than 3-fold, and LOX and Osm2 with a relative overexpression between 2- and 3-fold. Concerning the mixture of BP16 and flg15, it caused the overexpression of two genes, BCB and GluA.
2.4. Activity of Peptides in Planta
The effect of the peptide conjugate BP358 to reduce E. amylovora infections was tested in potted pear plants assays and compared with that of a mixture containing equal molar quantities of BP16 and flg15 (Figure 2). The incidence of the infections was determined. Monomers BP16 and flg15 were included for comparison purposes, and water and kasugamycin at 2 g/L, as a non-treated and positive control, respectively. All peptide treatments were performed at 125 µM. Two independent assays were carried out.
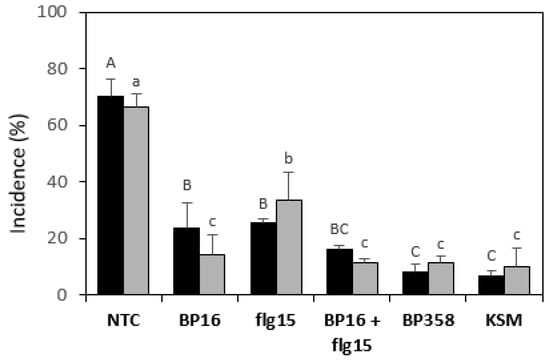
Figure 2.
Effect of the peptides on the incidence of E. amylovora infections in pear plants. Black bars correspond to assay one and grey bars to assay two. A non-treated control (NTC) and a reference treatment with kasugamycin (KSM) were included. The treatment with a mixture of BP16 and flg15 is labeled as BP16 + flg15. The confidence intervals for the means are indicated on top of the bars. Different letters (capital letters for assay one and lowercase letters for assay two) show significant differences between the treatments according to Duncan’s test (p < 0.05, ANOVA, LSD).
Treatment with BP16 and flg15 significantly reduced the incidence of fire blight in pear plants compared to the non-treated control. In one of the experiments, the peptide conjugate BP358 significantly decreased the incidence of the disease compared to the treatment with the monomers BP16 and flg15. In another experiment, BP358 significantly decreased the incidence compared to flg15, but they did not differ from BP16. Remarkably, the efficacy of BP358 did not differ significantly from kasugamycin. Results of the mixture of BP16 and flg15 did not differ from those of the monomers in one experiment and were improved compared to flg15 in the other one. In addition, it should also be pointed out that in both experiments, the efficacy of this mixture did not differ significantly from BP358 and from kasugamycin.
3. Discussion
Conjugation of two peptides to obtain sequences with a better biological profile is a widely used strategy. This approach has led to the design of interesting hybrid peptides with significant antimicrobial activity derived from cecropin A, melittin, lactoferricins, thanatin, cathelicidin, and LL-37, among others [30,31,32,33,34,37,51,52]. Within this field, we described peptide conjugates containing the antimicrobial peptide BP100 and fragments of cecropin A, melittin, and magainin [13]. These conjugates were more active than the monomers against plant pathogens such as E. amylovora, X. axonopodis pv. vesicatoria, and P. syringae pv. syringae. Interestingly, the hybridization of two peptides with different activity may be a promising approach since it would allow designing bifunctional peptides. In particular, we envisaged that the conjugation of an antimicrobial peptide with a plant defense elicitor peptide would provide sequences with both activities that would be useful for plant protection. Thus, in the present study, peptide conjugates BP358 (flg15-BP16) and BP359 (BP16-flg15), resulting from the combination of the antimicrobial peptide BP16 [11] and the plant defense elicitor peptide flg15 [25,26,54], were designed and tested against bacterial plant pathogens. In addition, the activity of these two conjugates was compared to that of a mixture of BP16 and flg15.
Peptides BP16 and flg15 were poorly active in vitro, and both the conjugation of these sequences and their order in the final peptide conjugate influenced the in vitro antibacterial activity against E. amylovora, P. syringae pv. actinidiae, P. syringae pv. syringae, X. arboricola pv. pruni, X. axonopodis pv. vesicatoria, and X. fragariae. Interestingly, the conjugation of BP16 and flg15 led to an enhancement of the activity, in particular, when BP16 was linked at the C-terminus of flg15. The resulting peptide conjugate BP358 (flg15-BP16) was more active (MIC between 1.6 and 6.2 μM) against E. amylovora, X. arboricola pv. pruni, X. axonopodis pv. vesicatoria, and X. fragariae than the antimicrobial peptide BP16 (MIC between 6.2 and 50 μM), and displayed the same MIC values as BP16 against P. syringae pv. actinidiae and P. syringae pv. syringae (MIC between 3.1 and 12.5 μM). In contrast, BP359, which included BP16 at the N-terminus, was only more active than BP16 against X. axonopodis pv. vesicatoria. The difference in activity observed between the two conjugates is in agreement with other studies, which report that the position of the monomers in the final conjugate is crucial for the activity [13,40,41,42,43].
Investigation of antibacterial activity also showed that a mixture containing equal molar quantities of both monomers BP16 and flg15 showed similar MIC values as BP16, and it was less active than the peptide conjugate BP358. This result suggests that these two monomers do not interfere with each other either when mixed or conjugated. Other peptide conjugates with higher activity than that of an equimolar mixture of the corresponding monomers have been described; a hybrid peptide derived from magainin 2 and PGLa [49], the peptide conjugate LFchimera, which incorporates lactoferrampin (265–284) and lactoferricin (17–30) [51,52], and hybrid peptides derived from parisin and BF2 or DesHDAP1 [43].
Concerning the toxicity, the conjugation of BP16 and flg15, the order of the monomers in the resulting peptide conjugates, or their mixture did not influence the hemolysis or the phytotoxicity. In fact, the monomers BP16 and flg15, the peptide conjugates BP358 and BP359, and the mixture of the monomers did not show cytotoxic activity to eukaryotic cells or toxicity to tobacco leaves. According to all these results, the conjugate BP358 had the best biological profile (MIC values between 1.6 and 12.5 μM and 5% hemolysis at 375 μM) and was selected for further studies.
The elicitation of plant defense was studied in tomato plants using genes related to the salicylic acid, jasmonic acid, or ethylene pathways, as well as with saline stress or wound damage [4,55]. As expected, tomato plants treated with flg15 overexpressed more genes and more intensely than the reference compound acibenzolar-S-methyl. The conjugation of flg15 with BP16 resulted in peptide BP358 that maintained the ability to act as a plant defense elicitor. Interestingly, BP358 triggered the overexpression of 6 out of 11 genes, sharing with flg15 the overexpression of genes Harp, GluA, Sub1, BPB, and Osm 2. The mixture of flg15 and BP16 had a lower elicitation activity promoting only the overexpression of genes GluA and BPB. This result suggests that these two peptides interfere with each other causing a reduction in the plant defense elicitation properties. As well as synergistic effects, antagonistic effects on defense induction have already been observed when using MAMPs mixtures, with a reduction in the reactive oxygen species (ROS) production in Arabidopsis thaliana of around 90% [56]. In addition, it cannot be discarded the possibility that BP16 blocks the binding site of the flagellin receptor FLS2 because it has been reported that other ligands different from flagellin bind to this receptor [57].
In the pear-E. amylovora pathosystem plant assays, both the peptide conjugate BP358 and the mixture of BP16 and flg15 are suitable options to reduce the effects of the disease. It is worth mentioning that the monomers displayed higher activity when tested in planta than in vitro. Especially remarkable is the case of flg15 that was not active in vitro whereas in planta caused a reduction in incidence of 50–64% in comparison to the non-treated control. Although the monomers turned out to be more active, BP358 and the mixture of BP16 and flg15 resulted in even more effective protection of plants from E. amylovora infections. BP358 exhibited a slightly higher efficacy than the mixture, and both were, in general, as effective as the antibiotic kasugamycin. The higher activity of BP358 could be attributed to its higher plant defense elicitation properties; this fact would also explain the improved activity shown by flg15 in the assays in planta.
Based on the above considerations, both peptide conjugation and the use of mixtures have shown to be feasible strategies to improve the biological activity of peptides as agents to control plant diseases. On the one hand, the peptide conjugate BP538 displayed high antimicrobial activity in planta and also promoted the overexpression of a high number of genes related to plant defense response. The use of peptide conjugates in plant protection is advantageous because their production can be achieved using microbial or plant biofactories. On the other hand, the mixture of peptides flg15 and BP16 also displayed high activity in planta and, since they are short peptide sequences, their chemical synthesis can be easily performed [13]. Moreover, treatments using peptide mixtures may reduce the development of bacterial resistance. In fact, Rolff et al. demonstrated that the combination of the antimicrobial peptides pexiganan and melittin slowed the evolution of resistance in Staphylococcus aureus compared to the use of individual antibiotics or antimicrobial peptides [47,58].
4. Materials and Methods
4.1. General Methods
Manual solid-phase synthesis was performed in polypropylene syringes (2 or 5 mL) fitted with a polyethylene porous disk. Solvents and soluble reagents were removed by suction. Most chemicals were from commercial suppliers Merck (Madrid, Spain), Iris Biotech GmbH (Marktredwitz, Germany), Carlo Erba (Sabadell, Spain), and used without further purification.
Peptides were analyzed using standard analytical high-performance liquid chromatography (HPLC) conditions with a Dionex liquid chromatography instrument composed of a UV/Vis Dionex UVD170U detector (Thermo Fisher Scientific, Sunnyvale, CA, USA), a P680 Dionex bomb, an ASI-100 Dionex automatic injector, and CHROMELEON 6.60 software (Thermo Fisher Scientific, Sunnyvale, CA, USA). Detection was performed at a wavelength of 220 nm. Solvent A was 0.1% aqueous trifluoroacetic acid (TFA), and solvent B was 0.1% TFA in CH3CN. Analyses were carried out with a Kromasil 100 C18 (4.6 mm × 40 mm × 3.5 µm) column with a 2–100% B over 7 min at a flow rate of 1 mL/min.
All peptides were purified on a CombiFlash Rf200 automated flash chromatography system using RediSep Rf Gold reversed-phase C18 column packed with high-performance C18 derivatized silica (Teledyne ISCO, Lincoln, NE, USA).
Electrospray-ionization mass spectrometry (ESI-MS) analyses were performed at the Serveis Tècnics de Recerca of the University of Girona with an Esquire 6000 ESI ion Trap LC/MS (Bruker Daltonics, Billerica, MA, USA) instrument equipped with an electrospray ion source. The instrument was operated in the positive ESI(+) ion mode. Samples (5 µL) were introduced into the mass spectrometer ion source directly through an HPLC autosampler. The mobile phase (80:20 CH3CN/H2O at a flow rate of 100 µL/min) was delivered by a 1200 Series HPLC pump (Agilent, Santa Clara, CA, USA). Nitrogen was employed as both the drying and nebulizing gas.
High-resolution mass spectrometry (HRMS) data were recorded on a Bruker MicroTof-QIITM instrument (Bruker Daltonics, Billerica, MA, USA) using ESI ionization source at the Serveis Tècnics de Recerca of the University of Girona. Samples were introduced into the mass spectrometer ion source by direct infusion using a syringe pump and were externally calibrated using sodium formate. The instrument was operated in the positive ion mode.
4.2. General Procedure for the Solid-Phase Synthesis of Peptides
Peptides were synthesized manually by the solid-phase method using a standard Fmoc/tBu strategy. A Fmoc-Rink-MBHA resin (0.55 mmol/g) was used for the synthesis of BP16, a Fmoc-Rink-ChemMatrix resin (0.66 mmol/g) for the synthesis of the conjugate BP358 (flg15-BP16), and a PAC-ChemMatrix resin (0.66 mmol/g) for the synthesis of flg15 and the conjugate BP359 (BP16-flg15) (Company, City, State Abbrev. if USA or Canada, Country). Fmoc-Leu-OH, Fmoc-Lys(Boc)-OH, Fmoc-Phe-OH, Fmoc-Ile-OH, Fmoc-Ala-OH, Fmoc-Gly-OH, Fmoc-Gln(Trt)-OH, Fmoc-Arg(Pmc)-OH, Fmoc-Ser(tBu)-OH, Fmoc-Asp(OtBu)-OH, and Fmoc-Asn(Trt)-OH were used as amino acid derivatives. The coupling of the first amino acid (5 equiv.) onto the PAC-derivatized resins was performed in the presence of N,N’-diisopropylcarbodiimide (DIC) (5 equiv.), 4-dimethylaminopyridine (DMAP) (0.5 equiv.), and N,N’-diisopropylethylamine (DIEA) (1 equiv.) in N,N-dimethylformamide (DMF) at room temperature for 2 h while stirring. This treatment was repeated twice and, then the resin was washed with DMF (6 × 1 min) and CH2Cl2 (3 × 1 min) and dried with diethyl ether (3 × 2 min). The completion of the coupling was checked using a Fmoc test. Then, the resin was acetylated with acetic anhydride/pyridine/CH2Cl2 (1.35:1.35:18, 2 × 30 min) followed by washes with CH2Cl2 (3 × 2 min), DMF (3 × 2 min), MeOH (2 × 2 min), CH2Cl2 (2 × 2 min), and DMF (6 × 1 min). Peptide elongation was carried out through sequential Fmoc removal and coupling steps of the corresponding Fmoc-protected amino acid. Fmoc group removal was performed with piperidine/DMF (3:7, 2 + 10 min). Couplings of the Fmoc-amino acids (4 equiv.) were mediated by ethyl 2-ciano-2-(hydroxyimino)acetate (Oxyma) (4 equiv.), and DIC (4 equiv.) in DMF at room temperature for 1 h under stirring. The completion of the couplings was checked using the Kaiser test [59]. After each coupling and deprotection step, the resin was washed with DMF (6 × 1 min) and CH2Cl2 (2 × 1 min). Once the peptidyl sequence was completed, the resin was treated with piperidine/N-methyl-2-pyrrolidinone (NMP) (3:7, 2 + 10 + 10 min), washed with NMP (6 × 1 min), CH2Cl2 (3 × 1 min), and diethyl ether (3 × 2 min), and air-dried. Finally, the resulting resin was treated with TFA/H2O/triisopropylsilane (TIS) (95:2.5:2.5) for 2 h at room temperature. Following TFA evaporation and diethyl ether extraction, the crude peptide was dissolved in H2O, lyophilized, purified with a CombiFlash, analyzed by HPLC, and characterized by ESI-MS and HRMS.
4.3. Bacterial Strains and Growth Conditions
The following plant pathogenic bacterial strains were used as target bacteria for in vitro experiments: E. amylovora PMV6076 (Institut National de la Recherche Agronomique, Angers, France) and EPS101 (Institut de Tecnologia Agroalimentària, Universitat de Girona, Spain), P. syringae pv. actinidiae Psa3700.1.1 (Instituto Valenciano de Investigaciones Agrarias, Valencia, Spain), P. syringae pv. syringae EPS94 (Institut de Tecnologia Agroalimentària, Universitat de Girona, Spain), X. arboricola pv. pruni CFBP5563 (Collection Française de Bactéries associées aux Plantes, Angers, France), X. axonopodis pv. vesicatoria 2133-2, and X. fragariae Xf349-9A (Instituto Valenciano de Investigaciones Agrarias, Valencia, Spain). All bacteria, except for X. fragariae, were stored in Luria Bertani (LB) broth supplemented with glycerol (20%) and maintained at −80 °C. For X. fragariae, Medium B [60] was used instead of LB. E. amylovora, P. syringae pv. actinidiae, P. syringae pv. syringae, and X. arboricola pv. pruni, were scraped from the agar media after growing for 24 h at 25 °C, and X. axonopodis pv. vesicatoria, and X. fragariae after growing for 48 h at 25 °C. The cell material was suspended in sterile water to obtain a suspension of 108 CFU mL−1. E. amylovora EPS101 isolated from an infected shoot of a Conference pear tree in Lleida (Spain) was used for the in planta assays [61].
4.4. Antibacterial Activity
Lyophilized peptides were solubilized in sterile Milli-Q water to a final concentration of 1 mM and filter sterilized through a 0.22 m pore filter. Minimum inhibitory concentration (MIC) of the peptides was assessed as previously described [11] using a growth inhibition assay. Dilutions of the peptides were made to obtain a final concentration of 100, 50, 25, 12.5, 6.2, 3.1, and 1.6 μM, and in the case of the peptide mixture, an equimolar solution was used. Kanamycin sulfate (Sigma, St. Louis, MO, USA) was included for E. amylovora as a reference antibiotic. Twenty microlitres of each dilution were mixed in a microtiter plate well with 20 μL of the corresponding bacterial suspension (final concentration 107 CFU mL−1) and 160 μL of Tryptic Soy Broth (TSB) (BioMèrieux, Marcy-l’Étoile, France) to a total volume of 200 μL. Three replicates for each peptide and concentration were used. Bacterial growth was determined by optical density measurement at 600 nm (Bioscreen C, Labsystem, Helsinki, Finland). Microplates were incubated at 25 °C with 20 s shaking before hourly absorbance measurement for 48 h. The experiment was repeated twice. The MIC was taken as the lowest peptide concentration with no growth at the end of the experiment.
4.5. Analysis of Bactericidal Activity
The bactericidal activity against E. amylovora was determined at the MIC (highest value observed in the growth inhibition assay) for the monomers BP16 (50 μM) and flg15 (100 μM), the conjugate BP358 (6.2 μM), and an equimolar mixture of BP16 and flg15 (25 μM), and it was compared to an untreated control. TSB broth-grown cultures of E. amylovora inoculated at 107 CFU ml−1 were incubated at the corresponding peptide or mixture concentration. Aliquots of 100 μL were removed at 30-min intervals for 3 h and diluted 10-fold, and the dilutions were plated on LB agar plates. The CFU were counted after a 48-h incubation at 25 °C. The experiment consisted of three replicates per treatment. Values were expressed as log CFU ml−1 during the experiment.
4.6. Hemolytic Activity
The hemolytic activity of the peptides was evaluated by determining hemoglobin release from erythrocyte suspensions of horse blood (5% v/v) (Oxoid) as previously described [11]. Blood was centrifuged at 6000 g for 5 min, washed three times with TRIS buffer (10 mM TRIS, 150 mM NaCl, pH 7.2), and diluted ten-fold. Peptides were solubilized in TRIS buffer. To test the hemolytic activity of the peptide mixture, an equimolar solution was used. Solubilized peptides were mixed with horse erythrocytes, and the final concentrations tested were 375, 250, 150, and 50 µM. The mixture was incubated under continuous shaking for 1 h at 37 °C. Then, the tubes were centrifuged at 3500g for 10 min, 80 μL aliquots of the supernatant transferred to 100-well microplates (Bioscreen), diluted with 80 μL water, and the absorbance measured at 540 nm (Bioscreen). Complete hemolysis was obtained by the addition of melittin at 100 µM (Sigma-Aldrich Corporation, Madrid, Spain). The percentage of hemolysis (H) was calculated using Equation (1):
where Op is the density for a given peptide concentration, Ob is the buffer, and Om is the melittin positive control.
H = 100 × [(Op − Ob)/(Om − Ob)]
4.7. Effect of Peptide Infiltration on Tobacco Leaves
Peptides were evaluated for their effect upon infiltration on tobacco leaves as described previously [13]. Peptide solutions of 50, 150, and 250 µM were infiltrated (100 μL) into the mesophylls of fully expanded tobacco leaves. To test the effect of the peptide mixture, an equimolar solution was used. Six independent inoculations were carried out in a single leaf, and at least three independent inoculations were performed per peptide and concentration randomly distributed in different leaves and plants. Control infiltrations with water (negative control) or melittin (positive control) at the same molar concentration were performed. The appearance of symptoms on the leaves was followed for 48 h after infiltration and measured as a lesion diameter.
4.8. Effect of Peptide Treatment on Induction of Defense Gene Expression of Tomato Plants
Seeds of tomato cv. Rio Grande plants were sown in hydroponic seed plugs (rockwool), germinated and grown under controlled greenhouse conditions (25 ± 2 °C, 16 h light/15 ± 2 °C, 8 h dark, and 60% relative humidity (RH). Two-week-old seedlings (two cotyledons) were transplanted into rockwool plugs (7.5 × 7.5 × 6.5 cm, Gordan Iberica, Spain). The experimental design consisted of three replicates of three plants per treatment. After two weeks, tomato leaves were sprayed with aqueous solutions of the peptides BP16, flg15, and BP358, an equimolar mixture of BP16 and flg15, at 125 µM, or with acybenzolar-S-methyl at 300 mg/L (Syngenta, Basel, Switzerland) until the run-off point. Water-sprayed plants were used as untreated controls. Twenty-four hours after product application, leaf samples were collected and processed to extract RNA for RT-qPCR assays. Plant material was ground to a fine powder in liquid nitrogen, adding 2 acid-washed glass beads (Sigma, 150 ± 600 μm) to the sample using the Tissuelyzer II system (Qiagen, Hilden, Germany). Total RNA was extracted from leaves using PureLink Plant RNA Reagent (Invitrogen, Life Technologies) according to the manufacturer’s manual. The RNA was solubilized in RNAse free water and was routinely subjected to DNAse treatment (Ambion® Turbo DNA-free™, Invitrogen Life Technologies, Carlsbad, CA, USA) to remove any contaminant DNA. In each step, the RNA was quantified using a Nanodrop N-2000 spectrophotometer, and its integrity was verified by denaturing agarose gel electrophoresis. First-strand of complementary DNA (cDNA) was generated from leave RNA using reverse transcriptase (High Capacity cDNA Reverse Transcription Kit, Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s manual.
To test gene defense induction in the treated tomato plants, a quantitative PCR (qPCR) assay was performed. qPCR was carried out in a fluorometric thermal cycler (qPCR Quant Studio 5, Applied Biosystems) by using a Mix SYBR®Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) as previously described [4]. Melting curve analysis was performed after each amplification to verify amplification specificity. A constitutive gene (actin) was used as a reference control, and the following genes implicated in plant defense response were analyzed: pathogenesis-related protein-1 (PR1), harpin (Harp), polyphenol oxidase (PPO), subtilisin-like protease (Sub1), blue copper binding-protein (BCB), osmotin (Osm2), acidic β-1,3 endoglucanase (GluA), lipoxygenase (LOX), protein inhibitor II (PinII), dehydrin (Tas14), and early-ripening tomato (ERT3). Specific oligonucleotides genes [4,55] were designed and used for their quantification. The primer concentration was 100 nM for all the genes except for the GluA, Harp, PR1, and actin genes which concentration was 300 nM. A calibration curve was prepared using decimal dilutions of recombinant plasmid DNA (target sequences were cloned into a vector pSpark (Canvax, Córdoba, Spain) in Escherichia coli DH5α cells). The efficiency for each standard curve was calculated to check that the efficiency within amplifications was similar.
Relative quantification of gene expression was done using the ΔΔCt method as previously described [4,52]. Ct values obtained for each repetition treatment were used to estimate the fold change value of the endogenous reference gene (actin) and the target plant defense genes. These results were used to calculate the ratios of the plant defense genes (relative to the actin gene and for all treatments analyzed, including the control plants). The statistical significance of the results for the selected peptides was determined using the REST2009 Software (Qiagen) [62].
4.9. In Planta Assays
The efficacy of peptides in fire blight suppression was determined using whole-plant infection assays with pear plants (Pyrus communis cv. Conference, Agromillora, Iberica S.A, Spain) inoculated with E. amylovora. Three-year-old pear plants grown in 20 cm diameter plastic pots in the greenhouse were used. During winter, plants were left outside the greenhouse for chilling, and during early spring were pruned to leave 3 or 4 shoots and were forced to bud in the greenhouse. Plants were fertilized once a week with a 200-ppm N/P/K solution (20:10:20) and were used when the shoots were about 3 or 4 cm long and had 5 or 6 young leaves per shoot. Pear plants were sprayed with a hand sprayer with 6 mL of an aqueous suspension of peptides BP358, BP16, or flg15 or with an equimolar mixture of BP16 and flg15. Peptides were assayed at 125 µM. After 48 h, treated plants were spray inoculated until the run-off point (6 mL) with a suspension of E. amylovora at 108 CFU/mL mixed with diatomaceous earth at 1 mg/mL. Plants were incubated under controlled greenhouse conditions (25 ± 2 °C, 16 h light/15 ± 2 °C, 8 h dark, and 60% RH). The experimental design consisted of three replicates of three plants per treatment. Water-sprayed plants were used as untreated control and plants treated with kasugamycin 8% at 2 g/L (Lainco, Rubí, Spain) as a positive control. Two independent experiments were performed. Incidence of infections was determined per each replicate after 5 days of pathogen inoculation as the percentage of leaves with symptoms [63]. The effect of peptide treatments on plant material infection was determined using analysis of variance (ANOVA) with the general linear model (GLM) procedure of the statistical analysis system (SAS) (Version 8.2, SAS Institute, Cary, NC, USA). Means were separated using Duncan’s test (p < 0.05).
5. Conclusions
In conclusion, this manuscript shows that the conjugation or the mixture of the plant elicitor peptide flg15 with the antimicrobial peptide BP16 could be suitable approaches to fight plant diseases caused by E. amylovora. It is worth highlighting the peptide conjugate BP358, incorporating BP16 at the C-terminus of flg15, that exhibited high activity in planta as well as the capacity to induce plant defense responses. In fact, to the best of our knowledge, this is the first example of a peptide conjugate with activity against plant pathogens resulting from the combination of two monomers with different biological activities. Remarkably, in the pear-E. amylovora pathosystem, BP358 was as effective as the antibiotic kasugamycin, being a promising alternative tool to be included in plant disease management strategies.
Supplementary Materials
The following are available online, HPLC, ESI-MS, and HRMS of peptides BP16 and flg15, and of peptide conjugates BP358 and BP359.
Author Contributions
Conceptualization, J.F., E.B., M.P., L.F., E.M. and A.B. (Anna Bonaterra); methodology, P.C.-F., C.C., À.O., A.B. (Aina Baró), J.F. and E.B.; data validation, P.C.-F., J.F., E.B., M.P., L.F., E.M. and A.B. (Anna Bonaterra); formal analysis, P.C.-F., E.B., M.P., L.F., E.M. and A.B. (Anna Bonaterra); investigation, P.C.-F., C.C., À.O., A.B. (Aina Baró), J.F. and E.B.; resources, E.B., M.P., L.F., E.M. and A.B. (Anna Bonaterra); data curation, J.F., E.B., M.P., L.F., E.M. and A.B. (Anna Bonaterra); writing—original draft preparation, P.C.-F.; writing—review and editing, P.C.-F., E.B., M.P., L.F., E.M. and A.B. (Anna Bonaterra); visualization, E.B., M.P., L.F., E.M. and A.B. (Anna Bonaterra); supervision, E.B., M.P., L.F., E.M. and A.B. (Anna Bonaterra); project administration, E.B., M.P., L.F., E.M. and A.B. (Anna Bonaterra); funding acquisition, E.B., M.P., L.F., E.M. and A.B. (Anna Bonaterra). All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by MINECO/FEDER, UE, grant number AGL2015-69876-C2-2-R, by the Universitat de Girona, grant number MPCUdG2016/038, and by MCIU/AEI/FEDER, UE, grant number RTI2018-099410-B-C22.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable
Acknowledgments
The authors acknowledge Serveis Tècnics de Recerca at the University of Girona for the mass spectrometry analysis.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are available from the authors.
References
- Oh, C.S.; Beer, S.V. Molecular genetics of Erwinia amylovora involved in the development of fire blight. FEMS Microbiol. Lett. 2005, 253, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Ferre, R.; Badosa, E.; Feliu, L.; Planas, M.; Montesinos, E.; Bardají, E. Inhibition of plant-pathogenic bacteria by short synthetic cecropin A-melittin hybrid peptides. Appl. Environ. Microbiol. 2006, 72, 3302–3308. [Google Scholar] [CrossRef] [PubMed]
- Aćimović, S.G.; Zeng, Q.; McGhee, G.C.; Sundin, G.W.; Wise, J.C. Control of fire blight (Erwinia amylovora) on apple trees with trunk-injected plant resistance inducers and antibiotics and assessment of induction of pathogenesis-related protein genes. Front. Plant Sci. 2015, 6, 1–10. [Google Scholar]
- Badosa, E.; Montesinos, L.; Camó, C.; Ruz, L.; Cabrefiga, J.; Francés, J.; Gascón, B.; Planas, M.; Feliu, L.; Montesinos, E. Control of fire blight infections with synthetic peptides that elicit plant defense responses. J. Plant Pathol. 2017, 99, 65–73. [Google Scholar]
- Sundin, G.W.; Bender, C.L. Ecological and genetic analysis of copper and streptomycin resistance in Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 1993, 59, 1018–1024. [Google Scholar] [CrossRef] [PubMed]
- Vidaver, A.K. Uses of antimicrobials in plant agriculture. Clin. Infect. Dis. 2002, 34, 107–110. [Google Scholar] [CrossRef]
- Ordax, M.; Marco-Noales, E.; López, M.M.; Biosca, E.G. Survival strategy of Erwinia amylovora against copper: Induction of the viable-but-nonculturable state. Appl. Environ. Microbiol. 2006, 72, 3482–3488. [Google Scholar] [CrossRef]
- Johnson, K.B.; Temple, T.N. Evaluation of strategies for fire blight control in organic pome fruit without antibiotics. Plant Dis. 2013, 97, 402–409. [Google Scholar] [CrossRef]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef]
- Sundin, G.W.; Wang, N. Antibiotic resistance in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 2018, 56, 161–180. [Google Scholar] [CrossRef]
- Badosa, E.; Ferre, R.; Planas, M.; Feliu, L.; Besalú, E.; Cabrefiga, J.; Bardají, E.; Montesinos, E. A library of linear undecapeptides with bactericidal activity against phytopathogenic bacteria. Peptides 2007, 28, 2276–2285. [Google Scholar] [CrossRef] [PubMed]
- Sarojini, V. Antimicrobial peptides for fire blight control. In Small Wonders: Peptides for Disease Control; ACS Symposium Series; Rajasekaran, K., Cary, J.W., Jaynes, J.M., Montesinos, E., Eds.; American Chemical Society: Washington, DC, USA, 2012; Volume 1095, pp. 397–414. [Google Scholar]
- Badosa, E.; Moiset, G.; Montesinos, L.; Talleda, M.; Bardají, E.; Feliu, L.; Planas, M.; Montesinos, E. Derivatives of the antimicrobial peptide BP100 for expression in plant systems. PLoS ONE 2013, 8, e85515. [Google Scholar] [CrossRef]
- Cameron, A.; De Zoysa, G.H.; Sarojini, V. Antimicrobial peptides against Pseudomonas syringae pv. actinidiae and Erwinia amylovora: Chemical synthesis, secondary structure, efficacy, and mechanistic investigations. Biopolymers 2014, 102, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Separovic, F.; O’Brien-Simpson, N.; Wade, J.D. Chemically modified and conjugated antimicrobial peptides against superbugs. Chem. Soc. Rev. 2021, 50, 4932–4973. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Björn, C.; Ekblom, J. Antimicrobial peptides as therapeutic agents: Opportunities and challenges. Crit. Rev. Biotechnol. 2020, 40, 978–992. [Google Scholar] [CrossRef]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef]
- Keymanesh, K.; Soltani, S.; Sardari, S. Application of antimicrobial peptides in agriculture and food industry. World J. Microbiol. Biotechnol. 2009, 25, 933–944. [Google Scholar] [CrossRef]
- Montesinos, E.; Badosa, E.; Cabrefiga, J.; Planas, M.; Feliu, L.; Bardají, E. Antimicrobial peptides for plant disease control from discovery to application. In Small Wonders: Peptides for Disease Control; ACS Symposium Series; Rajasekaran, K., Cary, J.W., Jaynes, J.M., Montesinos, E., Eds.; American Chemical Society: Washington, DC, USA, 2012; Volume 1095, pp. 235–261. [Google Scholar]
- Ciumac, D.; Gong, H.; Hu, X.; Lu, J.R. Membrane targeting cationic antimicrobial peptides. J. Colloid Interface Sci. 2019, 537, 163–185. [Google Scholar] [CrossRef]
- Choi, H.W.; Klessig, D.F. DAMPs, MAMPs, and NAMPs in plant innate immunity. BMC Plant Biol. 2016, 16, 232. [Google Scholar] [CrossRef]
- Abdul Malik, N.A.; Kumar, I.S.; Nadarajah, K. Elicitor and receptor molecules: Orchestrators of plant defense and immunity. Int. J. Mol. Sci. 2020, 21, 963. [Google Scholar] [CrossRef]
- Johnson, K.B.; Temple, T.N. Induction of systemic acquired resistance aids restoration of tree health in field-grown pear and apple diseased with fire blight. Plant Dis. 2017, 101, 1263–1268. [Google Scholar] [CrossRef] [PubMed]
- Felix, G.; Duran, J.D.; Volko, S.; Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999, 18, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Robatzek, S.; Bittel, P.; Chinchilla, D.; Köchner, P.; Felix, G.; Shiu, S.H.; Boller, T. Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant Mol. Biol. 2007, 64, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, A.; Neundorf, I. Design and application of antimicrobial peptide conjugates. Int. J. Mol. Sci. 2016, 17, 701. [Google Scholar] [CrossRef]
- Lorenzon, E.N.; Piccoli, J.P.; Santos-Filho, N.A.; Cilli, E.M. Dimerization of antimicrobial peptides: A promising strategy to enhance antimicrobial peptide activity. Protein Pept. Lett. 2019, 26, 98–107. [Google Scholar] [CrossRef]
- Wang, C.; Yang, C.; Chen, Y.C.; Ma, L.; Huang, K. Rational design of hybrid peptides: A novel drug design approach. Curr. Med. Sci. 2019, 39, 349–355. [Google Scholar] [CrossRef]
- Wei, X.B.; Wu, R.J.; Si, D.Y.; Liao, X.D.; Zhang, L.L.; Zhang, R.J. Novel hybrid peptide cecropin A (1-8)-LL37 (17-30) with potential antibacterial activity. Int. J. Mol. Sci. 2016, 17, 983. [Google Scholar] [CrossRef]
- Dong, N.; Li, X.R.; Xu, X.Y.; Lv, Y.F.; Li, Z.Y.; Shan, A.S.; Wang, J.L. Characterization of bactericidal efficiency, cell selectivity, and mechanism of short interspecific hybrid peptides. Amino Acids 2018, 50, 453–468. [Google Scholar] [CrossRef]
- Al Tall, Y.; Abualhaijaa, A.; Alsaggar, M.; Almaaytah, A.; Masadeh, M.; Alzoubi, K.H. Design and characterization of a new hybrid peptide from LL-37 and BMAP-27. Infect. Drug Resist. 2019, 12, 1035–1045. [Google Scholar] [CrossRef]
- Tian, L.; Zhang, D.; Su, P.; Wei, Y.; Wang, Z.; Wang, P.X.; Dai, C.J.; Gong, G.L. Design, recombinant expression, and antibacterial activity of a novel hybrid magainin-thanatin antimicrobial peptide. Prep. Biochem. Biotech. 2019, 49, 427–434. [Google Scholar] [CrossRef]
- Klubthawee, N.; Adisakwattana, P.; Hanpithakpong, W.; Somsri, S.; Aunpad, R. A novel, rationally designed, hybrid antimicrobial peptide, inspired by cathelicidin and aurein, exhibits membrane-active mechanisms against Pseudomonas aeruginosa. Sci. Rep. 2020, 10, 9117. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, J.; Cui, Q.; Jia, B.-Y.; Pei, Z.-H.; Odah, K.A.; Wang, Y.-M.; Dong, W.-L.; Kong, L.-C.; Ma, H.-X. Design and characterization of a novel hybrid antimicrobial peptide OM19R based on oncocin and MDAP-2. Int. J. Pept. Res. Ther. 2020, 26, 1839–1846. [Google Scholar] [CrossRef]
- Zhang, L.; Wei, X.; Zhang, R.; Petitte, J.N.; Si, D.; Li, Z.; Cheng, J.; Du, M. Design and development of a novel peptide for treating intestinal inflammation. Front. Immunol. 2019, 10, 1841. [Google Scholar] [CrossRef]
- Ahmad, B.; Hanif, Q.; Wei, X.; Zhang, L.; Sabir, N.; Li, Z.; Cheng, J.; Khan, S.A.; Basit, A.; Shahid, M.; et al. In vitro impact of yeast expressed hybrid peptide CATH-2TP5 as a prophylactic measure toward sepsis and inflammation. Front. Bioeng. Biotechnol. 2020, 8, 454. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.; Zhou, T.; Zhang, J.; Xu, J.; Guo, X.; Hu, H.; Zhang, X.; Hu, M.; Li, J.; Yang, W.; et al. Enhanced cell selectivity of hybrid peptides with potential antimicrobial activity and immunomodulatory effect. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129532. [Google Scholar] [CrossRef]
- Che, Y.Z.; Li, Y.R.; Zou, H.S.; Zou, L.F.; Zhang, B.; Chen, G.Y. A novel antimicrobial protein for plant protection consisting of a Xanthomonas oryzae harpin and active domains of cecropin A and melittin. Microb. Biotechnol. 2011, 4, 777–793. [Google Scholar] [CrossRef]
- Granoth, R.; Vadai, E.; Burstein, Y.; Fridkin, M.; Tzehoval, E. Tuftsin-THF-γ2 chimeric peptides: Potential novel immomodulators. Immunopharmacology 1997, 37, 43–52. [Google Scholar] [CrossRef]
- Tiwari, S.K.; Sutyak Noll, K.; Cavera, V.L.; Chikindas, M.L. Improved antimicrobial activities of synthetic-hybrid bacteriocins designed from enterocin E50-52 and pediocin PA-1. Appl. Environ. Microbiol. 2015, 81, 1661–1667. [Google Scholar] [CrossRef]
- Horn, M.; Neundorf, I. Design of a novel cell-permeable chimeric peptide to promote wound healing. Sci. Rep. 2018, 8, 16279. [Google Scholar] [CrossRef]
- Wade, H.M.; Darling, L.E.O.; Elmore, D.E. Hybrids made from antimicrobial peptides with different mechanisms of action show enhanced membrane permeabilization. Biochim. Biophys. Acta Biomembr. 2019, 1861, 182980. [Google Scholar] [CrossRef]
- Sheard, D.E.; O’Brien-Simpson, N.M.; Wade, J.D.; Separovic, F. Combating bacterial resistance by combination of antibiotics with antimicrobial peptides. Pure Appl. Chem. 2019, 91, 199–209. [Google Scholar] [CrossRef]
- Yan, H.; Hancock, R.E.W. Synergistic interactions between mammalian antimicrobial defense peptides. Antimicrob. Agents Chemother. 2001, 45, 1558–1560. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Baeder, D.Y.; Regoes, R.R.; Rolff, J. Combination effects of antimicrobial peptides. Antimicrob. Agents Chemother. 2016, 60, 1717–1724. [Google Scholar] [CrossRef]
- Amso, Z.; Hayouka, Z. Antimicrobial random peptide cocktails: A new approach to fight pathogenic bacteria. Chem. Commun. 2019, 55, 2007–2014. [Google Scholar] [CrossRef]
- Remington, J.M.; Liao, C.; Sharafi, M.; Ste Marie, E.J.; Ferrell, J.B.; Hondal, R.J.; Wargo, M.J.; Schneebeli, S.T.; Li, J. Aggregation state of synergistic antimicrobial peptides. J. Phys. Chem. Lett. 2020, 11, 9501–9506. [Google Scholar] [CrossRef]
- Nishida, M.; Imura, Y.; Yamamoto, M.; Kobayashi, S.; Yano, Y.; Matsuzaki, K. Interaction of a magainin-PGLa hybrid peptide with membranes: Insight into the mechanism of synergism. Biochemistry 2007, 46, 14284–14290. [Google Scholar] [CrossRef] [PubMed]
- Haney, E.F.; Hunter, H.N.; Matsuzaki, K.; Vogel, H.J. Solution NMR studies of amphibian antimicrobial peptides: Linking structure to function? Biochim. Biophys. Acta 2009, 1788, 1639–1655. [Google Scholar] [CrossRef] [PubMed]
- Bolscher, J.; Nazmi, K.; van Marle, J.; van ‘t Hof, W.; Veerman, E. Chimerization of lactoferricin and lactoferrampin peptides strongly potentiates the killing activity against Candida albicans. Biochem. Cell Biol. 2012, 90, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Haney, E.F.; Nazmi, K.; Bolscher, J.G.; Vogel, H.J. Structural and biophysical characterization of an antimicrobial peptide chimera comprised of lactoferricin and lactoferrampin. Biochim. Biophys. Acta 2012, 1818, 762–775. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Li, D.; Zhang, R.; Shuai, L.; Schulze, B.; Kroth, P.G.; Zhan, D.; Wang, D. Defense responses in female gametophytes of Saccharina japonica (Phaeophyta) induced by flg22-derived peptides. J. Appl. Phycol. 2016, 28, 1793–1801. [Google Scholar] [CrossRef][Green Version]
- Oliveras, À.; Baró, A.; Montesinos, L.; Badosa, E.; Montesinos, E.; Feliu, L.; Planas, M. Antimicrobial activity of linear lipopeptides derived from BP100 towards plant pathogens. PLoS ONE 2018, 13, e0201571. [Google Scholar] [CrossRef]
- Aslam, S.N.; Erbs, G.; Morrissey, K.L.; Newman, M.A.; Chinchilla, D.; Boller, T.; Molinaro, A.; Jackson, R.W.; Cooper, R.M. Microbe-associated molecular pattern (MAMP) signatures, synergy, size and charge: Influences on perception or mobility and host defence responses. Mol. Plant Pathol. 2009, 10, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Danna, C.H.; Millet, Y.A.; Koller, T.; Han, S.W.; Bent, A.F.; Ronald, P.C.; Ausubel, F.M. The Arabidopsis flagellin receptor FLS2 mediates the perception of Xanthomonas Ax21 secreted peptides. Proc. Natl. Acad. Sci. USA 2011, 108, 9286–9291. [Google Scholar] [CrossRef]
- Dobson, A.J.; Purves, J.; Kamysz, W.; Rolff, J. Comparing selection on S. aureus between antimicrobial peptides and common antibiotics. PLoS ONE 2013, 8, e76521. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, E.; Colescott, R.L.; Bossinger, C.D.; Cook, P.I. Color test for detection of free terminal amino groups in the solid-phase synthesis of peptides. Anal. Biochem. 1970, 34, 595–598. [Google Scholar] [CrossRef]
- Hazel, W.; Civerolo, E. Procedures for growth and inoculation of Xanthomonas fragariae, causal organism of angular leaf spot of strawberry. Plant Dis. 1980, 64, 178–181. [Google Scholar] [CrossRef]
- Cabrefiga, J.; Montesinos, E. Analysis of aggressiveness of Erwinia amylovora using disease-dose and time relationships. Phytopathology 2005, 95, 1430–1437. [Google Scholar] [CrossRef][Green Version]
- Pfaffl, M.W. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Güell, I.; Cabrefiga, J.; Badosa, E.; Ferre, R.; Talleda, M.; Bardají, E.; Planas, M.; Feliu, L.; Montesinos, E. Improvement of the efficacy of linear undecapeptides against plant-pathogenic bacteria by incorporation of D-amino acids. Appl. Environ. Microbiol. 2011, 77, 2667–2675. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
