Establishing a Metabolite Extraction Method to Study the Metabolome of Blastocystis Using NMR
Abstract
1. Introduction
2. Materials and Methods
2.1. Blastocystis Culture
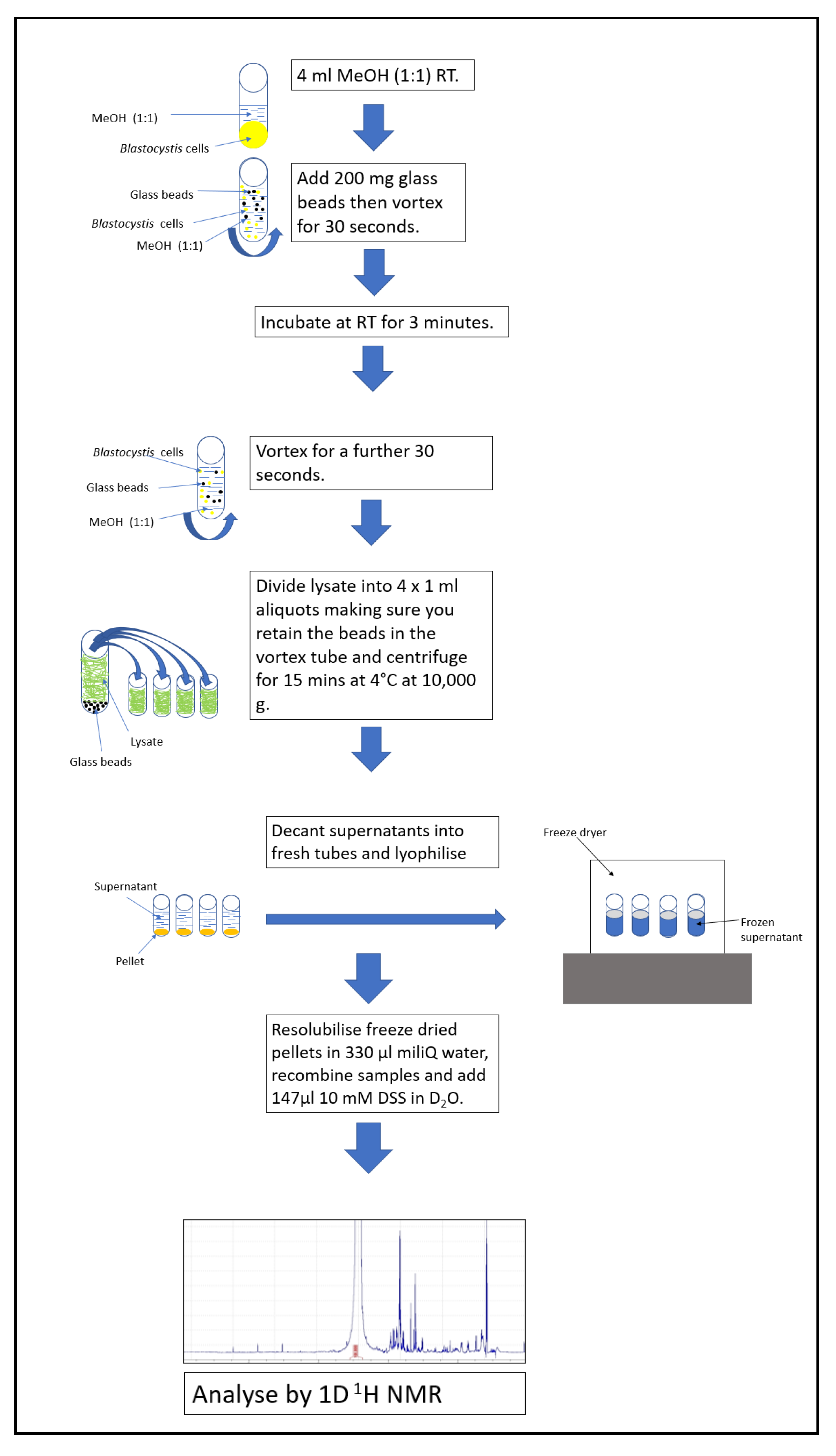
2.2. Cell lysis and Metabolite Extraction
2.3. Preparation for 1H NMR Acquisition
2.4. Analysis of Aqueous Extracts by 1H NMR Spectroscopy
2.5. Processing and Analysis of 1H NMR Data
3. Results
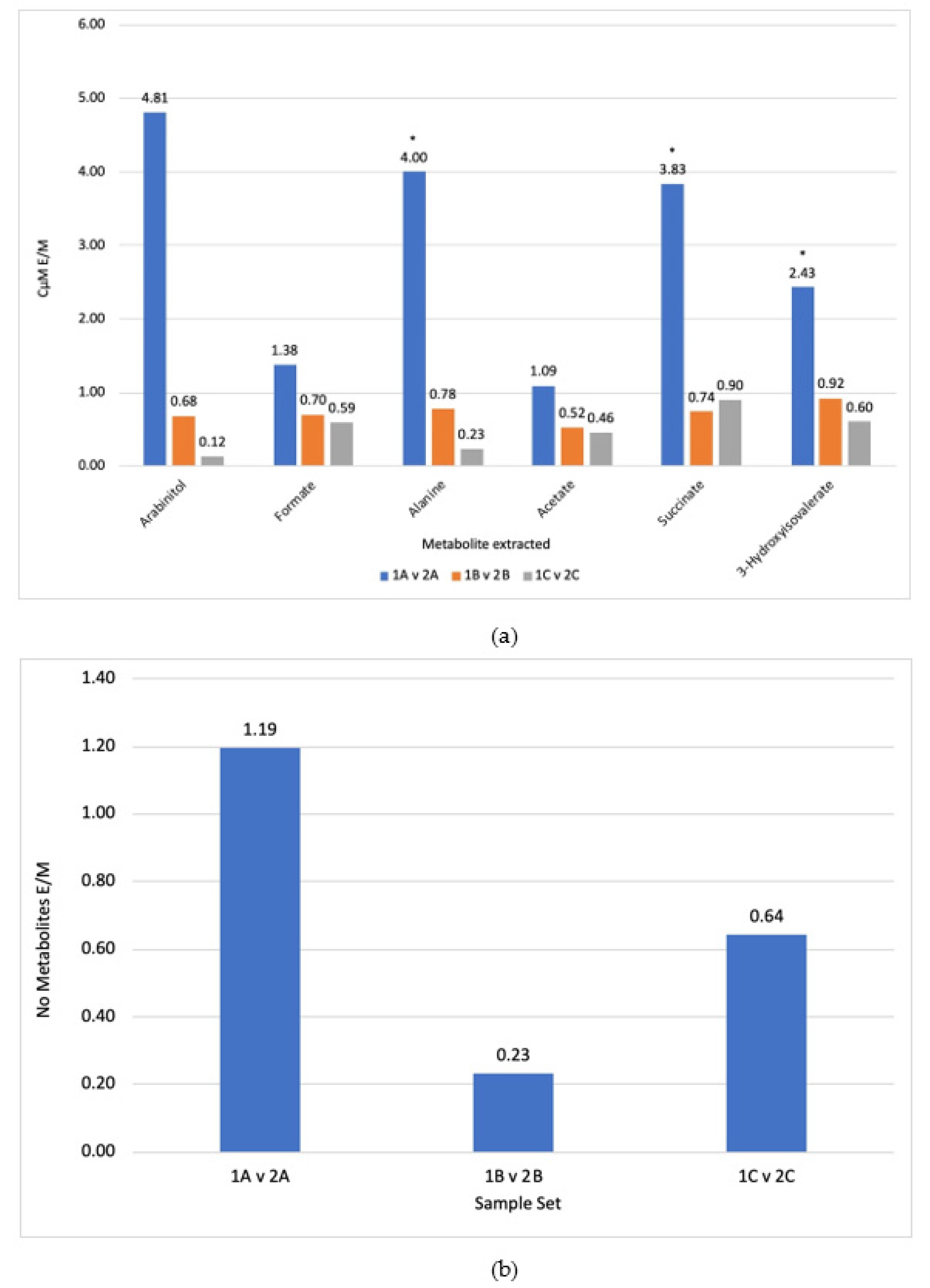
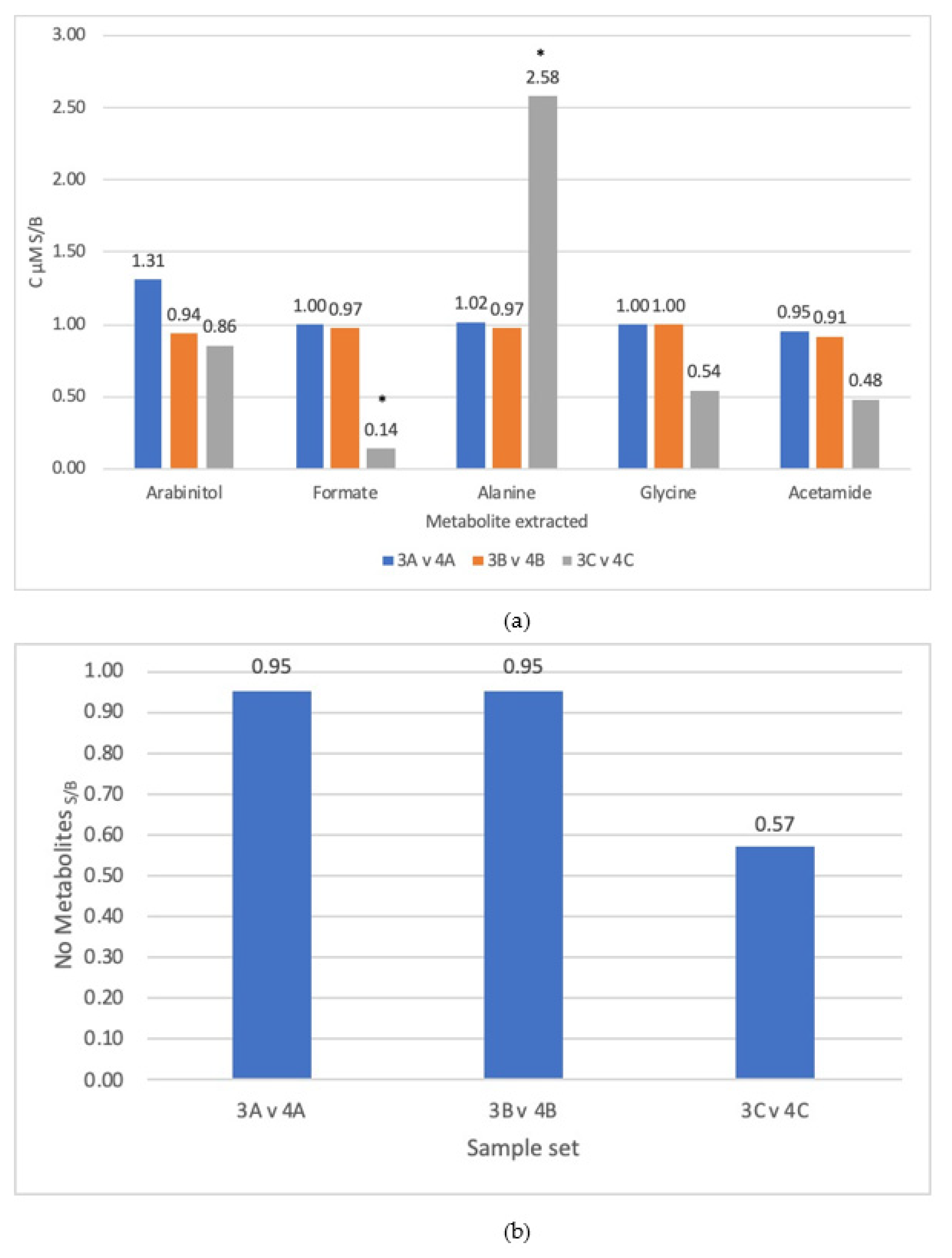
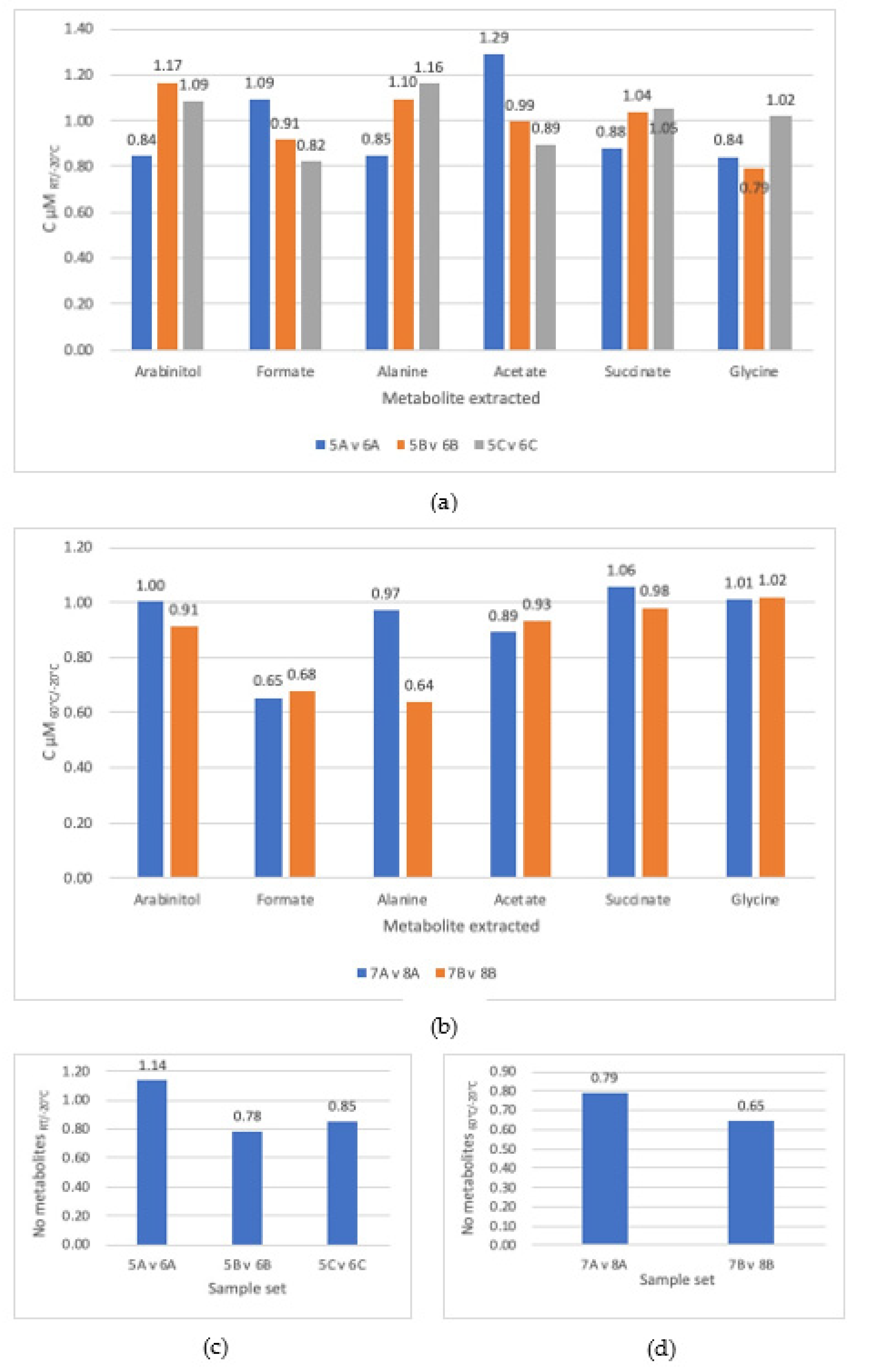
3.1. Comparison of Steps
3.2. Extraction Solvent
3.3. Lysis Method
3.4. Incubation Temperature
4. Discussion
4.1. Methanol Was Determined to Be the Optimal Extraction Solvent
4.2. Bead Bashing Was Determined to Be the Optimal Lysis Method
4.3. Temperature Was Not an Important Factor in Metabolite Extraction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Sample Availability
References
- Stechmann, A.; Hamblin, K.; Pérez-Brocal, V.; Gaston, D.; Richmond, G.S.S.; van der Giezen, M.; Clark, C.G.; Roger, A.J. Organelles in Blastocystis That Blur the Distinction between Mitochondria and Hydrogenosomes. Curr. Biol. 2008, 18, 580–585. [Google Scholar] [CrossRef]
- Tan, K.S.W. New Insights on Classification, Identification, and Clinical Relevance of Blastocystis spp. Clin. Microbiol. Rev. 2008, 21, 639–665. [Google Scholar] [CrossRef]
- Denoeud, F.; Roussel, M.; Noel, B.; Wawrzyniak, I.; Da Silva, C.; Diogon, M.; Viscogliosi, E.; Brochier-Armanet, C.; Couloux, A.; Poulain, J.; et al. Genome Sequence of the Stramenopile Blastocystis, a Human Anaerobic Parasite. Genome Biol. 2011, 12, R29. [Google Scholar] [CrossRef]
- Pérez-Brocal, V.; Clark, C.G. Analysis of Two Genomes from the Mitochondrion-Like Organelle of the Intestinal Parasite Blastocystis: Complete Sequences, Gene Content, and Genome Organization. Mol. Biol. Evol. 2008, 25, 2475–2482. [Google Scholar] [CrossRef][Green Version]
- Wawrzyniak, I.; Roussel, M.; Diogon, M.; Couloux, A.; Texier, C.; Tan, K.S.W.; Vivarès, C.P.; Delbac, F.; Wincker, P.; El Alaoui, H. Complete Circular DNA in the Mitochondria-like Organelles of Blastocystis hominis. Int. J. Parasitol. 2008, 38, 1377–1382. [Google Scholar] [CrossRef]
- Lantsman, Y.; Tan, K.S.W.; Morada, M.; Yarlett, N. Biochemical Characterization of Amitochondrial-like Organelle from Blastocystis Sp. Subtype 7. Microbiology 2008, 154, 2757–2766. [Google Scholar] [CrossRef] [PubMed]
- Vermathen, M.; Müller, J.; Furrer, J.; Müller, N.; Vermathen, P. 1H HR-MAS NMR Spectroscopy to Study the Metabolome of the Protozoan Parasite Giardia lamblia. Talanta 2018, 188, 429–441. [Google Scholar] [CrossRef]
- Li, W.; Lee, R.E.B.; Lee, R.E.; Li, J. Methods for Acquisition and Assignment of Multidimensional High-Resolution Magic Angle Spinning NMR of Whole Cell Bacteria. Anal. Chem. 2005, 77, 5785–5792. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.A.; Jüppner, J.; Bajdzienko, K.; Giavalisco, P. Protocol: A Fast, Comprehensive and Reproducible One-Step Extraction Method for the Rapid Preparation of Polar and Semi-Polar Metabolites, Lipids, Proteins, Starch and Cell Wall Polymers from a Single Sample. Plant Methods 2016, 12, 1–15. [Google Scholar] [CrossRef]
- Geier, F.M.; Want, E.J.; Leroi, A.M.; Bundy, J.G. Cross-Platform Comparison of Caenorhabditis elegans Tissue Extraction Strategies for Comprehensive Metabolome Coverage. Anal. Chem. 2011, 83, 3730–3736. [Google Scholar] [CrossRef]
- Beltran, A.; Suarez, M.; Rodríguez, M.A.; Vinaixa, M.; Samino, S.; Arola, L.; Correig, X.; Yanes, O. Assessment of Compatibility between Extraction Methods for NMR- and LC/MS-Based Metabolomics. Anal. Chem. 2012, 84, 5838–5844. [Google Scholar] [CrossRef]
- Bruno, C.; Patin, F.; Bocca, C.; Nadal-Desbarats, L.; Bonnier, F.; Reynier, P.; Emond, P.; Vourc’h, P.; Joseph-Delafont, K.; Corcia, P.; et al. The Combination of Four Analytical Methods to Explore Skeletal Muscle Metabolomics: Better Coverage of Metabolic Pathways or a Marketing Argument? J. Pharm. Biomed. Anal. 2018, 148, 273–279. [Google Scholar] [CrossRef]
- Fathi, F.; Brun, A.; Rott, K.H.; Cobra, P.F.; Tonelli, M.; Eghbalnia, H.R.; Caviedes-Vidal, E.; Karasov, W.H.; Markley, J.L. NMR-Based Identification of Metabolites in Polar and Non-Polar Extracts of Avian Liver. Metabolites 2017, 7, 61. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Cheng, M.L.; Chiang, M.H.; Wang, C.J.; Tsai, M.H.; Lin, G. Metabolomic Analysis Reveals Distinct Profiles in the Plasma and Urine Associated with IgE Reactions in Childhood Asthma. J. Clin. Med. 2020, 9, 887. [Google Scholar] [CrossRef] [PubMed]
- Leenders, J.; Grootveld, M.; Percival, B.; Gibson, M.; Casanova, F.; Wilson, P.B. Benchtop Low-Frequency 60 MHz NMR Analysis of Urine: A Comparative Metabolomics Investigation. Metabolites 2020, 10, 155. [Google Scholar] [CrossRef]
- Saude, E.; Sykes, B. Urine Stability for Metabolomic Studies: Effects of Preparation and Storage. Metabolomics 2007, 3, 19–27. [Google Scholar] [CrossRef]
- Gómez-Gallego, C.; Morales, J.M.; Monleón, D.; du Toit, E.; Kumar, H.; Linderborg, K.M.; Zhang, Y.; Yang, B.; Isolauri, E.; Salminen, S.; et al. Human Breast Milk NMR Metabolomic Profile across Specific Geographical Locations and Its Association with the Milk Microbiota. Nutrients 2018, 10, 1355. [Google Scholar] [CrossRef]
- Zheng, J.; Mandal, R.; Wishart, D.S. A Sensitive, High-Throughput LC-MS/MS Method for Measuring Catecholamines in Low Volume Serum. Anal. Chim. Acta 2018, 1037, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Giraudeau, P. NMR-Based Metabolomics and Fluxomics: Developments and Future Prospects. Analyst 2020, 145, 2457–2472. [Google Scholar] [CrossRef]
- Markley, J.L.; Brüschweiler, R.; Edison, A.S.; Eghbalnia, H.R.; Powers, R.; Raftery, D.; Wishart, D.S. The Future of NMR-Based Metabolomics. Curr. Opin. Biotechnol. 2017, 43, 34–40. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Raftery, D. Can NMR Solve Some Significant Challenges in Metabolomics? J. Magn. Reson. 2015, 260, 144–160. [Google Scholar] [CrossRef]
- Takis, P.G.; Ghini, V.; Tenori, L.; Turano, P.; Luchinat, C. Uniqueness of the NMR Approach to Metabolomics. TrAC Trends Anal. Chem. 2019, 120, 115300. [Google Scholar] [CrossRef]
- Vignoli, A.; Ghini, V.; Meoni, G.; Licari, C.; Takis, P.G.; Tenori, L.; Turano, P.; Luchinat, C. High-Throughput Metabolomics by 1D NMR. Angew. Chem. Int. Ed. 2019, 58, 968–994. [Google Scholar] [CrossRef]
- Palmioli, A.; Ceresa, C.; Tripodi, F.; La Ferla, B.; Nicolini, G.; Airoldi, C. On-Cell Saturation Transfer Difference NMR Study of Bombesin Binding to GRP Receptor. Bioorganic Chem. 2020, 99, 103861. [Google Scholar] [CrossRef] [PubMed]
- Findeisen, M.; Brand, T.; Berger, S. A 1H-NMR Thermometer Suitable for Cryoprobes. Magn. Reson. Chem. 2007, 45, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Raiford, D.S.; Fisk, C.L.; Becker, E.D. Calibration of Methanol and Ethylene Glycol Nuclear Magnetic Resonance Thermometers. Anal. Chem. 1979, 51, 2050–2051. [Google Scholar] [CrossRef]
- Wu, P.S.C.; Otting, G. Rapid Pulse Length Determination in High-Resolution NMR. J. Magn. Reson. 2005, 176, 115–119. [Google Scholar] [CrossRef]
- Snytnikova, O.A.; Khlichkina, A.A.; Sagdeev, R.Z.; Tsentalovich, Y.P. Evaluation of Sample Preparation Protocols for Quantitative NMR-Based Metabolomics. Metabolomics 2019, 15. [Google Scholar] [CrossRef]
- Anwar, M.A.; Vorkas, P.A.; Li, J.V.; Shalhoub, J.; Want, E.J.; Davies, A.H.; Holmes, E. Optimization of Metabolite Extraction of Human Vein Tissue for Ultra Performance Liquid Chromatography-Mass Spectrometry and Nuclear Magnetic Resonance-Based Untargeted Metabolic Profiling. Analyst 2015, 140, 7586–7597. [Google Scholar] [CrossRef] [PubMed]
| Experiment No. | Batch No. | Extraction Solvent | Lysis Method | Incubation Temp |
|---|---|---|---|---|
| 1 | 1 | 4 mL EtOH (3:1) −20 °C | Sonication 3 × 30 s | 3 min −20 °C |
| 2 | 4 mL MeOH (1:1) −20 °C | |||
| 2 | 1 | 4 mL MeOH (1:1) −20 °C | Bead Bashing–200 mg beads vortex 30 s | 3 min −20 °C |
| 2 | 4 mL MeOH (1:1) −20 °C | Sonication 3 × 30 s | ||
| 3 | 1 | 4 mL MeOH (1:1) −20 °C | Sonication 3 × 30 s | 3 min −20 °C |
| 2 | 4 mL MeOH (1:1) RT | 3 min RT | ||
| 4 | 1 | 4 mL MeOH (1:1) 60 °C | Sonication 3 × 30 s | 3 min 60 °C |
| 2 | 4 mL MeOH (1:1) RT | 3 min RT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Newton, J.M.; Betts, E.L.; Yiangou, L.; Ortega Roldan, J.; Tsaousis, A.D.; Thompson, G.S. Establishing a Metabolite Extraction Method to Study the Metabolome of Blastocystis Using NMR. Molecules 2021, 26, 3285. https://doi.org/10.3390/molecules26113285
Newton JM, Betts EL, Yiangou L, Ortega Roldan J, Tsaousis AD, Thompson GS. Establishing a Metabolite Extraction Method to Study the Metabolome of Blastocystis Using NMR. Molecules. 2021; 26(11):3285. https://doi.org/10.3390/molecules26113285
Chicago/Turabian StyleNewton, Jamie M., Emma L. Betts, Lyto Yiangou, Jose Ortega Roldan, Anastasios D. Tsaousis, and Gary S. Thompson. 2021. "Establishing a Metabolite Extraction Method to Study the Metabolome of Blastocystis Using NMR" Molecules 26, no. 11: 3285. https://doi.org/10.3390/molecules26113285
APA StyleNewton, J. M., Betts, E. L., Yiangou, L., Ortega Roldan, J., Tsaousis, A. D., & Thompson, G. S. (2021). Establishing a Metabolite Extraction Method to Study the Metabolome of Blastocystis Using NMR. Molecules, 26(11), 3285. https://doi.org/10.3390/molecules26113285






